Beneficial Effects of Lactobacillus plantarum Strains on Non-Alcoholic Fatty Liver Disease in High Fat/High Fructose Diet-Fed Rats
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of L. plantarum Strains
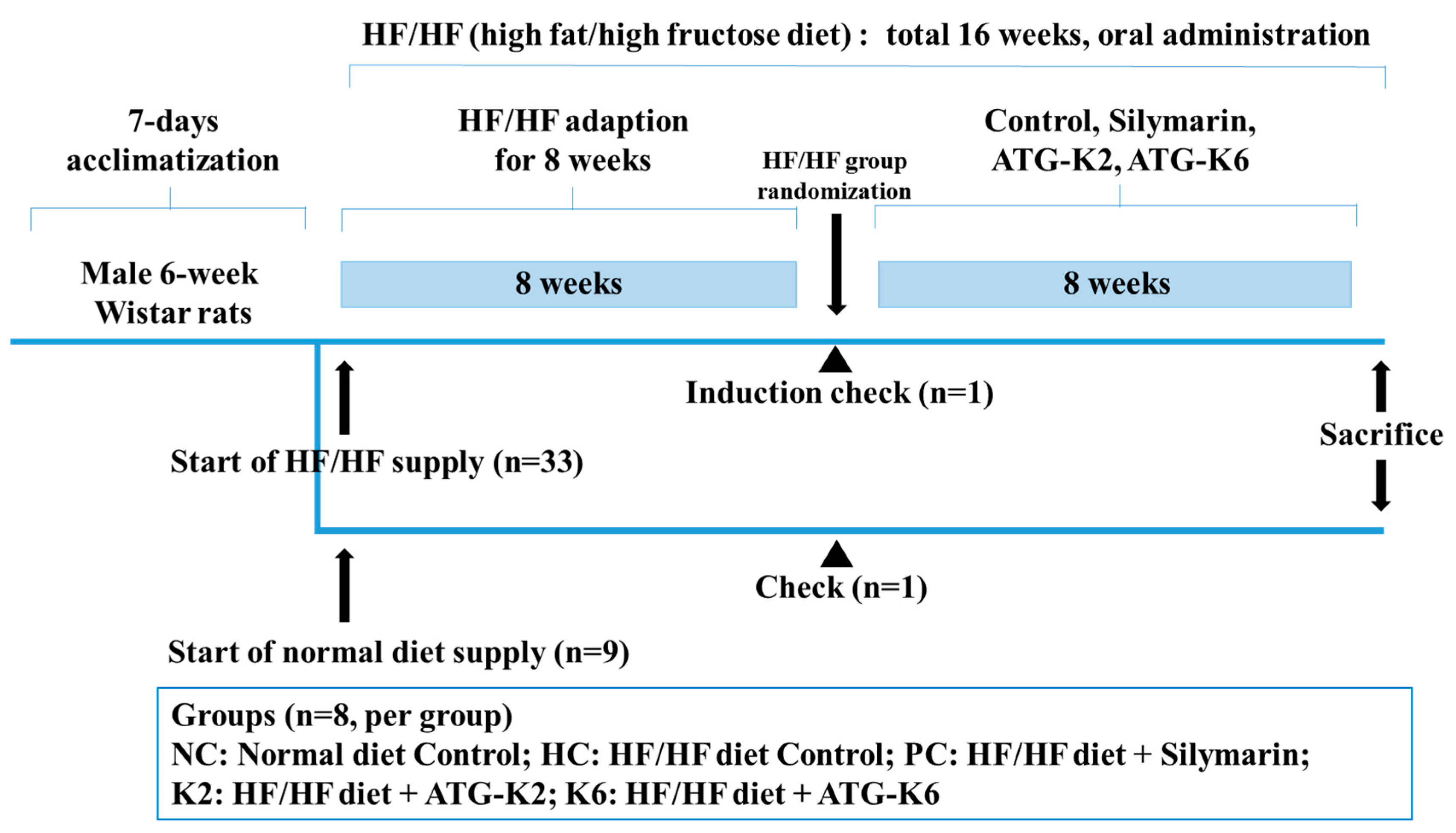
2.2. Animals and Treatment
2.3. Histological Analysis and Oil-Red O Staining
2.4. Measurement of Serum Biochemistry Markers
2.5. Measurement of Liver Biochemistry Markers
2.6. Quantitative Reverse Transcription PCR (qRT-PCR)
2.7. Protein Extraction and Western Blotting
2.8. Fecal Microbiota Profiling
2.9. Statistical Analysis
3. Results
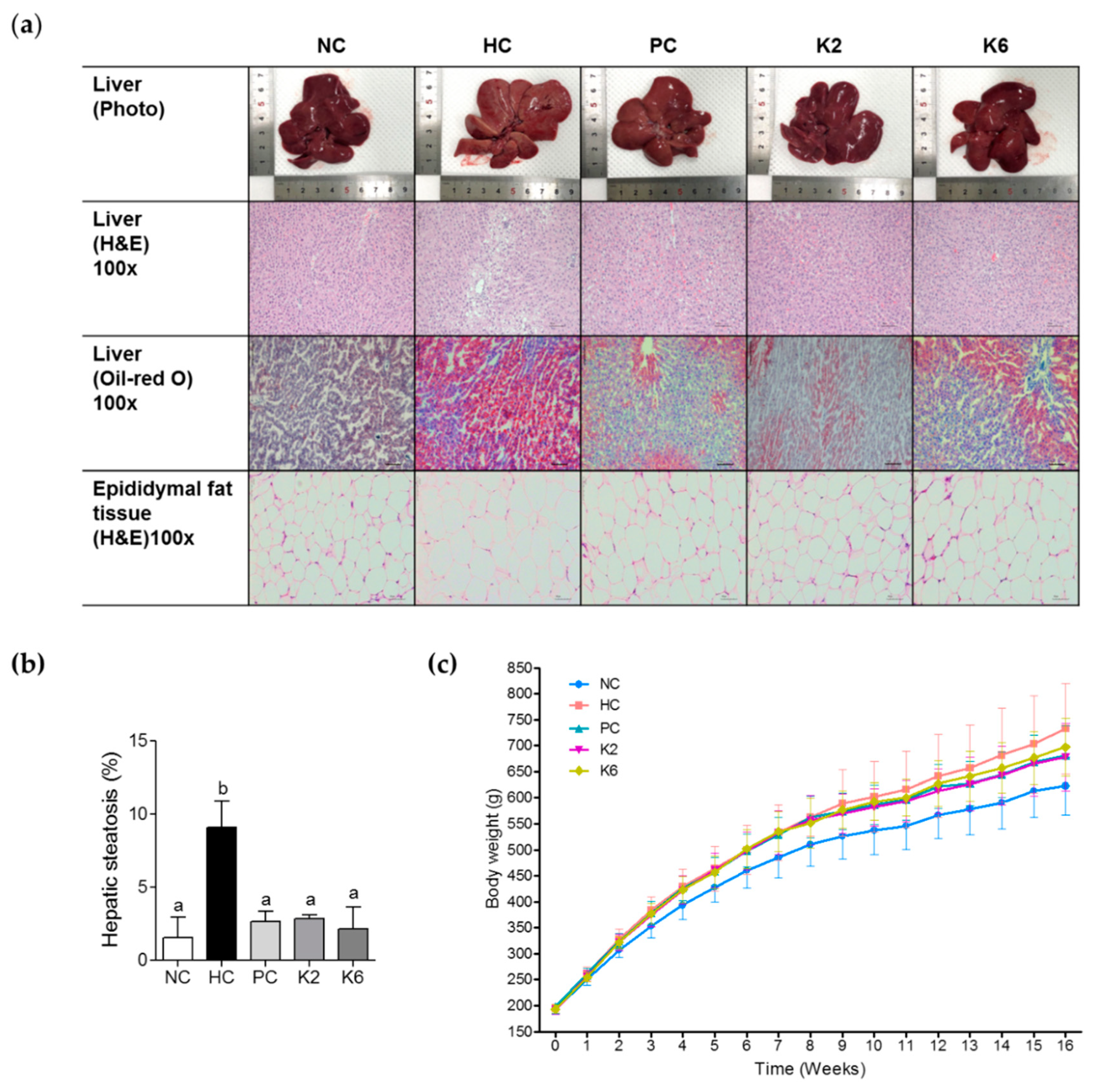
3.1. Effect of the L. plantarum K2 and K6 on Hepatic Steatosis and Body Weight in the Rat Model of HF/HF Diet-Induced NAFLD
3.2. The effects of L. plantarum K2 and K6 on Serum Biochemical Parameters
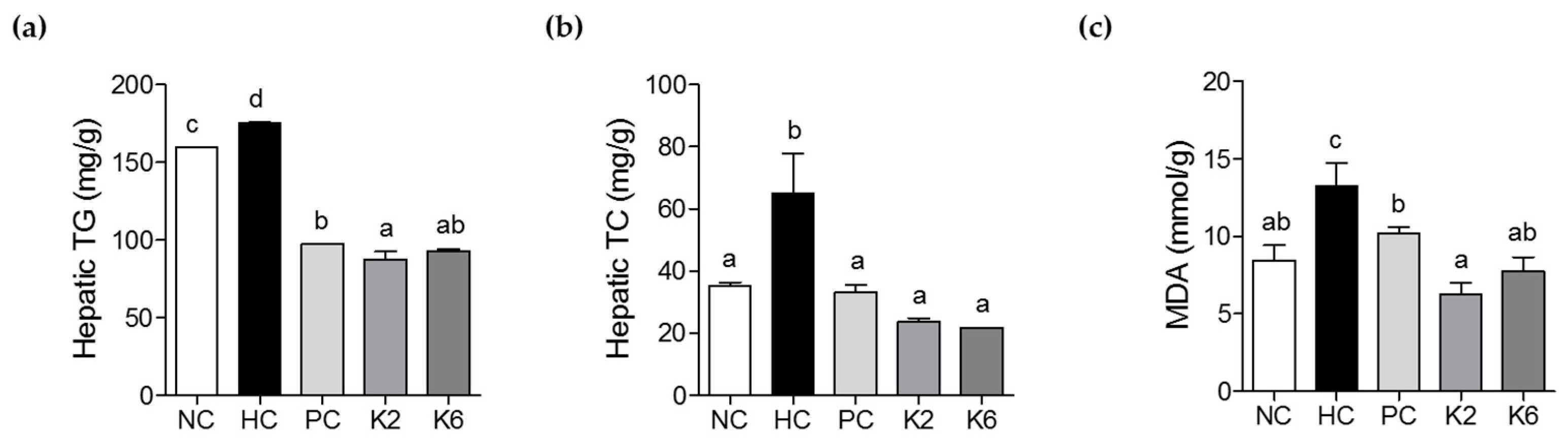
3.3. The Effects of L. plantarum K2 and K6 on Hepatic Biochemical Parameters
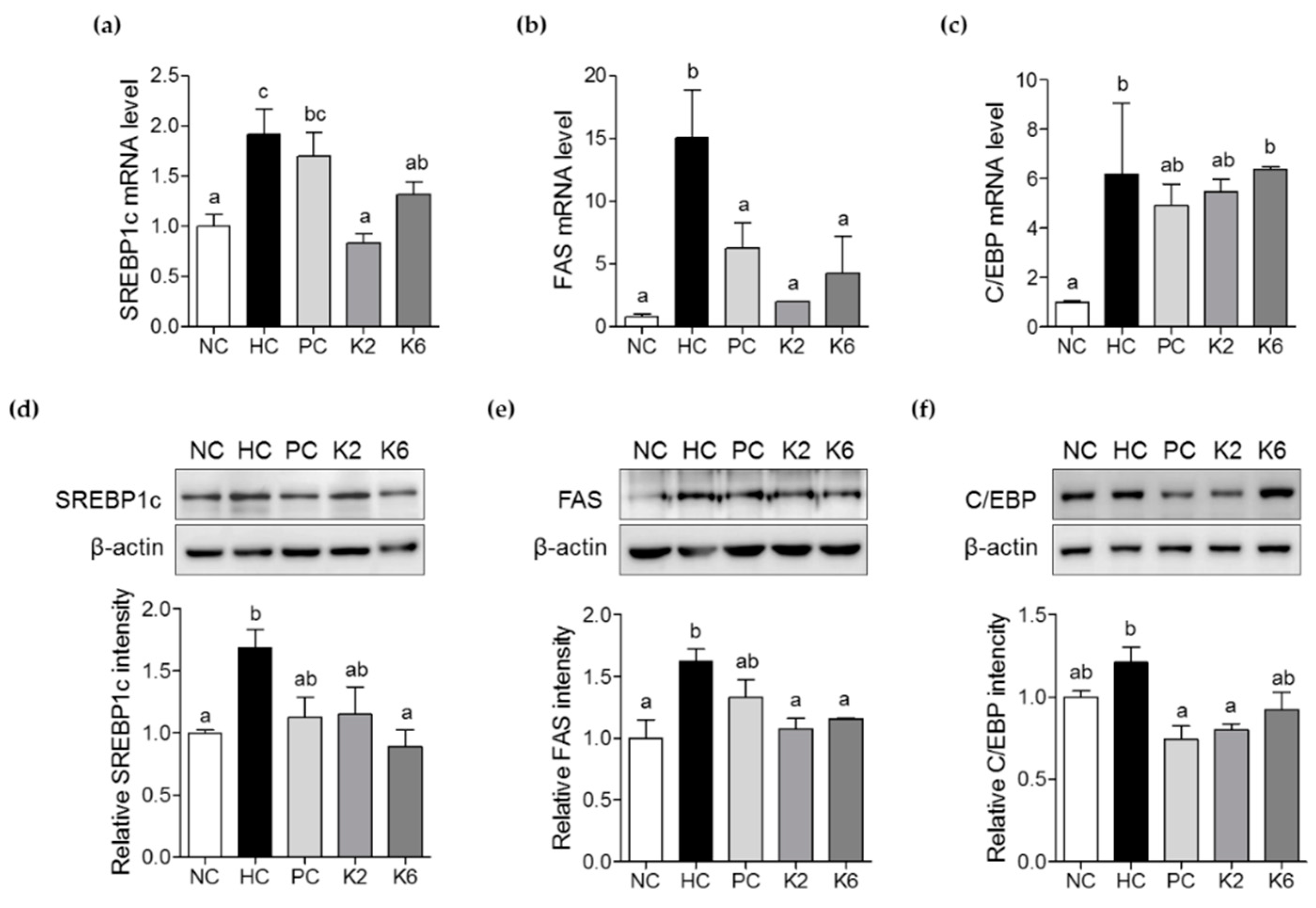
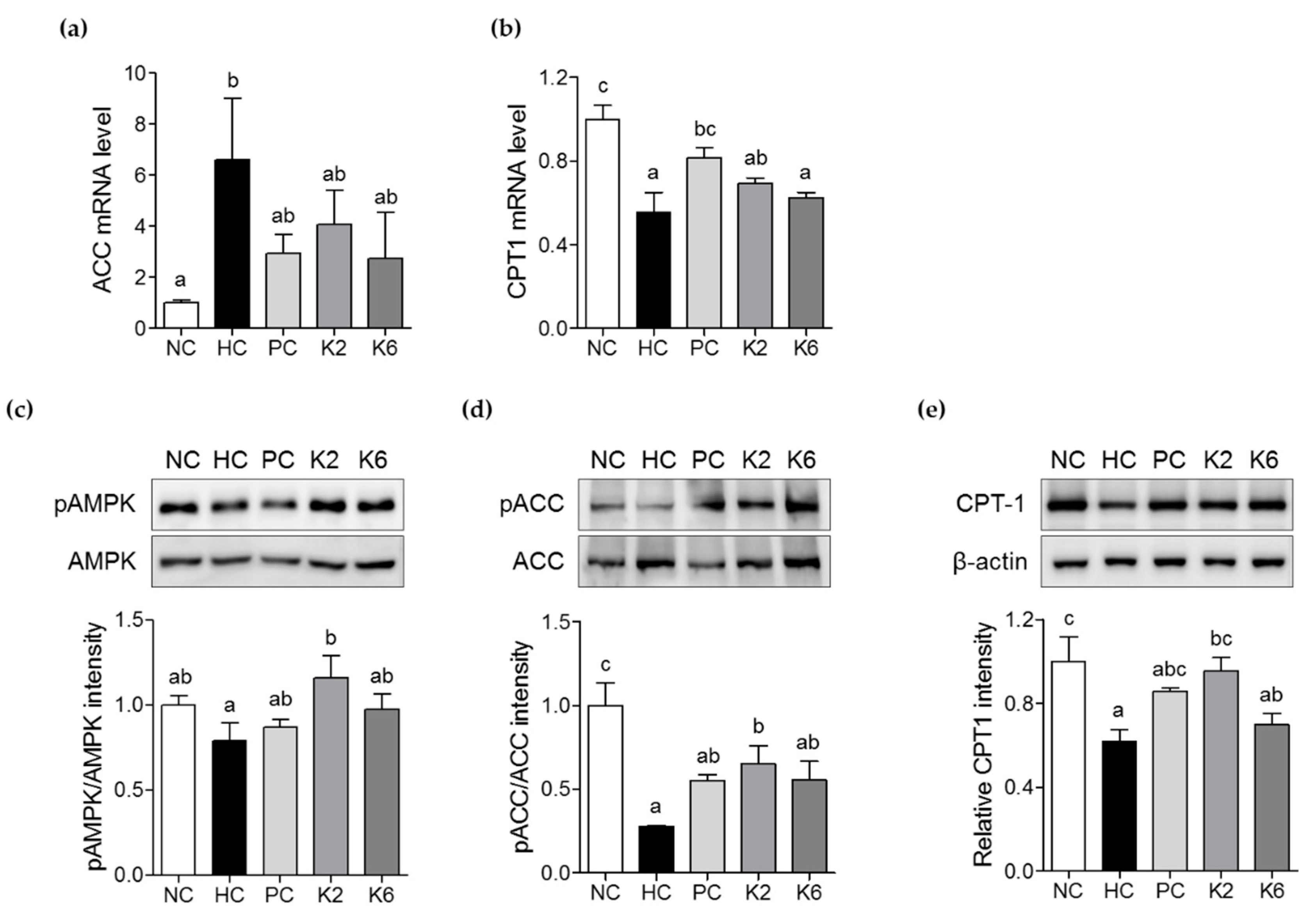
3.4. The Effects of L. plantarum K2 and K6 on Lipogenesis-Related Gene Expression in the Liver
3.5. The Effects of L. plantarum K2 and K6 on Lipid Metabolism in the Liver
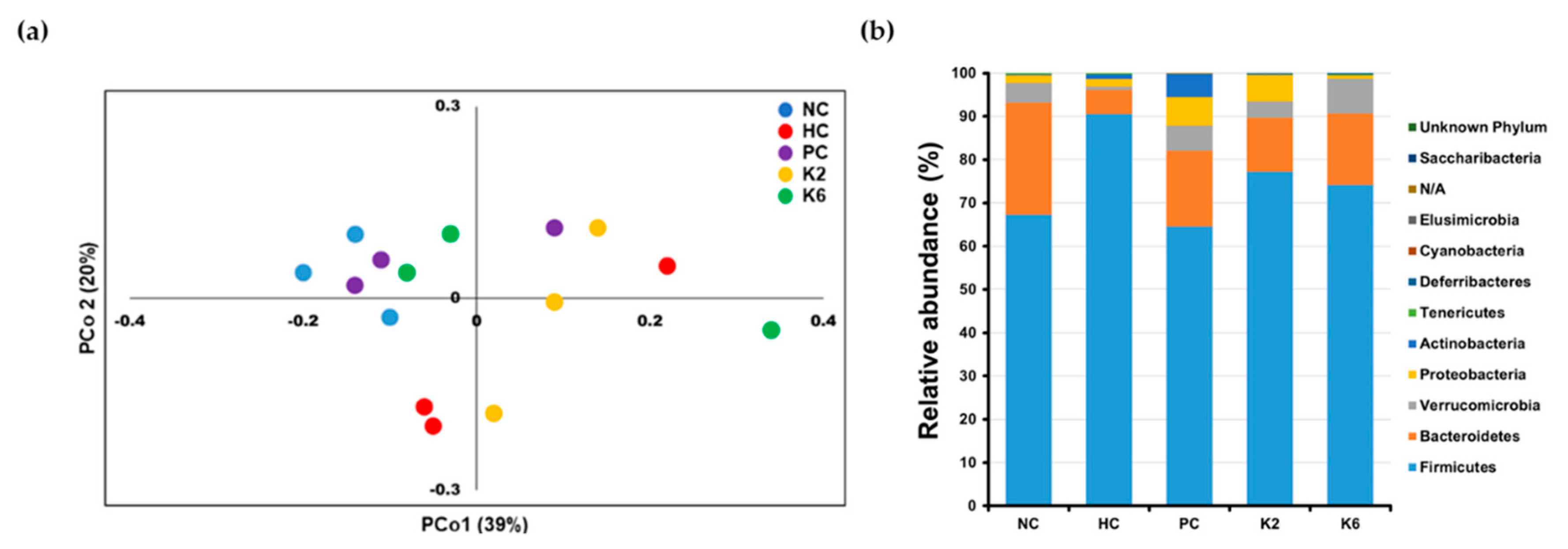
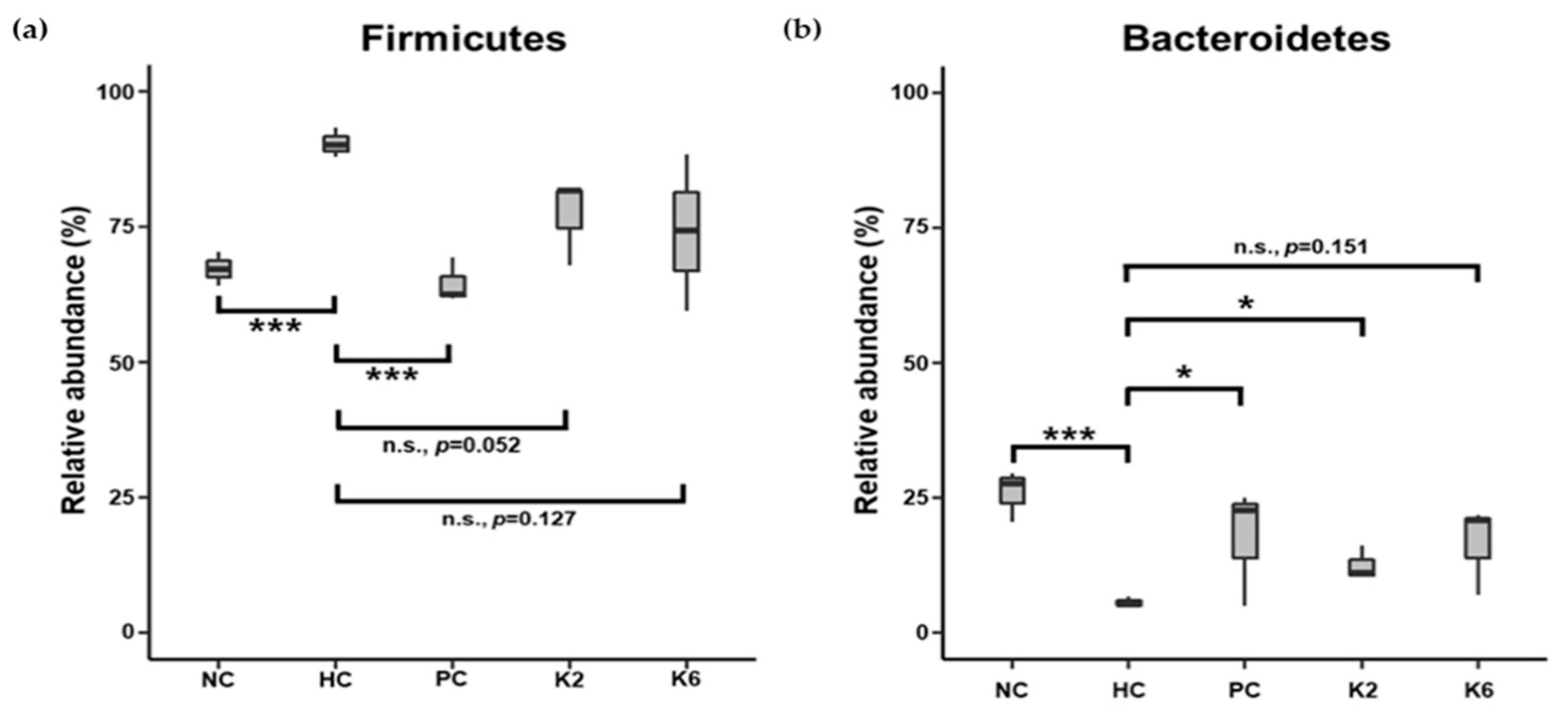
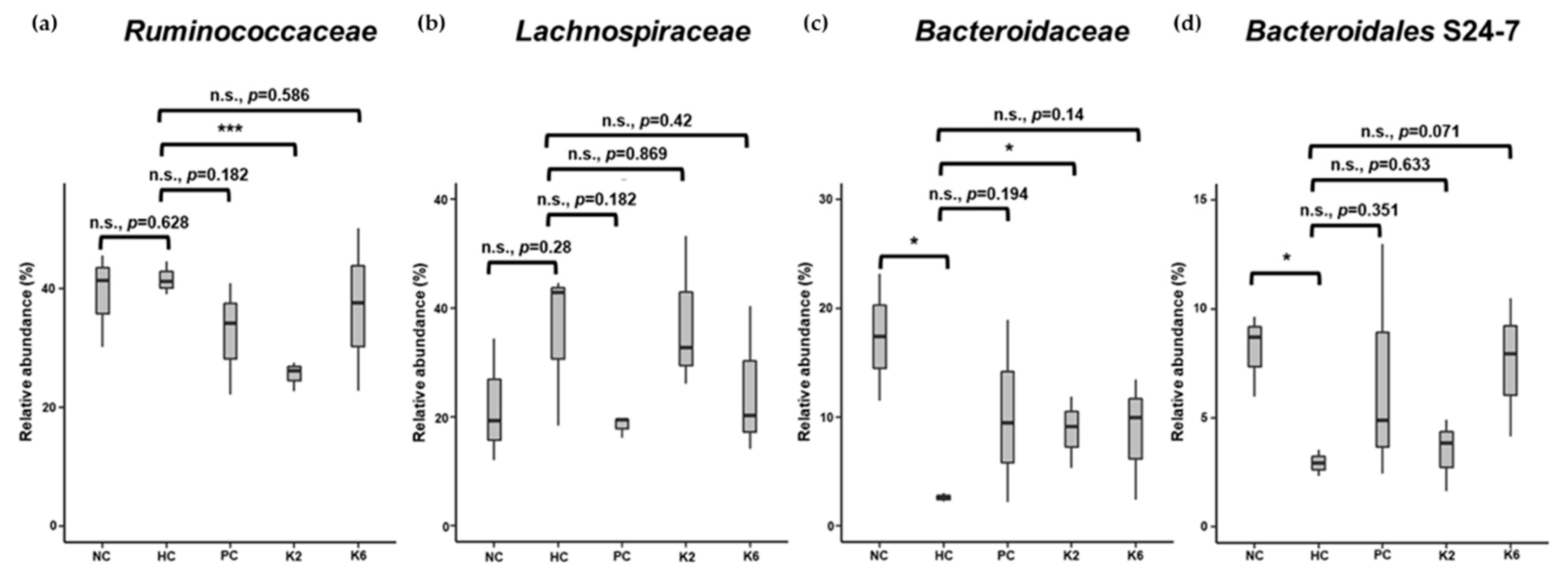
3.6. The Effects of L. plantarum Administration on Fecal Microbiota Composition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Samuel, V.T.; Shulman, G.I. Nonalcoholic Fatty Liver Disease as a Nexus of Metabolic and Hepatic Diseases. Cell Metab. 2018, 27, 22–41. [Google Scholar] [CrossRef]
- Machado, M.; Marques-Vidal, P.; Cortez-Pinto, H. Hepatic histology in obese patients undergoing bariatric surgery. J. Hepatol. 2006, 45, 600–606. [Google Scholar] [CrossRef]
- Cobbina, E.; Akhlaghi, F. Non-alcoholic fatty liver disease (NAFLD) - pathogenesis, classification, and effect on drug metabolizing enzymes and transporters. Drug Metab. Rev. 2017, 49, 197–211. [Google Scholar] [CrossRef]
- Bedossa, P. Pathology of non-alcoholic fatty liver disease. Liver Int. Off. J. Int. Assoc. Study Liver 2017, 37, 85–89. [Google Scholar] [CrossRef]
- Adams, L.A.; Lymp, J.F.; St Sauver, J.; Sanderson, S.O.; Lindor, K.D.; Feldstein, A.; Angulo, P. The natural history of nonalcoholic fatty liver disease: A population-based cohort study. Gastroenterology 2005, 129, 113–121. [Google Scholar] [CrossRef]
- Ter Horst, K.W.; Serlie, M.J. Fructose Consumption, Lipogenesis, and Non-Alcoholic Fatty Liver Disease. Nutrients 2017, 981. [Google Scholar] [CrossRef]
- Herman, M.A.; Samuel, V.T. The Sweet Path to Metabolic Demise: Fructose and Lipid Synthesis. Trends Endocrinol. Metab. 2016, 27, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Softic, S.; Cohen, D.E.; Kahn, C.R. Role of Dietary Fructose and Hepatic De Novo Lipogenesis in Fatty Liver Disease. Dig. Dis. Sci. 2016, 61, 1282–1293. [Google Scholar] [CrossRef] [PubMed]
- Jensen, T.; Abdelmalek, M.F.; Sullivan, S.; Nadeau, K.J.; Green, M.; Roncal, C.; Nakagawa, T.; Kuwabara, M.; Sato, Y.; Kang, D.H.; et al. Fructose and sugar: A major mediator of non-alcoholic fatty liver disease. J. Hepatol. 2018, 68, 1063–1075. [Google Scholar] [CrossRef] [PubMed]
- Salomone, F.; Godos, J.; Zelber-Sagi, S. Natural antioxidants for non-alcoholic fatty liver disease: Molecular targets and clinical perspectives. Liver Int. 2016, 36, 5–20. [Google Scholar] [CrossRef]
- Cacciapuoti, F.; Scognamiglio, A.; Palumbo, R.; Forte, R.; Cacciapuoti, F. Silymarin in non alcoholic fatty liver disease. World J. Hepatol. 2013, 5, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.W.; Lin, S.X.; Shen, Z.H.; Luo, W.W.; Wang, X.Y. Systematic Review with Meta-Analysis: The Effects of Probiotics in Nonalcoholic Fatty Liver Disease. Gastroenterol. Res. Pract. 2019, 2019, 1484598. [Google Scholar] [CrossRef] [PubMed]
- Aller, R.; De Luis, D.A.; Izaola, O.; Conde, R.; Gonzalez Sagrado, M.; Primo, D.; De La Fuente, B.; Gonzalez, J. Effect of a probiotic on liver aminotransferases in nonalcoholic fatty liver disease patients: A double blind randomized clinical trial. Eur. Rev. Med. Pharm. Sci. 2011, 15, 1090–1095. [Google Scholar]
- Famouri, F.; Shariat, Z.; Hashemipour, M.; Keikha, M.; Kelishadi, R. Effects of Probiotics on Nonalcoholic Fatty Liver Disease in Obese Children and Adolescents. J. Pediatr. Gastroenterol. Nutr. 2017, 64, 413–417. [Google Scholar] [CrossRef]
- Seddik, H.A.; Bendali, F.; Gancel, F.; Fliss, I.; Spano, G.; Drider, D. Lactobacillus plantarum and Its Probiotic and Food Potentialities. Probiotics Antimicrob. Proteins 2017, 9, 111–122. [Google Scholar] [CrossRef]
- Zhao, Z.; Wang, C.; Zhang, L.; Zhao, Y.; Duan, C.; Zhang, X.; Gao, L.; Li, S. Lactobacillus plantarum NA136 improves the non-alcoholic fatty liver disease by modulating the AMPK/Nrf2 pathway. Appl. Microbiol. Biotechnol. 2019, 103, 5843–5850. [Google Scholar] [CrossRef]
- Li, C.; Nie, S.P.; Zhu, K.X.; Ding, Q.; Li, C.; Xiong, T.; Xie, M.Y. Lactobacillus plantarum NCU116 improves liver function, oxidative stress and lipid metabolism in rats with high fat diet induced non-alcoholic fatty liver disease. Food Funct. 2014, 5, 3216–3223. [Google Scholar] [CrossRef]
- Beck, B.R.; Park, G.S.; Lee, Y.H.; Im, S.; Jeong, D.Y.; Kang, J. Whole Genome Analysis of Lactobacillus plantarum Strains Isolated From Kimchi and Determination of Probiotic Properties to Treat Mucosal Infections by Candida albicans and Gardnerella vaginalis. Front. Microbiol. 2019, 10, 433. [Google Scholar] [CrossRef]
- Burton, D.T.; Cooper, K.R.; Goodfellow, W.L., Jr.; Rosenblatt, D.H. Uptake, elimination, and metabolism of 14C-picric acid and 14C-picramic acid in the American oyster (Crassostrea virginica). Arch. Environ. Contam. Toxicol. 1984, 13, 653–663. [Google Scholar] [CrossRef]
- Lin, M.Y.; Yen, C.L. Antioxidative ability of lactic acid bacteria. J. Agric. Food Chem. 1999, 47, 1460–1466. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef]
- Park, M.; Yoo, J.H.; Lee, Y.S.; Lee, H.J. Lonicera caerulea Extract Attenuates Non-Alcoholic Fatty Liver Disease in Free Fatty Acid-Induced HepG2 Hepatocytes and in High Fat Diet-Fed Mice. Nutrients 2019, 11, 494. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2013, 42, D643–D648. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011, 5, 169. [Google Scholar] [CrossRef]
- Rivera-Espinoza, Y.; Gallardo-Navarro, Y. Non-dairy probiotic products. Food Microbiol. 2010, 27, 1–11. [Google Scholar] [CrossRef]
- Park, S.S.; Lee, Y.J.; Kang, H.; Yang, G.; Hong, E.J.; Lim, J.Y.; Oh, S.; Kim, E. Lactobacillus amylovorus KU4 ameliorates diet-induced obesity in mice by promoting adipose browning through PPARgamma signaling. Sci. Rep. 2019, 9, 20152. [Google Scholar] [CrossRef]
- John, G.K.; Mullin, G.E. The Gut Microbiome and Obesity. Curr. Oncol. Rep. 2016, 18, 45. [Google Scholar] [CrossRef]
- Ye, H.; Li, Q.; Zhang, Z.; Sun, M.; Zhao, C.; Zhang, T. Effect of a novel potential probiotic Lactobacillus paracasei Jlus66 isolated from fermented milk on nonalcoholic fatty liver in rats. Food Funct. 2017, 8, 4539–4546. [Google Scholar] [CrossRef]
- Al-Muzafar, H.M.; Amin, K.A. Probiotic mixture improves fatty liver disease by virtue of its action on lipid profiles, leptin, and inflammatory biomarkers. BMC Complement. Altern. Med. 2017, 17, 43. [Google Scholar] [CrossRef]
- Oh, R.C.; Hustead, T.R.; Ali, S.M.; Pantsari, M.W. Mildly Elevated Liver Transaminase Levels: Causes and Evaluation. Am. Fam. Physician 2017, 96, 709–715. [Google Scholar]
- Fraser, A.; Longnecker, M.P.; Lawlor, D.A. Prevalence of elevated alanine aminotransferase among US adolescents and associated factors: NHANES 1999-2004. Gastroenterology 2007, 133, 1814–1820. [Google Scholar] [CrossRef]
- Liu, Z.; Que, S.; Xu, J.; Peng, T. Alanine aminotransferase-old biomarker and new concept: A review. Int. J. Med. Sci. 2014, 11, 925–935. [Google Scholar] [CrossRef]
- Masarone, M.; Rosato, V.; Dallio, M.; Gravina, A.G.; Aglitti, A.; Loguercio, C.; Federico, A.; Persico, M. Role of Oxidative Stress in Pathophysiology of Nonalcoholic Fatty Liver Disease. Oxidative Med. Cell. Longev. 2018, 2018, 9547613. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.L.; Norhaizan, M.E.; Liew, W.P.; Sulaiman Rahman, H. Antioxidant and Oxidative Stress: A Mutual Interplay in Age-Related Diseases. Front. Pharmacol. 2018, 9, 1162. [Google Scholar] [CrossRef]
- Padurariu, M.; Ciobica, A.; Hritcu, L.; Stoica, B.; Bild, W.; Stefanescu, C. Changes of some oxidative stress markers in the serum of patients with mild cognitive impairment and Alzheimer’s disease. Neurosci. Lett. 2010, 469, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Jiang, L.; Li, Y.; Bai, G.; Zhao, J.; Zhang, M.; Zhang, J. Hydrogen protects against liver injury during CO2 pneumoperitoneum in rats. Oncotarget 2018, 9, 2631–2645. [Google Scholar] [CrossRef] [PubMed]
- Osburn, W.O.; Kensler, T.W. Nrf2 signaling: An adaptive response pathway for protection against environmental toxic insults. Mutat. Res. 2008, 659, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Ma, J.Z.; Zeng, Q.; Cechova, S.; Gantz, A.; Nievergelt, C.; O’Connor, D.; Lipkowitz, M.; Le, T.H. Loss of GSTM1, a NRF2 target, is associated with accelerated progression of hypertensive kidney disease in the African American Study of Kidney Disease (AASK). Am. J. Physiol. Ren. Physiol. 2013, 304, F348–F355. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, Y.; Wang, Y.; Fu, A.; Gong, L.; Li, W.; Li, Y. Bacillus amyloliquefaciens SC06 alleviates the oxidative stress of IPEC-1 via modulating Nrf2/Keap1 signaling pathway and decreasing ROS production. Appl. Microbiol. Biotechnol. 2017, 101, 3015–3026. [Google Scholar] [CrossRef]
- Bogdanova, K.; Poczatkova, H.; Uherkova, L.; Riegrova, D.; Rypka, M.; Feher, J.; Marchesini, G.; Vesely, J. Non-alcoholic fatty liver disease (NAFLD)-a novel common aspect of the metabolic syndrome. Biomed. Pap. Med. Fac. Univ. PalackyOlomoucCzechoslov. 2006, 150, 101–104. [Google Scholar] [CrossRef]
- Jeon, T.I.; Osborne, T.F. SREBPs: Metabolic integrators in physiology and metabolism. Trends Endocrinol. Metab. 2012, 23, 65–72. [Google Scholar] [CrossRef]
- Wong, C.P.; Kaneda, T.; Morita, H. Plant natural products as an anti-lipid droplets accumulation agent. J. Nat. Med. 2014, 68, 253–266. [Google Scholar] [CrossRef]
- Kang, J.H.; Yun, S.I.; Park, M.H.; Park, J.H.; Jeong, S.Y.; Park, H.O. Anti-obesity effect of Lactobacillus gasseri BNR17 in high-sucrose diet-induced obese mice. PLoS ONE 2013, 8, e54617. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Pan, M.; Huang, R.; Tian, X.; Tao, X.; Shah, N.P.; Wei, H.; Wan, C. Modulation of the small intestinal microbial community composition over short-term or long-term administration with Lactobacillus plantarum ZDY2013. J. Dairy Sci. 2016, 99, 6913–6921. [Google Scholar] [CrossRef] [PubMed]
- Arslan, N. Obesity, fatty liver disease and intestinal microbiota. World J. Gastroenterol. WJG 2014, 20, 16452. [Google Scholar] [CrossRef] [PubMed]
- Ritze, Y.; Bárdos, G.; Claus, A.; Ehrmann, V.; Bergheim, I.; Schwiertz, A.; Bischoff, S.C. Lactobacillus rhamnosus GG protects against non-alcoholic fatty liver disease in mice. PLoS ONE 2014, 9, e80169. [Google Scholar] [CrossRef] [PubMed]
- Mouzaki, M.; Comelli, E.M.; Arendt, B.M.; Bonengel, J.; Fung, S.K.; Fischer, S.E.; McGilvray, I.D.; Allard, J.P. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology 2013, 58, 120–127. [Google Scholar] [CrossRef]


| Gene | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| SREBP-1c | CCC TGC GAA GTG CTC ACA A | GCG TTT CTA CCA CTT CAG GTT TCA |
| FAS | GCT GCT ACA AAC AGG ACC ATC AC | TCT TGC TGG CCT CCA CTG AC |
| C/EBPα | GCC AAG AAG TCG GTG GAT AA | CCT TGA CCA AGG AGC TCT CA |
| ACC | CAA TCC TCG GCA CAT GGA GA | GCT CAG CCA AGC GGA TGT AGA |
| CPT-1 | CCA TCT CTT CTG CCT CTA TGT | GTC AGG GTT TTT CTC AAA GTC |
| β-actin | GAT TAC TGC CCT GGC TCC TA | TCA TCG TAC TCC TGC TTG CT |
| Groups | Initial Body Weight | Body Weight after Randomization (A) | Terminal Body Weight (B) | Body Weight Gain (B-A) |
|---|---|---|---|---|
| NC | 194.0 ± 8.4 | 485.5 ± 38.9 a | 622.8 ± 55.6 a | 137.2 ± 30.8 a |
| HC | 194.0 ± 7.6 | 522.4 ± 29.5 b | 733.3 ± 87.2 b | 210.9 ± 63.1 b |
| PC | 522.4 ± 28.6 b | 680.4 ± 57.5 ab | 158.0 ± 35.9 a | |
| K2 | 521.9 ± 31.1 b | 677.9 ± 64.8 ab | 156.0 ± 37.7 a | |
| K6 | 523.6 ± 27.1 b | 697.1 ± 55.6 b | 173.5 ± 30.9 a |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, E.-J.; Lee, Y.-S.; Kim, S.M.; Park, G.-S.; Lee, Y.H.; Jeong, D.Y.; Kang, J.; Lee, H.-J. Beneficial Effects of Lactobacillus plantarum Strains on Non-Alcoholic Fatty Liver Disease in High Fat/High Fructose Diet-Fed Rats. Nutrients 2020, 12, 542. https://doi.org/10.3390/nu12020542
Park E-J, Lee Y-S, Kim SM, Park G-S, Lee YH, Jeong DY, Kang J, Lee H-J. Beneficial Effects of Lactobacillus plantarum Strains on Non-Alcoholic Fatty Liver Disease in High Fat/High Fructose Diet-Fed Rats. Nutrients. 2020; 12(2):542. https://doi.org/10.3390/nu12020542
Chicago/Turabian StylePark, Eun-Jung, You-Suk Lee, Sung Min Kim, Gun-Seok Park, Yong Hyun Lee, Do Yeun Jeong, Jihee Kang, and Hae-Jeung Lee. 2020. "Beneficial Effects of Lactobacillus plantarum Strains on Non-Alcoholic Fatty Liver Disease in High Fat/High Fructose Diet-Fed Rats" Nutrients 12, no. 2: 542. https://doi.org/10.3390/nu12020542
APA StylePark, E.-J., Lee, Y.-S., Kim, S. M., Park, G.-S., Lee, Y. H., Jeong, D. Y., Kang, J., & Lee, H.-J. (2020). Beneficial Effects of Lactobacillus plantarum Strains on Non-Alcoholic Fatty Liver Disease in High Fat/High Fructose Diet-Fed Rats. Nutrients, 12(2), 542. https://doi.org/10.3390/nu12020542






