Biological Role of Unsaturated Fatty Acid Desaturases in Health and Disease
Abstract
1. Introduction
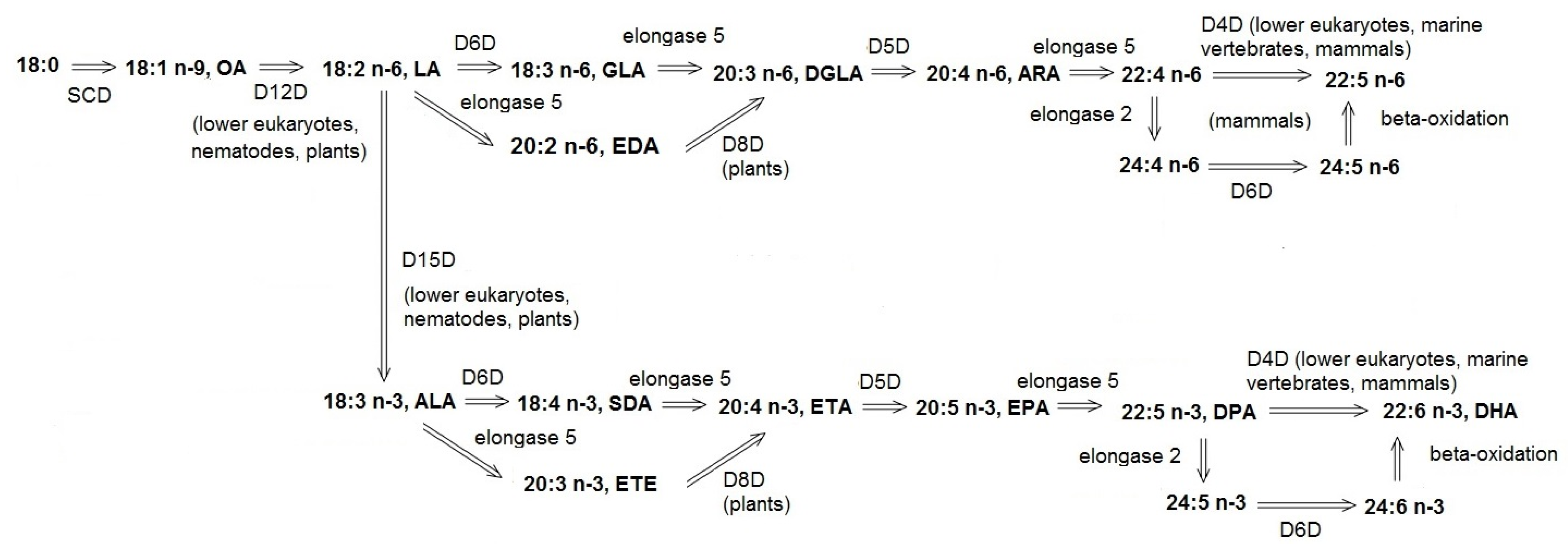
2. Activity of Unsaturated Fatty Acid Desaturases
3. Role of Unsaturated Fatty Acid Desaturases
3.1. Role of Unsaturated Fatty Acid Desaturases in Microorganisms
3.2. Role of Unsaturated Fatty Acid Desaturases in Plants
3.3. Role of Unsaturated Fatty Acid Desaturases in Humans and Other Mammals
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ADHD | Attention-Deficit Hyperactivity Disorder |
| ALA | alpha-linolenic acid, 18:3 n-3 |
| ARA | arachidonic acid, 20:4 n-6 |
| CAD | coronary artery disease |
| CD | Crohn’s disease |
| CLA | conjugated linoleic acid, trans-11, cis-13 18:2 |
| D5D | delta-5 desaturase |
| D6D | delta-6 desaturase |
| D12D | delta-12 desaturase |
| D15D | delta-15 desaturase |
| DGLA | dihomo-gamma-linolenic acid, 20:3 n-6 |
| DHA | docosahexaenoic acid, 22:6 n-3 |
| DPA | docosapentaenoic acid, 22:5 n-3 |
| EDA | eicosadienoic acid, 20:2 n-6 |
| ETA | eicosatetraenoic acid, 20:4 n-3 |
| ETE | eicosatrienoic acid, 20:3 n-3 |
| EPA | eicosapentaenoic acid, 20:5 n-3 |
| FA | fatty acid |
| GLA | gamma-linolenic acid, 18:3 n-6 |
| LA | linoleic acid, 18:2 n-6 |
| MUFA | monounsaturated fatty acids |
| NAD | nicotinamide adenine dinucleotide |
| NADH | reduced nicotinamide adenine dinucleotide |
| NAFLD | non-alcoholic fatty liver disease |
| OA | oleic acid, 18:1 n-9 |
| PUFA | polyunsaturated fatty acids |
| SCD | stearoyl-CoA desaturase |
| SDA | stearidonic acid, 18:4 n-3 |
| SNP | single nucleotide polymorphism |
| VA | trans-vaccenic acid, trans-11 18:1 |
References
- Das, U.N. Biological significance of essential fatty acids. J. Assoc. Physicians India 2006, 54, 309–319. [Google Scholar] [PubMed]
- Calder, P.C. Functional Roles of Fatty Acids and Their Effects on Human Health. J. Parenter. Enter. Nutr. 2015, 39, 18S–32S. [Google Scholar] [CrossRef] [PubMed]
- Zárate, R.; El Jaber-Vazdekis, N.; Tejera, N.; Pérez, J.A.; Rodríguez, C. Significance of long chain polyunsaturated fatty acids in human health. Clin. Transl. Med. 2017, 6, 25. [Google Scholar] [CrossRef] [PubMed]
- Nagy, K.; Tiuca, I.-D. Importance of Fatty Acids in Physiopathology of Human Body. In Fatty Acids; InTech: London, UK, 2017. [Google Scholar]
- Massey, K.A.; Nicolaou, A. Lipidomics of polyunsaturated-fatty-acid-derived oxygenated metabolites. Biochem. Soc. Trans. 2011, 39, 1240–1246. [Google Scholar] [CrossRef]
- Burdge, G.C. Is essential fatty acid interconversion an important source of PUFA in humans? Br. J. Nutr. 2019, 121, 615–624. [Google Scholar] [CrossRef]
- Davidson, J.; Rotondo, D.; Rizzo, M.; Leaver, H. Therapeutic implications of disorders of cell death signalling: Membranes, micro-environment, and eicosanoid and docosanoid metabolism. Br. J. Pharmacol. 2012, 166, 1193–1210. [Google Scholar] [CrossRef]
- de Bus, I.; Witkamp, R.; Zuilhof, H.; Albada, B.; Balvers, M. The role of n-3 PUFA-derived fatty acid derivatives and their oxygenated metabolites in the modulation of inflammation. Prostaglandins Other Lipid Mediat. 2019, 144, 106351. [Google Scholar] [CrossRef]
- Sijben, J.W.C.; Calder, P.C. Differential immunomodulation with long-chain n-3 PUFA in health and chronic disease. Proc. Nutr. Soc. 2007, 66, 237–259. [Google Scholar] [CrossRef]
- Marventano, S.; Galvano, F.; Buscemi, S.; Mistretta, A. A review of recent evidence in human studies of n-3 and n-6 PUFA intake on cardiovascular disease, cancer, and depressive disorders: Does the ratio really matter? Artic. Int. J. Food Sci. Nutr. 2015. [Google Scholar] [CrossRef]
- Kim, W.; Deik, A.; Gonzalez, C.; Gonzalez, M.E.; Fu, F.; Ferrari, M.; Churchhouse, C.L.; Florez, J.C.; Jacobs, S.B.R.; Clish, C.B.; et al. Polyunsaturated Fatty Acid Desaturation Is a Mechanism for Glycolytic NAD + Recycling. Cell Metab. 2019, 29, 856–870.e7. [Google Scholar] [CrossRef]
- Huang, J.-Z.; Jiang, X.-Z.; Xia, X.-F.; Yu, A.-Q.; Mao, R.-Y.; Chen, X.-F.; Tian, B.-Y. Cloning and Functional Identification of Delta5 Fatty Acid Desaturase Gene and Its 5′-Upstream Region from Marine Fungus Thraustochytrium sp. FJN-10. Mar. Biotechnol. 2011, 13, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, T.; Sakaguchi, K.; Hamaguchi, R.; Kobayashi, T.; Abe, E.; Hama, Y.; Hayashi, M.; Honda, D.; Okita, Y.; Sugimoto, S.; et al. Analysis of Δ12-fatty acid desaturase function revealed that two distinct pathways are active for the synthesis of PUFAs in T. aureum ATCC 34304. J. Lipid Res. 2012, 53, 1210–1222. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Yu, A.; Li, M.; Ou, X.; Xing, L.; Li, M. Identification of a novel C22-∆4-producing docosahexaenoic acid (DHA) specific polyunsaturated fatty acid desaturase gene from Isochrysis galbana and its expression in Saccharomyces cerevisiae. Biotechnol. Lett. 2012, 34, 2265–2274. [Google Scholar] [CrossRef] [PubMed]
- Martin-Montalvo, A.; Sun, Y.; Diaz-Ruiz, A.; Ali, A.; Gutierrez, V.; Palacios, H.H.; Curtis, J.; Siendones, E.; Ariza, J.; Abulwerdi, G.A.; et al. Cytochrome b 5 reductase and the control of lipid metabolism and healthspan. NPJ Aging Mech. Dis. 2016, 2. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, H.; Zhang, J.; Hu, Y.; Zhang, L.; Wu, X.; Su, X.; Li, T.; Zou, X.; Liang, B. The cytochrome b5 reductase HPO-19 is required for biosynthesis of polyunsaturated fatty acids in Caenorhabditis elegans. BBA - Mol. Cell Biol. Lipids 2016, 1861, 310–319. [Google Scholar] [CrossRef] [PubMed]
- Behrouzian, B.; Buist, P.H. Mechanism of fatty acid desaturation: a bioorganic perspective. Prostaglandins. Leukot. Essent. Fatty Acids 2003, 68, 107–112. [Google Scholar] [CrossRef]
- Los, D.A.; Murata, N. Structure and expression of fatty acid desaturases. Biochim. Biophys. Acta - Lipids Lipid Metab. 1998, 1394, 3–15. [Google Scholar] [CrossRef]
- Meesapyodsuk, D.; Qiu, X. The Front-end Desaturase: Structure, Function, Evolution and Biotechnological Use. Lipids 2012, 47, 227–237. [Google Scholar] [CrossRef]
- Ternes, P.; Franke, S.; Zähringer, U.; Sperling, P.; Heinz, E. Identification and characterization of a sphingolipid delta 4-desaturase family. J. Biol. Chem. 2002, 277, 25512–25518. [Google Scholar] [CrossRef]
- Sperling, P.; Ternes, P.; Zank, T.K.; Heinz, E. The evolution of desaturases. Prostaglandins. Leukot. Essent. Fatty Acids 2003, 68, 73–95. [Google Scholar] [CrossRef]
- Hucik, B.; Sarr, O.; Nakamura, M.T.; Dyck, D.J.; Mutch, D.M. Reduced delta-6 desaturase activity partially protects against high-fat diet-induced impairment in whole-body glucose tolerance. J. Nutr. Biochem. 2019, 67, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Park, W.J.; Kothapalli, K.S.D.; Lawrence, P.; Tyburczy, C.; Brenna, J.T. An alternate pathway to long-chain polyunsaturates: the FADS2 gene product Δ8-desaturates 20:2n-6 and 20:3n-3. J. Lipid Res. 2009, 50, 1195–1202. [Google Scholar] [CrossRef] [PubMed]
- Guillou, H.; Rioux, V.; Catheline, D.; Thibault, J.-N.; Bouriel, M.; Jan, S.; D’Andrea, S.; Legrand, P. Conversion of hexadecanoic acid to hexadecenoic acid by rat Δ6-desaturase. J. Lipid Res. 2003, 44, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Gordon, J.S.; Hsuan, C.; Stenn, K.; Prouty, S.M. Identification of the Δ-6 Desaturase of Human Sebaceous Glands: Expression and Enzyme Activity. J. Investig. Dermatol. 2003, 120, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Harauma, A.; Hatanaka, E.; Yasuda, H.; Nakamura, M.T.; Salem, N.; Moriguchi, T. Effects of arachidonic acid, eicosapentaenoic acid and docosahexaenoic acid on brain development using artificial rearing of delta-6-desaturase knockout mice. Prostaglandins, Leukot. Essent. Fat. Acids 2017, 127, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Sayanova, O.; Haslam, R.; Venegas Caleron, M.; Napier, J.A. Cloning and characterization of unusual fatty acid desaturases from Anemone leveillei: Identification of an acyl-coenzyme A C20 Delta5-desaturase responsible for the synthesis of sciadonic acid. Plant Physiol. 2007, 144, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.; Datla, N.; Mackenzie, S.L.; Qiu, X. Isolation and characterization of a Δ5 FA desaturase from Pythium irregulare by heterologous expression in Saccharomyces cerevisiae and oilseed crops. Lipids 2002, 37, 863–868. [Google Scholar] [CrossRef]
- Lamers, D.; Visscher, B.; Weusthuis, R.A.; Francke, C.; Wijffels, R.H.; Lokman, C. Overexpression of delta-12 desaturase in the yeast Schwanniomyces occidentalis enhances the production of linoleic acid. Bioresour. Technol. 2019, 289, 121672. [Google Scholar] [CrossRef]
- Cripps, C.; Borgeson, C.; Blomquist, G.J.; de Renobales, M. The Δ12-desaturase from the house cricket, Acheta domesticus (Orthoptera: Gryllidae): Characterization and form of the substrate. Arch. Biochem. Biophys. 1990, 278, 46–51. [Google Scholar] [CrossRef]
- Borgeson, C.E.; de Renobales, M.; Blomquist, G.J. Characterization of the Δ12 desaturase in the American cockroach, Periplaneta americana: The nature of the substrate. Biochim. Biophys. Acta - Lipids Lipid Metab. 1990, 1047, 135–140. [Google Scholar] [CrossRef]
- Brandstetter, B.; Ruther, J. An insect with a delta-12 desaturase, the jewel wasp Nasonia vitripennis, benefits from nutritional supply with linoleic acid. Sci. Nat. 2016, 103, 40. [Google Scholar] [CrossRef] [PubMed]
- Vessby, B.; Gustafsson, I.-B.; Tengblad, S.; Berglund, L. Indices of fatty acid desaturase activity in healthy human subjects: Effects of different types of dietary fat. Br. J. Nutr. 2013, 110, 871–879. [Google Scholar] [CrossRef] [PubMed]
- Joshi, K.; Gadgil, M.; Pandit, A.; Otiv, S.; Kothapalli, K.S.D.; Brenna, J.T. Dietary pattern regulates fatty acid desaturase 1 gene expression in Indian pregnant women to spare overall long chain polyunsaturated fatty acids levels. Mol. Biol. Rep. 2019, 46, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Ralston, J.C.; Matravadia, S.; Gaudio, N.; Holloway, G.P.; Mutch, D.M. Polyunsaturated fatty acid regulation of adipocyte FADS1 and FADS2 expression and function. Obesity 2015, 23, 725–728. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Mai, K.; Xu, W.; Yuan, Y.; Zhang, Y.; Ai, Q. Characterization, mRNA expression and regulation of δ6 fatty acyl desaturase (FADS2) by dietary n-3 long chain polyunsaturated fatty acid (LC-PUFA) levels in grouper larvae (Epinephelus coioides). Aquaculture 2014, 434, 212–219. [Google Scholar] [CrossRef]
- Wijendran, V.; Downs, I.; Srigley, C.T.; Kothapalli, K.S.D.; Park, W.J.; Blank, B.S.; Zimmer, J.P.; Butt, C.M.; Salem, N.; Brenna, J.T. Dietary arachidonic acid and docosahexaenoic acid regulate liver fatty acid desaturase (FADS) alternative transcript expression in suckling piglets. Prostaglandins Leukot. Essent. Fat. Acids 2013, 89, 345–350. [Google Scholar] [CrossRef]
- Geay, F.; Santigosa I Culi, E.; Corporeau, C.; Boudry, P.; Dreano, Y.; Corcos, L.; Bodin, N.; Vandeputte, M.; Zambonino-Infante, J.L.; Mazurais, D.; et al. Regulation of FADS2 expression and activity in European sea bass (Dicentrarchus labrax, L.) fed a vegetable diet. Comp. Biochem. Physiol. - B Biochem. Mol. Biol. 2010, 156, 237–243. [Google Scholar] [CrossRef]
- Brenner, R.R. Hormonal modulation of δ6 and δ5 desaturases: Case of diabetes. Prostaglandins Leukot. Essent. Fat. Acids 2003, 68, 151–162. [Google Scholar] [CrossRef]
- Childs, C.E.; Romeu-Nadal, M.; Burdge, G.C.; Calder, P.C. The Polyunsaturated Fatty Acid Composition of Hepatic and Plasma Lipids Differ by Both Sex and Dietary Fat Intake in Rats. J. Nutr. 2010, 140, 245–250. [Google Scholar] [CrossRef]
- McNamara, R.K.; Able, J.; Jandacek, R.; Rider, T.; Tso, P. Gender differences in rat erythrocyte and brain docosahexaenoic acid composition: Role of ovarian hormones and dietary omega-3 fatty acid composition. Psychoneuroendocrinology 2009, 34, 532–539. [Google Scholar] [CrossRef]
- Dong, Y.; Wang, S.; You, C.; Xie, D.; Jiang, Q.; Li, Y. Hepatocyte nuclear factor 4α (Hnf4α) is involved in transcriptional regulation of Δ6/Δ5 fatty acyl desaturase (fad) gene expression in marine teleost Siganus canaliculatus. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2019, 110353. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.; Fu, Z.; Wang, S.; You, C.; Monroig, Ó.; Tocher, D.R.; Li, Y. Characteristics of the fads2 gene promoter in marine teleost Epinephelus coioides and role of Sp1-binding site in determining promoter activit. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Zhao, J.; Chen, J.; Wang, S.; Liu, Y.; Zhang, Q.; You, C.; Monroig, Ó.; Tocher, D.R.; Li, Y. Cloning and characterization of ∆6/∆5 fatty acyl desaturase (Fad) gene promoter in the marine teleost Siganus canaliculatus. Gene 2018, 647, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Howard, T.D.; Mathias, R.A.; Seeds, M.C.; Herrington, D.M.; Hixson, J.E.; Shimmin, L.C.; Hawkins, G.A.; Sellers, M.; Ainsworth, H.C.; Sergeant, S.; et al. DNA Methylation in an Enhancer Region of the FADS Cluster Is Associated with FADS Activity in Human Liver. PLoS ONE 2014, 9, e97510. [Google Scholar] [CrossRef] [PubMed]
- Rahbar, E.; Ainsworth, H.C.; Howard, T.D.; Hawkins, G.A.; Ruczinski, I.; Mathias, R.; Seeds, M.C.; Sergeant, S.; Hixson, J.E.; Herrington, D.M.; et al. Uncovering the DNA methylation landscape in key regulatory regions within the FADS cluster. PLoS ONE 2017, 12, e0180903. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.; Nuccio, M.L.; Gross, L.M.; Thomas, T.L. Isolation of a delta 6-desaturase gene from the cyanobacterium Synechocystis sp. strain PCC 6803 by gain-of-function expression in Anabaena sp. strain PCC 7120. Plant Mol. Biol. 1993, 22, 293–300. [Google Scholar] [CrossRef]
- Michaelson, L.V.; Lazarus, C.M.; Griffiths, G.; Napier, J.A.; Stobart, A.K. Isolation of a Δ 5 -Fatty Acid Desaturase Gene from Mortierella alpina. J. Biol. Chem. 1998, 273, 19055–19059. [Google Scholar] [CrossRef]
- Wallis, J.G.; Browse, J. The Δ8-Desaturase ofEuglena gracilis:An Alternate Pathway for Synthesis of 20-Carbon Polyunsaturated Fatty Acids. Arch. Biochem. Biophys. 1999, 365, 307–316. [Google Scholar] [CrossRef]
- Diomandé, S.E.; Guinebretière, M.-H.; De Sarrau, B.; Nguyen-the, C.; Broussolle, V.; Brillard, J. Fatty acid profiles and desaturase-encoding genes are different in thermo- and psychrotolerant strains of the Bacillus cereus Group. BMC Res. Notes 2015, 8, 329. [Google Scholar] [CrossRef]
- Yoshida, K.; Hashimoto, M.; Hori, R.; Adachi, T.; Okuyama, H.; Orikasa, Y.; Nagamine, T.; Shimizu, S.; Ueno, A.; Morita, N.; et al. Bacterial Long-Chain Polyunsaturated Fatty Acids: Their Biosynthetic Genes, Functions, and Practical Use. Mar. Drugs 2016, 14, 94. [Google Scholar] [CrossRef]
- Siliakus, M.F.; van der Oost, J.; Kengen, S.W.M. Adaptations of archaeal and bacterial membranes to variations in temperature, pH and pressure. Extremophiles 2017, 21, 651–670. [Google Scholar] [CrossRef] [PubMed]
- Bale, N.J.; Rijpstra, W.I.C.; Sahonero-Canavesi, D.X.; Oshkin, I.Y.; Belova, S.E.; Dedysh, S.N.; Sinninghe Damsté, J.S. Fatty Acid and Hopanoid Adaption to Cold in the Methanotroph Methylovulum psychrotolerans. Front. Microbiol. 2019, 10, 589. [Google Scholar] [CrossRef] [PubMed]
- Okuyama, H.; Orikasa, Y.; Nishida, T.; Watanabe, K.; Morita, N. Bacterial Genes Responsible for the Biosynthesis of Eicosapentaenoic and Docosahexaenoic Acids and Their Heterologous Expression. Appl. Environ. Microbiol. 2007, 51, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Martins, D.; Custódio, L.; Barreira, L.; Pereira, H.; Ben-Hamadou, R.; Varela, J.; Abu-Salah, K.; Martins, D.A.; Custódio, L.; Barreira, L.; et al. Alternative Sources of n-3 Long-Chain Polyunsaturated Fatty Acids in Marine Microalgae. Mar. Drugs 2013, 11, 2259–2281. [Google Scholar] [CrossRef]
- Marella, E.R.; Holkenbrink, C.; Siewers, V.; Borodina, I. Engineering microbial fatty acid metabolism for biofuels and biochemicals. Curr. Opin. Biotechnol. 2018, 50, 39–46. [Google Scholar] [CrossRef]
- Bellou, S.; Triantaphyllidou, I.-E.; Aggeli, D.; Elazzazy, A.M.; Baeshen, M.N.; Aggelis, G. Microbial oils as food additives: Recent approaches for improving microbial oil production and its polyunsaturated fatty acid content. Curr. Opin. Biotechnol. 2016, 37, 24–35. [Google Scholar] [CrossRef]
- Winwood, R.J. Micro-organismes producteurs de lipides Recent developments in the commercial production of DHA and EPA rich oils from micro-algae. EDP Sci. 2013, 20. [Google Scholar]
- Chauton, M.S.; Reitan, K.I.; Norsker, N.H.; Tveterås, R.; Kleivdal, H.T. A techno-economic analysis of industrial production of marine microalgae as a source of EPA and DHA-rich raw material for aquafeed: Research challenges and possibilities. Aquaculture 2015, 436, 95–103. [Google Scholar] [CrossRef]
- Sahin, D.; Tas, E.; Altindag, U.H. Enhancement of docosahexaenoic acid (DHA) production from Schizochytrium sp. S31 using different growth medium conditions. AMB Express 2018, 8, 7. [Google Scholar] [CrossRef]
- Peltomaa, E.; Johnson, M.; Taipale, S. Marine Cryptophytes Are Great Sources of EPA and DHA. Mar. Drugs 2017, 16, 3. [Google Scholar] [CrossRef]
- Shanklin, J.; Cahoon, E.B. Desaturation and related modifications of fatty acids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 611–641. [Google Scholar] [CrossRef] [PubMed]
- Iba, K. Acclimative response to temperature stress in higher plants: Approaches of Gene Engineering for Temperature Tolerance. Annu. Rev. Plant Biol. 2002, 53, 225–245. [Google Scholar] [CrossRef] [PubMed]
- Gibson, S.; Arondel, V.; Iba, K.; Somerville, C. Cloning of a temperature-regulated gene encoding a chloroplast omega-3 desaturase from Arabidopsis thaliana. Plant Physiol. 1994, 106, 1615–1621. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Chen, M.; Zhu, M.; Jiang, Y.; Meng, J.; Zhao, D.; Tao, J. Cloning, Characterization, and Expression Analysis of Three FAD8 Genes Encoding a Fatty Acid Desaturase from Seeds of Paeonia ostii. Molecules 2018, 23, 929. [Google Scholar] [CrossRef] [PubMed]
- Hazel, J.R.; Eugene Williams, E. The role of alterations in membrane lipid composition in enabling physiological adaptation of organisms to their physical environment. Prog. Lipid Res. 1990, 29, 167–227. [Google Scholar] [CrossRef]
- Kodama, H.; Hamada, T.; Horiguchi, G.; Nishimura, M.; Iba, K. Genetic Enhancement of Cold Tolerance by Expression of a Gene for Chloroplast [omega]-3 Fatty Acid Desaturase in Transgenic Tobacco. Plant Physiol. 1994, 105, 601–605. [Google Scholar] [CrossRef]
- Murakami, Y.; Tsuyama, M.; Kobayashi, Y.; Kodama, H.; Iba, K. Trienoic fatty acids and plant tolerance of high temperature. Science 2000, 287, 476–479. [Google Scholar] [CrossRef]
- Zhang, J.-T.; Zhu, J.-Q.; Zhu, Q.; Liu, H.; Gao, X.-S.; Zhang, H.-X. Fatty acid desaturase-6 (Fad6) is required for salt tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2009, 390, 469–474. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, H.; Sun, J.; Li, B.; Zhu, Q.; Chen, S.; Zhang, H. Arabidopsis Fatty Acid Desaturase FAD2 Is Required for Salt Tolerance during Seed Germination and Early Seedling Growth. PLoS ONE 2012, 7, e30355. [Google Scholar] [CrossRef]
- Hernández, M.L.; Sicardo, M.D.; Martínez-Rivas, J.M. Differential Contribution of Endoplasmic Reticulum and Chloroplast ω-3 Fatty Acid Desaturase Genes to the Linolenic Acid Content of Olive (Olea europaea) Fruit. Plant Cell Physiol. 2016, 57, 138–151. [Google Scholar] [CrossRef]
- Lakra, N.; Mahmood, S.; Marwal, A.; Sudheep, N.M.; Anwar, K. Bioengineered Plants Can Be an Alternative Source of Omega-3 Fatty Acids for Human Health. In Plant and Human Health, Volume 2; Springer International Publishing: Cham, Switzerland, 2019; pp. 361–382. [Google Scholar]
- Qiu, X.; Hong, H.; Datla, N.; MacKenzie, S.L.; Taylor, D.C.; Thomas, T.L. Expression of borage Δ6 desaturase in Saccharomyces cerevisiae and oilseed crops. Can. J. Bot. 2002, 80, 42–49. [Google Scholar] [CrossRef]
- Sayanova, O.; Smith, M.A.; Lapinskas, P.; Stobart, A.K.; Dobson, G.; Christie, W.W.; Shewry, P.R.; Napier, J.A. Expression of a borage desaturase cDNA containing an N-terminal cytochrome b5 domain results in the accumulation of high levels of delta6-desaturated fatty acids in transgenic tobacco. Proc. Natl. Acad. Sci. USA 1997, 94, 4211–4216. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-López, N.; Haslam, R.P.; Venegas-Calerón, M.; Larson, T.R.; Graham, I.A.; Napier, J.A.; Sayanova, O. The synthesis and accumulation of stearidonic acid in transgenic plants: A novel source of ‘heart-healthy’ omega-3 fatty acids. Plant Biotechnol. J. 2009, 7, 704–716. [Google Scholar] [CrossRef] [PubMed]
- Okuzaki, A.; Ogawa, T.; Koizuka, C.; Kaneko, K.; Inaba, M.; Imamura, J.; Koizuka, N. CRISPR/Cas9-mediated genome editing of the fatty acid desaturase 2 gene in Brassica napus. Plant Physiol. Biochem. 2018, 131, 63–69. [Google Scholar] [CrossRef]
- Shrestha, P.; Zhou, X.-R.; Vibhakaran Pillai, S.; Petrie, J.; de Feyter, R.; Singh, S. Comparison of the Substrate Preferences of ω3 Fatty Acid Desaturases for Long Chain Polyunsaturated Fatty Acids. Int. J. Mol. Sci. 2019, 20, 3058. [Google Scholar] [CrossRef]
- Cheng, B.; Wu, G.; Vrinten, P.; Falk, K.; Bauer, J.; Qiu, X. Towards the production of high levels of eicosapentaenoic acid in transgenic plants: The effects of different host species, genes and promoters. Transgenic Res. 2010, 19, 221–229. [Google Scholar] [CrossRef]
- Damude, H.G.; Kinney, A.J. Engineering Oilseed Plants for a Sustainable, Land-Based Source of Long Chain Polyunsaturated Fatty Acids. Lipids 2007, 42, 179–185. [Google Scholar] [CrossRef]
- Ruiz-Lopez, N.; Haslam, R.P.; Usher, S.L.; Napier, J.A.; Sayanova, O. Reconstitution of EPA and DHA biosynthesis in Arabidopsis: Iterative metabolic engineering for the synthesis of n−3 LC-PUFAs in transgenic plants. Metab. Eng. 2013, 17, 30–41. [Google Scholar] [CrossRef]
- Usher, S.; Haslam, R.P.; Ruiz-Lopez, N.; Sayanova, O.; Napier, J.A. Field trial evaluation of the accumulation of omega-3 long chain polyunsaturated fatty acids in transgenic Camelina sativa: Making fish oil substitutes in plants. Metab. Eng. Commun. 2015, 2, 93–98. [Google Scholar] [CrossRef]
- Marwal, A.; Anwar, K. Bioengineered Plants Can Be an Alternative Source of Omega-3 Fatty Acids for Human Health: Phytochemistry and Molecular Aspects. In Plant and Human Health; Springer: Cham, Switzerland, 2019. [Google Scholar]
- Coppens, P.; da Silva, M.F.; Pettman, S. European regulations on nutraceuticals, dietary supplements and functional foods: A framework based on safety. Toxicology 2006, 221, 59–74. [Google Scholar] [CrossRef]
- Starr, R.R. Too Little, Too Late: Ineffective Regulation of Dietary Supplements in the United States. Am. J. Public Health 2015, 105, 478. [Google Scholar] [CrossRef] [PubMed]
- Resnik, D.B. Proportionality in Public Health Regulation: The Case of Dietary Supplements. Food Ethics 2017. [Google Scholar] [CrossRef] [PubMed]
- Salvador, A.M.; García-Maldonado, E.; Gallego-Narbón, A.; Zapatera, B.; Vaquero, M.P. Fatty Acid Profile and Cardiometabolic Markers in Relation with Diet Type and Omega-3 Supplementation in Spanish Vegetarians. Nutrients 2019, 11, 1659. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Huang, T.; Weng, X.; Shou, T.; Wang, Q.; Zhou, X.; Hu, Q.; Li, D. Plasma n-3 and n-6 fatty acids and inflammatory markers in Chinese vegetarians. Lipids Health Dis. 2014, 13, 151. [Google Scholar] [CrossRef] [PubMed]
- Sanders, T.A.B. Polyunsaturated Fatty Acid Status in Vegetarians. In Vegetarian and Plant-Based Diets in Health and Disease Prevention; Elsevier Inc.: Amsterdam, the Netherlands, 2017; pp. 667–681. ISBN 9780128039694. [Google Scholar]
- Tanaka, T.; Shen, J.; Abecasis, G.R.; Kisialiou, A.; Ordovas, J.M.; Guralnik, J.M.; Singleton, A.; Bandinelli, S.; Cherubini, A.; Arnett, D.; et al. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genet. 2009, 5, e1000338. [Google Scholar] [CrossRef] [PubMed]
- Merino, D.M.; Johnston, H.; Clarke, S.; Roke, K.; Nielsen, D.; Badawi, A.; El-Sohemy, A.; Ma, D.W.L.; Mutch, D.M. Polymorphisms in FADS1 and FADS2 alter desaturase activity in young Caucasian and Asian adults. Mol. Genet. Metab. 2011, 103, 171–178. [Google Scholar] [CrossRef]
- de Toro-Martín, J.; Guénard, F.; Rudkowska, I.; Lemieux, S.; Couture, P.; Vohl, M.-C.C. A common variant in ARHGEF10 alters delta-6 desaturase activity and influence susceptibility to hypertriglyceridemia. J. Clin. Lipidol. 2018, 12, 311–320.e3. [Google Scholar] [CrossRef]
- Xie, L.; Innis, S.M. Association of Fatty Acid Desaturase Gene Polymorphisms with Blood Lipid Essential Fatty Acids and Perinatal Depression among Canadian Women: A Pilot Study. J. Nutrigenet. Nutrigenomics 2009, 2, 243–250. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Kothapalli, K.S.D.; Brenna, J.T. Desaturase and elongase-limiting endogenous long-chain polyunsaturated fatty acid biosynthesis. Curr. Opin. Clin. Nutr. Metab. Care 2016, 19, 103–110. [Google Scholar] [CrossRef]
- Dumont, J.; Huybrechts, I.; Spinneker, A.; Gottrand, F.; Grammatikaki, E.; Bevilacqua, N.; Vyncke, K.; Widhalm, K.; Kafatos, A.; Molnar, D.; et al. FADS1 Genetic Variability Interacts with Dietary α-Linolenic Acid Intake to Affect Serum Non-HDL–Cholesterol Concentrations in European Adolescents. J. Nutr. 2011, 141, 1247–1253. [Google Scholar] [CrossRef]
- Meldrum, S.J.; Li, Y.; Zhang, G.; Heaton, A.E.M.; D’vaz, N.; Manz, J.; Reischl, E.; Berthold; Koletzko, V.; Prescott, S.L.; et al. Can polymorphisms in the fatty acid desaturase (FADS) gene cluster alter the effects of fish oil supplementation on plasma and erythrocyte fatty acid profiles? An exploratory study. Eur. J. Nutr. 2018, 57, 2583–2594. [Google Scholar] [CrossRef]
- Czajkowska, M.; Brzęk, P.; Dobrzyń, P. A novel polymorphism in the fatty acid desaturase 2 gene (Fads2): A possible role in the basal metabolic rate. PLoS ONE 2019, 14, e0213138. [Google Scholar] [CrossRef]
- Garcia, C.; Guillocheau, E.; Richard, L.; Drouin, G.; Catheline, D.; Legrand, P.; Rioux, V. Conversion of dietary trans-vaccenic acid to trans11,cis13-conjugated linoleic acid in the rat lactating mammary gland by Fatty Acid Desaturase 3-catalyzed methyl-end Δ13-desaturation. Biochem. Biophys. Res. Commun. 2018, 505, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Garcia, C.; Duby, C.; Catheline, D.; Toral, P.G.; Bernard, L.; Legrand, P.; Rioux, V. Synthesis of the suspected trans-11,cis-13 conjugated linoleic acid isomer in ruminant mammary tissue by FADS3-catalyzed Δ13-desaturation of vaccenic acid. J. Dairy Sci. 2017, 100, 783–796. [Google Scholar] [CrossRef] [PubMed]
- de la Garza Puentes, A.; Montes Goyanes, R.; Chisaguano Tonato, A.M.; Torres-Espínola, F.J.; Arias García, M.; de Almeida, L.; Bonilla Aguirre, M.; Guerendiain, M.; Castellote Bargalló, A.I.; Segura Moreno, M.; et al. Association of maternal weight with FADS and ELOVL genetic variants and fatty acid levels- The PREOBE follow-up. PLoS ONE 2017, 12, e0179135. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Athinarayanan, S.; Jiang, G.; Chalasani, N.; Zhang, M.; Liu, W. Fatty Acid Desaturase 1 Gene Polymorphisms Control Human Hepatic Lipid Composition. Hepatology 2014. [Google Scholar]
- Kwak, J.H.; Paik, J.K.; Kim, O.Y.; Jang, Y.; Lee, S.H.; Ordovas, J.M.; Lee, J.H. FADS gene polymorphisms in Koreans: Association with ω6 polyunsaturated fatty acids in serum phospholipids, lipid peroxides, and coronary artery disease. Atherosclerosis 2011, 214, 94–100. [Google Scholar] [CrossRef]
- Li, S.W.; Lin, K.; Ma, P.; Zhang, Z.L.; Zhou, Y.D.; Lu, S.Y.; Zhou, X.; Liu, S.M. FADS Gene Polymorphisms Confer the Risk of Coronary Artery Disease in a Chinese Han Population through the Altered Desaturase Activities: Based on High-Resolution Melting Analysis. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-W.; Wang, J.; Yang, Y.; Liu, Z.-J.; Cheng, L.; Liu, H.-Y.; Ma, P.; Luo, W.; Liu, S.-M. Polymorphisms in FADS1 and FADS2 alter plasma fatty acids and desaturase levels in type 2 diabetic patients with coronary artery disease. J. Transl. Med. 2016, 14, 79. [Google Scholar] [CrossRef]
- Liu, F.; Li, Z.; Lv, X.; Ma, J. Dietary n-3 Polyunsaturated Fatty Acid Intakes Modify the Effect of Genetic Variation in Fatty Acid Desaturase 1 on Coronary Artery Disease. PLoS ONE 2015, 10, e0121255. [Google Scholar] [CrossRef]
- Kanai, M.; Akiyama, M.; Takahashi, A.; Matoba, N.; Momozawa, Y.; Ikeda, M.; Iwata, N.; Ikegawa, S.; Hirata, M.; Matsuda, K.; et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet. 2018, 50, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Stahl, E.A.; Breen, G.; Forstner, A.J.; McQuillin, A.; Ripke, S.; Trubetskoy, V.; Mattheisen, M.; Wang, Y.; Coleman, J.R.I.; Gaspar, H.A.; et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 2019, 51, 793–803. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M.; Takahashi, A.; Kamatani, Y.; Okahisa, Y.; Kunugi, H.; Mori, N.; Sasaki, T.; Ohmori, T.; Okamoto, Y.; Kawasaki, H.; et al. A genome-wide association study identifies two novel susceptibility loci and trans population polygenicity associated with bipolar disorder. Mol. Psychiatry 2018, 23, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Mychaleckyj, J.C.; Nayak, U.; Colgate, E.R.; Zhang, D.; Carstensen, T.; Ahmed, S.; ahmed, T.; Mentzer, A.J.; Alam, M.; Kirkpatrick, B.D.; et al. Multiplex genomewide association analysis of breast milk fatty acid composition extends the phenotypic association and potential selection of FADS1 variants to arachidonic acid, a critical infant micronutrient. J. Med. Genet. 2018, 55, 459–468. [Google Scholar]
- Wei, Q.; Yu, D.; Liu, M.; Wang, M.; Zhao, M.; Liu, M.; Jia, W.; Ma, H.; Fang, J.; Xu, W.; et al. Genome-wide association study identifies three susceptibility loci for laryngeal squamous cell carcinoma in the Chinese population. Nat. Genet. 2014, 46, 1110–1114. [Google Scholar] [CrossRef]
- Giri, A.; Hellwege, J.N.; Keaton, J.M.; Park, J.; Qiu, C.; Warren, H.R.; Torstenson, E.S.; Kovesdy, C.P.; Sun, Y.V.; Wilson, O.D.; et al. Trans-ethnic association study of blood pressure determinants in over 750,000 individuals. Nat. Genet. 2019, 51, 51–62. [Google Scholar] [CrossRef]
- van Setten, J.; Verweij, N.; Mbarek, H.; Niemeijer, M.N.; Trompet, S.; Arking, D.E.; Brody, J.A.; Gandin, I.; Grarup, N.; Hall, L.M.; et al. Genome-wide association meta-analysis of 30,000 samples identifies seven novel loci for quantitative ECG traits. Eur. J. Hum. Genet. 2019, 27, 952–962. [Google Scholar] [CrossRef]
- Eijgelsheim, M.; Newton-Cheh, C.; Sotoodehnia, N.; de bakker, P.I.W.; Müller, M.; Morrison, A.C.; Smith, A.V.; Isaacs, A.; Sanna, S.; Dörr, M.; et al. Genome-wide association analysis identifies multiple loci related to resting heart rate. Hum. Mol. Genet. 2010, 19, 3885–3894. [Google Scholar] [CrossRef]
- Den Hoed, M.; Eijgelsheim, M.; Esko, T.; Brundel, B.J.J.M.; Peal, D.S.; Evans, D.M.; Nolte, I.M.; Segrè, A.V.; Holm, H.; Handsaker, R.E.; et al. Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat. Genet. 2013, 45, 621–631. [Google Scholar] [CrossRef]
- Surakka, I.; Horikoshi, M.; Mägi, R.; Sarin, A.P.; Mahajan, A.; Lagou, V.; Marullo, L.; Ferreira, T.; Miraglio, B.; Timonen, S.; et al. The impact of low-frequency and rare variants on lipid levels. Nat. Genet. 2015, 47, 589–597. [Google Scholar] [CrossRef]
- Teslovich, T.M.; Musunuru, K.; Smith, A.V.; Edmondson, A.C.; Stylianou, I.M.; Koseki, M.; Pirruccello, J.P.; Ripatti, S.; Chasman, D.I.; Willer, C.J.; et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2010, 466, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Willer, C.J.; Schmidt, E.M.; Sengupta, S.; Peloso, G.M.; Gustafsson, S.; Kanoni, S.; Ganna, A.; Chen, J.; Buchkovich, M.L.; Mora, S.; et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013, 45, 1274–1285. [Google Scholar] [PubMed]
- Kathiresan, S.; Willer, C.J.; Peloso, G.M.; Demissie, S.; Musunuru, K.; Schadt, E.E.; Kaplan, L.; Bennett, D.; Li, Y.; Tanaka, T.; et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 2009, 41, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Waterworth, D.M.; Ricketts, S.L.; Song, K.; Chen, L.; Zhao, J.H.; Ripatti, S.; Aulchenko, Y.S.; Zhang, W.; Yuan, X.; Lim, N.; et al. Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 2264–2276. [Google Scholar] [CrossRef]
- Spracklen, C.N.; Chen, P.; Kim, Y.J.; Wang, X.; Cai, H.; Li, S.; Long, J.; Wu, Y.; Wang, Y.X.; Takeuchi, F.; et al. Association analyses of East Asian individuals and trans-ancestry analyses with European individuals reveal new loci associated with cholesterol and triglyceride levels. Hum. Mol. Genet. 2017, 26, 1770–1784. [Google Scholar] [CrossRef]
- Bentley, A.R.; Sung, Y.J.; Brown, M.R.; Winkler, T.W.; Kraja, A.T.; Ntalla, I.; Schwander, K.; Chasman, D.I.; Lim, E.; Deng, X.; et al. Multi-ancestry genome-wide gene–smoking interaction study of 387,272 individuals identifies new loci associated with serum lipids. Nat. Genet. 2019, 51, 636–648. [Google Scholar] [CrossRef]
- De Vries, P.S.; Brown, M.R.; Bentley, A.R.; Sung, Y.J.; Winkler, T.W.; Ntalla, I.; Schwander, K.; Kraja, A.T.; Guo, X.; Franceschini, N.; et al. Multiancestry Genome-Wide Association Study of Lipid Levels Incorporating Gene-Alcohol Interactions. Am. J. Epidemiol. 2019, 188, 1033–1054. [Google Scholar] [CrossRef]
- Kulminski, A.M.; Huang, J.; Loika, Y.; Arbeev, K.G.; Bagley, O.; Yashkin, A.; Duan, M.; Culminskaya, I. Strong impact of natural-selection-free heterogeneity in genetics of age-related phenotypes. Aging (Albany. NY). 2018, 10, 492–514. [Google Scholar] [CrossRef]
- Wojcik, G.L.; Graff, M.; Nishimura, K.K.; Tao, R.; Haessler, J.; Gignoux, C.R.; Highland, H.M.; Patel, Y.M.; Sorokin, E.P.; Avery, C.L.; et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature 2019, 570, 514–518. [Google Scholar] [CrossRef]
- Hoffmann, T.J.; Theusch, E.; Haldar, T.; Ranatunga, D.K.; Jorgenson, E.; Medina, M.W.; Kvale, M.N.; Kwok, P.Y.; Schaefer, C.; Krauss, R.M.; et al. A large electronic-health-record-based genome-wide study of serum lipids. Nat. Genet. 2018, 50, 401–413. [Google Scholar] [CrossRef]
- Sabatti, C.; Service, S.K.; Hartikainen, A.L.; Pouta, A.; Ripatti, S.; Brodsky, J.; Jones, C.G.; Zaitlen, N.A.; Varilo, T.; Kaakinen, M.; et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat. Genet. 2009, 41, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Klarin, D.; Damrauer, S.M.; Cho, K.; Sun, Y.V.; Teslovich, T.M.; Honerlaw, J.; Gagnon, D.R.; DuVall, S.L.; Li, J.; Peloso, G.M.; et al. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nat. Genet. 2018, 50, 1514–1523. [Google Scholar] [CrossRef] [PubMed]
- Kilpeläinen, T.O.; Bentley, A.R.; Noordam, R.; Sung, Y.J.; Schwander, K.; Winkler, T.W.; Jakupović, H.; Chasman, D.I.; Manning, A.; Ntalla, I.; et al. Multi-ancestry study of blood lipid levels identifies four loci interacting with physical activity. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Wessel, J.; Chu, A.Y.; Willems, S.M.; Wang, S.; Yaghootkar, H.; Brody, J.A.; Dauriz, M.; Hivert, M.F.; Raghavan, S.; Lipovich, L.; et al. Low-frequency and rare exome chip variants associate with fasting glucose and type 2 diabetes susceptibility. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Manning, A.K.; Hivert, M.F.; Scott, R.A.; Grimsby, J.L.; Bouatia-Naji, N.; Chen, H.; Rybin, D.; Liu, C.T.; Bielak, L.F.; Prokopenko, I.; et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 2012, 44, 659–669. [Google Scholar] [CrossRef]
- Dupuis, J.; Langenberg, C.; Prokopenko, I.; Saxena, R.; Soranzo, N.; Jackson, A.U.; Wheeler, E.; Glazer, N.L.; Bouatia-Naji, N.; Gloyn, A.L.; et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010, 42, 105–116. [Google Scholar] [CrossRef]
- Dorajoo, R.; Sun, Y.; Han, Y.; Ke, T.; Burger, A.; Chang, X.; Low, H.Q.; Guan, W.; Lemaitre, R.N.; Khor, C.C.; et al. A genome-wide association study of n-3 and n-6 plasma fatty acids in a Singaporean Chinese population. Genes Nutr. 2015, 10, 1–11. [Google Scholar] [CrossRef]
- Marklund, M.; Morris, A.P.; Mahajan, A.; Ingelsson, E.; Lindgren, C.M.; Lind, L.; Risérus, U. Genome-wide association studies of estimated fatty acid desaturase activity in serum and adipose tissue in elderly individuals: Associations with insulin sensitivity. Nutrients 2018, 10, 1791. [Google Scholar] [CrossRef]
- Kichaev, G.; Bhatia, G.; Loh, P.R.; Gazal, S.; Burch, K.; Freund, M.K.; Schoech, A.; Pasaniuc, B.; Price, A.L. Leveraging Polygenic Functional Enrichment to Improve GWAS Power. Am. J. Hum. Genet. 2019, 104, 65–75. [Google Scholar] [CrossRef]
- Astle, W.J.; Elding, H.; Jiang, T.; Allen, D.; Ruklisa, D.; Mann, A.L.; Mead, D.; Bouman, H.; Riveros-Mckay, F.; Kostadima, M.A.; et al. The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell 2016, 167, 1415–1429.e19. [Google Scholar] [CrossRef]
- Hong, K.W.; Jin, H.S.; Song, D.; Kwak, H.K.; Kim, S.S.; Kim, Y. Genome-wide association study of serum albumin:globulin ratio in Korean populations. J. Hum. Genet. 2013, 58, 174–177. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jonsson, S.; Sveinbjornsson, G.; De Lapuente Portilla, A.L.; Swaminathan, B.; Plomp, R.; Dekkers, G.; Ajore, R.; Ali, M.; Bentlage, A.E.H.; Elmér, E.; et al. Identification of sequence variants influencing immunoglobulin levels. Nat. Genet. 2017, 49, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- Pilling, L.C.; Atkins, J.L.; Duff, M.O.; Beaumont, R.N.; Jones, S.E.; Tyrrell, J.; Kuo, C.L.; Ruth, K.S.; Tuke, M.A.; Yaghootkar, H.; et al. Red blood cell distribution width: Genetic evidence for aging pathways in 116,666 volunteers. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Xu, M.; Zhang, B.; Liang, J.; Chen, P.; Lee, J.Y.; Johnson, T.A.; Li, H.; Yang, X.; Dai, J.; et al. Meta-analysis of genome-wide association studies of adult height in East Asians identifies 17 novel loci. Hum. Mol. Genet. 2015, 24, 1791–1800. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, R.N.; Tanaka, T.; Tang, W.; Manichaikul, A.; Foy, M.; Kabagambe, E.K.; Nettleton, J.A.; King, I.B.; Weng, L.C.; Bhattacharya, S.; et al. Genetic loci associated with plasma phospholipid N-3 fatty acids: A Meta-Analysis of Genome-Wide association studies from the charge consortium. PLoS Genet. 2011, 7. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.; Steffen, B.T.; Lemaitre, R.N.; Wu, J.H.Y.; Tanaka, T.; Manichaikul, A.; Foy, M.; Rich, S.S.; Wang, L.; Nettleton, J.A.; et al. Genome-Wide association study of plasma n6 polyunsaturated fatty acids within the cohorts for heart and aging research in genomic epidemiology consortium. Circ. Cardiovasc. Genet. 2014, 7, 321–331. [Google Scholar] [CrossRef]
- Pividori, M.; Schoettler, N.; Nicolae, D.L.; Ober, C.; Im, H.K. Shared and distinct genetic risk factors for childhood-onset and adult-onset asthma: Genome-wide and transcriptome-wide studies. Lancet Respir. Med. 2019, 7, 509–522. [Google Scholar] [CrossRef]
- Yap, C.X.; Sidorenko, J.; Wu, Y.; Kemper, K.E.; Yang, J.; Wray, N.R.; Robinson, M.R.; Visscher, P.M. Dissection of genetic variation and evidence for pleiotropy in male pattern baldness. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef]
- Hagenaars, S.P.; Hill, W.D.; Harris, S.E.; Ritchie, S.J.; Davies, G.; Liewald, D.C.; Gale, C.R.; Porteous, D.J.; Deary, I.J.; Marioni, R.E. Genetic prediction of male pattern baldness. PLOS Genet. 2017, 13, e1006594. [Google Scholar] [CrossRef]
- Schmit, S.L.; Edlund, C.K.; Schumacher, F.R.; Gong, J.; Harrison, T.A.; Huyghe, J.R.; Qu, C.; Melas, M.; Van Den Berg, D.J.; Wang, H.; et al. Novel common genetic susceptibility loci for colorectal cancer. J. Natl. Cancer Inst. 2019, 111, 146–157. [Google Scholar] [CrossRef]
- Tanikawa, C.; Kamatani, Y.; Takahashi, A.; Momozawa, Y.; Leveque, K.; Nagayama, S.; Mimori, K.; Mori, M.; Ishii, H.; Inazawa, J.; et al. GWAS identifies two novel colorectal cancer loci at 16q24.1 and 20q13.12. Carcinogenesis 2018, 39, 652–660. [Google Scholar] [CrossRef] [PubMed]
- Law, P.J.; Timofeeva, M.; Fernandez-Rozadilla, C.; Broderick, P.; Studd, J.; Fernandez-Tajes, J.; Farrington, S.; Svinti, V.; Palles, C.; Orlando, G.; et al. Association analyses identify 31 new risk loci for colorectal cancer susceptibility. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- McKay, J.D.; Hung, R.J.; Han, Y.; Zong, X.; Carreras-Torres, R.; Christiani, D.C.; Caporaso, N.E.; Johansson, M.; Xiao, X.; Li, Y.; et al. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat. Genet. 2017, 49, 1126–1132. [Google Scholar] [CrossRef] [PubMed]
- Verweij, N.; Leach, I.M.; Van Den Boogaard, M.; Van Veldhuisen, D.J.; Christoffels, V.M.; Hillege, H.L.; Van Gilst, W.H.; Barnett, P.; De Boer, R.A.; Van Der Harst, P. Genetic Determinants of P Wave Duration and PR Segment. Circ. Cardiovasc. Genet. 2014, 7, 475–481. [Google Scholar] [CrossRef]
- van der Harst, P.; van Setten, J.; Verweij, N.; Vogler, G.; Franke, L.; Maurano, M.T.; Wang, X.; Mateo Leach, I.; Eijgelsheim, M.; Sotoodehnia, N.; et al. 52 Genetic Loci Influencing Myocardial Mass. J. Am. Coll. Cardiol. 2016, 68, 1435–1448. [Google Scholar] [CrossRef]
- Arking, D.E.; Pulit, S.L.; Crotti, L.; Van Der Harst, P.; Munroe, P.B.; Koopmann, T.T.; Sotoodehnia, N.; Rossin, E.J.; Morley, M.; Wang, X.; et al. Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat. Genet. 2014, 46, 826–836. [Google Scholar] [CrossRef]
- Andaleon, A.; Mogil, L.S.; Wheeler, H.E. Genetically regulated gene expression underlies lipid traits in Hispanic cohorts. PLoS ONE 2019, 14, e0220827. [Google Scholar] [CrossRef]
- Aulchenko, Y.S.; Ripatti, S.; Lindqvist, I.; Boomsma, D.; Heid, I.M.; Pramstaller, P.P.; Penninx, B.W.J.H.; Janssens, A.C.J.W.; Wilson, J.F.; Spector, T.; et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat. Genet. 2009, 41, 47–55. [Google Scholar]
- Han, Y.; Pei, Y.; Liu, Y.; Zhang, L.; Wu, S.; Tian, Q.; Chen, X.; Shen, H.; Zhu, X.; Papasian, C.J.; et al. Bivariate genome-wide association study suggests fatty acid desaturase genes and cadherin DCHS2 for variation of both compressive strength index and appendicular lean mass in males. Bone 2012, 51, 1000–1007. [Google Scholar] [CrossRef]
- Hu, Y.; Tanaka, T.; Zhu, J.; Guan, W.; Wu, J.H.Y.; Psaty, B.M.; McKnight, B.; King, I.B.; Sun, Q.; Richard, M.; et al. Discovery and fine-mapping of loci associated with MUFAs through trans-ethnic meta-analysis in Chinese and European populations. J. Lipid Res. 2017, 58, 974–981. [Google Scholar] [CrossRef]
- Chen, P.; Takeuchi, F.; Lee, J.Y.; Li, H.; Wu, J.Y.; Liang, J.; Long, J.; Tabara, Y.; Goodarzi, M.O.; Pereira, M.A.; et al. Multiple nonglycemic genomic loci are newly associated with blood level of glycated hemoglobin in East Asians. Diabetes 2014, 63, 2551–2562. [Google Scholar] [CrossRef]
- Wheeler, E.; Leong, A.; Liu, C.T.; Hivert, M.F.; Strawbridge, R.J.; Podmore, C.; Li, M.; Yao, J.; Sim, X.; Hong, J.; et al. Impact of common genetic determinants of Hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: A transethnic genome-wide meta-analysis. PLoS Med. 2017, 14. [Google Scholar] [CrossRef] [PubMed]
- Benyamin, B.; Esko, T.; Ried, J.S.; Radhakrishnan, A.; Vermeulen, S.H.; Traglia, M.; Gögele, M.; Anderson, D.; Broer, L.; Podmore, C.; et al. Novel loci affecting iron homeostasis and their effects in individuals at risk for hemochromatosis. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Kemp, J.P.; Morris, J.A.; Medina-Gomez, C.; Forgetta, V.; Warrington, N.M.; Youlten, S.E.; Zheng, J.; Gregson, C.L.; Grundberg, E.; Trajanoska, K.; et al. Identification of 153 new loci associated with heel bone mineral density and functional involvement of GPC6 in osteoporosis. Nat. Genet. 2017, 49, 1468–1475. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K. Identification of 613 new loci associated with heel bone mineral density and a polygenic risk score for bone mineral density, osteoporosis and fracture. PLoS ONE 2018, 13, e0200785. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Z.; Van Sommeren, S.; Huang, H.; Ng, S.C.; Alberts, R.; Takahashi, A.; Ripke, S.; Lee, J.C.; Jostins, L.; Shah, T.; et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015, 47, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Jostins, L.; Ripke, S.; Weersma, R.K.; Duerr, R.H.; McGovern, D.P.; Hui, K.Y.; Lee, J.C.; Philip Schumm, L.; Sharma, Y.; Anderson, C.A.; et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012, 491, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Okada, Y.; Wu, D.; Trynka, G.; Raj, T.; Terao, C.; Ikari, K.; Kochi, Y.; Ohmura, K.; Suzuki, A.; Yoshida, S.; et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 2014, 506, 376–381. [Google Scholar] [CrossRef]
- Laufer, V.A.; Tiwari, H.K.; Reynolds, R.J.; Danila, M.I.; Wang, J.; Edberg, J.C.; Kimberly, R.P.; Kottyan, L.C.; Harley, J.B.; Mikuls, T.R.; et al. Genetic influences on susceptibility to rheumatoid arthritis in African-Americans. Hum. Mol. Genet. 2019, 28, 858–874. [Google Scholar] [CrossRef]
- Dashti, H.S.; Jones, S.E.; Wood, A.R.; Lane, J.M.; van Hees, V.T.; Wang, H.; Rhodes, J.A.; Song, Y.; Patel, K.; Anderson, S.G.; et al. Genome-wide association study identifies genetic loci for self-reported habitual sleep duration supported by accelerometer-derived estimates. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef]
- Hicks, A.A.; Pramstaller, P.P.; Johansson, Å.; Vitart, V.; Rudan, I.; Ugocsai, P.; Aulchenko, Y.; Franklin, C.S.; Liebisch, G.; Erdmann, J.; et al. Genetic determinants of circulating sphingolipid concentrations in European populations. PLoS Genet. 2009, 5. [Google Scholar] [CrossRef] [PubMed]
- Mozaffarian, D.; Kabagambe, E.K.; Johnson, C.O.; Lemaitre, R.N.; Manichaikul, A.; Sun, Q.; Foy, M.; Wang, L.; Wiener, H.; Irvin, M.R.; et al. Genetic loci associated with circulating phospholipid trans fatty acids: A meta-analysis of genome-wide association studies from the CHARGE consortium. Am. J. Clin. Nutr. 2015, 101, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Izadi, F. Single Nucleotide Polymorphism as Risk Variants in Crohn‘s Disease. Govaresh 2018, 23, 183–192. [Google Scholar]
- Jezernik, G.; Potočnik, U. Comprehensive genetic study of fatty acids helps explain the role of noncoding inflammatory bowel disease associated SNPs and fatty acid metabolism in disease pathogenesis. Prostaglandins Leukot. Essent. Fat. Acids 2018, 130, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Qiao, S.; Shen, W.; Lin, Z.; Guo, Z.; Gong, J.; Ge, Y.; Shui, G.; Li, Y.; Zhu, W. Disturbance of Fatty Acid Desaturation Mediated by FADS2 in Mesenteric Adipocytes Contributes to Chronic Inflammation of Crohn’s Disease. Lancet 2019. [Google Scholar]
- Ito, Z.; Uchiyama, K.; Odahara, S.; Takami, S.; Saito, K.; Kobayashi, H.; Koido, S.; Kubota, T.; Ohkusa, T.; Saruta, M. Fatty Acids as Useful Serological Markers for Crohn’s Disease. Dig. Dis. 2017. [Google Scholar] [CrossRef]
- Turner, D.; Zlotkin, S.; Shah, P.; Griffiths, A. Omega 3 fatty acids (fish oil) for maintenance of remission in Crohn’s disease. In Cochrane Database of Systematic Reviews; Turner, D., Ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2007. [Google Scholar]
- Costea, I.; MacK, D.R.; Lemaitre, R.N.; Israel, D.; Marcil, V.; Ahmad, A.; Amre, D.K. Interactions between the dietary polyunsaturated fatty acid ratio and genetic factors determine susceptibility to pediatric Crohn’s disease. Gastroenterology 2014, 146. [Google Scholar] [CrossRef]
- Lindoso, L.; Venkateswaran, S.; Kugathasan, S. PUFAs and IBD. Inflamm. Bowel Dis. 2017, 23, 1905–1907. [Google Scholar] [CrossRef]
- Naughton, S.S.; Mathai, M.L.; Hryciw, D.H.; McAinch, A.J. Linoleic acid and the pathogenesis of obesity. Prostaglandins Other Lipid Mediat. 2016, 125, 90–99. [Google Scholar] [CrossRef]
- Yary, T.; Voutilainen, S.; Tuomainen, T.-P.; Ruusunen, A.; Nurmi, T.; Virtanen, J.K. Omega-6 polyunsaturated fatty acids, serum zinc, delta-5- and delta-6-desaturase activities and incident metabolic syndrome. J. Hum. Nutr. Diet. 2017, 30, 506–514. [Google Scholar] [CrossRef]
- Warensjö, E.; Rosell, M.; Hellenius, M.-L.; Vessby, B.; De Faire, U.; Risérus, U. Associations between estimated fatty acid desaturase activities in serum lipids and adipose tissue in humans: links to obesity and insulin resistance. Lipids Health Dis. 2009, 8, 37. [Google Scholar] [CrossRef] [PubMed]
- Elbein, S.C.; Kern, P.A.; Rasouli, N.; Yao-Borengasser, A.; Sharma, N.K.; Das, S.K. Global Gene Expression Profiles of Subcutaneous Adipose and Muscle From Glucose-Tolerant, Insulin-Sensitive, and Insulin-Resistant Individuals Matched for BMI. Diabetes 2011, 60, 1019–1029. [Google Scholar] [CrossRef] [PubMed]
- Fujita, H.; Hara, K.; Shojima, N.; Horikoshi, M.; Iwata, M.; Hirota, Y.; Tobe, K.; Seino, S.; Kadowaki, T. Variations with modest effects have an important role in the genetic background of type 2 diabetes and diabetes-related traits. J. Hum. Genet. 2012, 57, 776–779. [Google Scholar] [CrossRef] [PubMed]
- Dupuis, J. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010, 42, 105–116. [Google Scholar]
- Powell, D.R.; Gay, J.P.; Smith, M.; Wilganowski, N.; Harris, A.; Holland, A.; Reyes, M.; Kirkham, L.; Kirkpatrick, L.L.; Zambrowicz, B.; et al. Fatty acid desaturase 1 knockout mice are lean with improved glycemic control and decreased development of atheromatous plaque. Diabetes. Metab. Syndr. Obes. 2016, 9, 185–199. [Google Scholar] [CrossRef]
- Baugh, S.D.P.; Pabba, P.K.; Barbosa, J.; Coulter, E.; Desai, U.; Gay, J.P.; Gopinathan, S.; Han, Q.; Hari, R.; Kimball, S.D.; et al. Design, synthesis, and in vivo activity of novel inhibitors of delta-5 desaturase for the treatment of metabolic syndrome. Bioorg. Med. Chem. Lett. 2015, 25, 3836–3839. [Google Scholar] [CrossRef]
- Yashiro, H.; Takagahara, S.; Tamura, Y.O.; Miyahisa, I.; Matsui, J.; Suzuki, H.; Ikeda, S.; Watanabe, M. A Novel Selective Inhibitor of Delta-5 Desaturase Lowers Insulin Resistance and Reduces Body Weight in Diet-Induced Obese C57BL/6J Mice. PLoS ONE 2016, 11, e0166198. [Google Scholar] [CrossRef]
- Stoffel, W.; Hammels, I.; Jenke, B.; Binczek, E.; Schmidt-Soltau, I.; Brodesser, S.; Odenthal, M.; Thevis, M. Obesity resistance and deregulation of lipogenesis in Δ6-fatty acid desaturase (FADS 2) deficiency. EMBO Rep. 2014, 15, 110–120. [Google Scholar] [CrossRef]
- Fedor, D.; Kelley, D.S. Prevention of insulin resistance by n-3 polyunsaturated fatty acids. Curr. Opin. Clin. Nutr. Metab. Care 2009, 12, 138–146. [Google Scholar] [CrossRef]
- Cavaliere, G.; Trinchese, G.; Bergamo, P.; De Filippo, C.; Mattace Raso, G.; Gifuni, G.; Putti, R.; Moni, B.H.; Canani, R.B.; Meli, R.; et al. Polyunsaturated Fatty Acids Attenuate Diet Induced Obesity and Insulin Resistance, Modulating Mitochondrial Respiratory Uncoupling in Rat Skeletal Muscle. PLoS ONE 2016, 11, e0149033. [Google Scholar] [CrossRef]
- Wanders, A.J.; Blom, W.A.M.; Zock, P.L.; Geleijnse, J.M.; Brouwer, I.A.; Alssema, M. Plant-derived polyunsaturated fatty acids and markers of glucose metabolism and insulin resistance: A meta-analysis of randomized controlled feeding trials. BMJ Open Diabetes Res. Care 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Jump, D.B.; Botolin, D.; Wang, Y.; Xu, J.; Christian, B.; Demeure, O. Fatty Acid Regulation of Hepatic Gene Transcription. J. Nutr. 2005, 135, 2503–2506. [Google Scholar] [CrossRef] [PubMed]
- Schenk, S.; Saberi, M.; Olefsky, J.M. Insulin sensitivity: Modulation by nutrients and inflammation. J. Clin. Investig. 2008, 118, 2992–3002. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Matute, P.; Pérez-Echarri, N.; Martínez, J.A.; Marti, A.; Moreno-Aliaga, M.J. Eicosapentaenoic acid actions on adiposity and insulin resistance in control and high-fat-fed rats:Rrole of apoptosis, adiponectinand tumour necrosis factor-α. Br. J. Nutr. 2007, 97, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Spahis, S.; Alvarez, F.; Dubois, J.; Ahmed, N.; Peretti, N.; Levy, E. Plasma fatty acid composition in French-Canadian children with non-alcoholic fatty liver disease: Effect of n-3 PUFA supplementation. Prostaglandins Leukot. Essent. Fat. Acids 2015, 99, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Feng, R.; Luo, C.; Li, C.; Du, S.; Okekunle, A.P.; Li, Y.; Chen, Y.; Zi, T.; Niu, Y. Free fatty acids profile among lean, overweight and obese non-alcoholic fatty liver disease patients: A case - Control study. Lipids Health Dis. 2017, 16. [Google Scholar] [CrossRef]
- Walle, P.; Takkunen, M.; Männistö, V.; Vaittinen, M.; Lankinen, M.; Kärjä, V.; Käkelä, P.; Ågren, J.; Tiainen, M.; Schwab, U.; et al. Fatty acid metabolism is altered in non-alcoholic steatohepatitis independent of obesity. Metabolism 2016, 65, 655–666. [Google Scholar] [CrossRef]
- Walle, P.; Männistö, V.; de Mello, V.D.; Vaittinen, M.; Perfilyev, A.; Hanhineva, K.; Ling, C.; Pihlajamäki, J. Liver DNA methylation of FADS2 associates with FADS2 genotype. Clin. Epigenetics 2019, 11, 10. [Google Scholar] [CrossRef]
- Puri, P.; Wiest, M.M.; Cheung, O.; Mirshahi, F.; Sargeant, C.; Min, H.-K.; Contos, M.J.; Sterling, R.K.; Fuchs, M.; Zhou, H.; et al. The plasma lipidomic signature of nonalcoholic steatohepatitis. Hepatology 2009, 50, 1827–1838. [Google Scholar] [CrossRef]
- Nobili, V.; Alisi, A.; Liu, Z.; Liang, T.; Crudele, A.; Raponi, M.; Lin, J.; Chalasani, N.P.; Liu, W. In a pilot study, reduced fatty acid desaturase 1 function was associated with nonalcoholic fatty liver disease and response to treatment in children. Pediatr. Res. 2018, 84, 696–703. [Google Scholar] [CrossRef]
- He, Z.; Zhang, R.; Jiang, F.; Zhang, H.; Zhao, A.; Xu, B.; Jin, L.; Wang, T.; Jia, W.; Jia, W.; et al. FADS1-FADS2 genetic polymorphisms are associated with fatty acid metabolism through changes in DNA methylation and gene expression. Clin. Epigenetics 2018, 10, 113. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.; Anstee, Q.M.; Marietti, M.; Hardy, T.; Henry, L.; Eslam, M.; George, J.; Bugianesi, E. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Wong, V.W.-S.; Adams, L.A.; de Lédinghen, V.; Wong, G.L.-H.; Sookoian, S. Noninvasive biomarkers in NAFLD and NASH — current progress and future promise. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 461–478. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhao, Z.; Liu, S.; Xiao, Y.; Miao, M.; Dong, Q.; Xin, Y. Association of Nonalcoholic Fatty Liver Disease and Coronary Artery Disease with FADS2 rs3834458 Gene Polymorphism in the Chinese Han Population. Gastroenterol. Res. Pract. 2019. [Google Scholar] [CrossRef] [PubMed]
- Röhrig, F.; Schulze, A. Fatty acid synthase The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 2016. [Google Scholar]
- Vriens, K.; Christen, S.; Parik, S.; Broekaert, D.; Yoshinaga, K.; Talebi, A.; Dehairs, J.; Escalona-Noguero, C.; Schmieder, R.; Cornfield, T.; et al. Evidence for an alternative fatty acid desaturation pathway increasing cancer plasticity. Nature 2019, 566, 403–406. [Google Scholar] [CrossRef]
- Zhang, B.; Jia, W.H.; Matsuda, K.; Kweon, S.S.; Matsuo, K.; Xiang, Y.B.; Shin, A.; Jee, S.H.; Kim, D.H.; Cai, Q.; et al. Large-scale genetic study in east Asians identifies six new loci associated with colorectal cancer risk. Nat. Genet. 2014, 46, 533–542. [Google Scholar] [CrossRef]
- Pender-Cudlip, M.C.; Krag, K.J.; Martini, D.; Yu, J.; Guidi, A.; Skinner, S.S.; Zhang, Y.; Qu, X.; He, C.; Xu, Y.; et al. Delta-6-desaturase activity and arachidonic acid synthesis are increased in human breast cancer tissue. Cancer Sci. 2013, 104, 760–764. [Google Scholar] [CrossRef]
- Konstorum, A.; Lynch, M.L.; Torti, S.V.; Torti, F.M.; Laubenbacher, R.C. A Systems Biology Approach to Understanding the Pathophysiology of High-Grade Serous Ovarian Cancer: Focus on Iron and Fatty Acid Metabolism. Omi. A J. Integr. Biol. 2018, 22, 502–513. [Google Scholar] [CrossRef]
- Zhao, H.; Langerød, A.; Ji, Y.; Nowels, K.W.; Nesland, J.M.; Tibshirani, R.; Bukholm, I.K.; Kåresen, R.; Botstein, D.; Børresen-Dale, A.-L.; et al. Different Gene Expression Patterns in Invasive Lobular and Ductal Carcinomas of the Breast. Mol. Biol. Cell 2004, 15, 2523–2536. [Google Scholar] [CrossRef]
- He, C.; Qu, X.; Wan, J.; Rong, R.; Huang, L.; Cai, C.; Zhou, K.; Gu, Y.; Qian, S.Y.; Kang, J.X. Inhibiting Delta-6 Desaturase Activity Suppresses Tumor Growth in Mice. PLoS ONE 2012, 7, e47567. [Google Scholar] [CrossRef] [PubMed]
- Faubert, B.; Li, K.Y.; Cai, L.; Hensley, C.T.; Kim, J.; Zacharias, L.G.; Yang, C.; Do, Q.N.; Doucette, S.; Burguete, D.; et al. Lactate Metabolism in Human Lung Tumors. Cell 2017, 171, 358–371.e9. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Innis, S.M. Genetic Variants of the FADS1 FADS2 Gene Cluster Are Associated with Altered (n-6) and (n-3) Essential Fatty Acids in Plasma and Erythrocyte Phospholipids in Women during Pregnancy and in Breast Milk during Lactation. J. Nutr. 2008, 138, 2222–2228. [Google Scholar] [CrossRef] [PubMed]
- Muc, M.; Kreiner-Møller, E.; Larsen, J.; Madura; Birch, S.; Pedersen, S.; Bisgaard, H.; Lauritzen, L. Maternal fatty acid desaturase genotype correlates with infant immune responses at 6 months. Br. J. Nutr. 2015, 114, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Liu, G.-L.; Li, X.; Chen, X.-Y.; Wu, Y.-X.; Cui, C.-C.; Zhang, X.; Yang, G.; Xie, L. Association of polyunsaturated fatty acids in breast milk with fatty acid desaturase gene polymorphisms among Chinese lactating mothers. Prostaglandins, Leukot. Essent. Fat. Acids 2016, 109, 66–71. [Google Scholar] [CrossRef]
- Caspi, A.; Williams, B.; Kim-Cohen, J.; Craig, I.W.; Milne, B.J.; Poulton, R.; Schalkwyk, L.C.; Taylor, A.; Werts, H.; Moffitt, T.E. Moderation of breastfeeding effects on the IQ by genetic variation in fatty acid metabolism. Proc. Natl. Acad. Sci. USA 2007, 104, 18860–18865. [Google Scholar] [CrossRef]
- Gould, J.F.; Smithers, L.G. Prenatal n-3 Long-Chain Polyunsaturated Fatty Acids and Children’s Executive Functions. Omega Fat. Acids Brain Neurol. Heal. 2019, 83–105. [Google Scholar]
- Badiou, S.; Tuaillon, E.; Viljoen, J.; Escudié, J.B.; Cristol, J.P.; Newell, M.L.; Van de Perre, P.; Neveu, D. Association between breast milk fatty acids and HIV-1 transmission through breastfeeding. Prostaglandins, Leukot. Essent. Fat. Acids 2016, 105, 35–42. [Google Scholar] [CrossRef]
- Andersen, K.R.; Harsløf, L.B.S.; Schnurr, T.M.; Hansen, T.; Hellgren, L.; Michaelsen, K.F.; Lauritzen, L. A study of associations between early DHA status and fatty acid desaturase (FADS) SNP and developmental outcomes in children of obese mothers. Br. J. Nutr. 2017, 117, 278–286. [Google Scholar] [CrossRef]
- Brookes, K.J.; Chen, W.; Xu, X.; Taylor, E.; Asherson, P. Association of Fatty Acid Desaturase Genes with Attention-Deficit/Hyperactivity Disorder. Biol. Psychiatry 2006, 60, 1053–1061. [Google Scholar] [CrossRef]
- Nishida, I.; Murata, N. Chilling Sensitivity In Plants And Cyanobacteria: The Crucial Contribution of Membrane Lipids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 541–568. [Google Scholar] [CrossRef] [PubMed]
- Healy-Stoffel, M.; Levant, B. N-3 (Omega-3) Fatty Acids: Effects on Brain Dopamine Systems and Potential Role in the Etiology and Treatment of Neuropsychiatric Disorders. CNS Neurol. Disord. Drug Targets 2018, 17, 216–232. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Wang, D.; Sun, X.; Yan, J.; Ren, B.; Lu, Q.; Liu, Y.; Zeng, J.; Huang, N.; Xie, Q.; et al. Alterations of eicosanoids and related mediators in patients with schizophrenia meta-analysis View project Alterations of eicosanoids and related mediators in patients with schizophrenia. J. Psychiatr. Res. 2018, 102, 168–178. [Google Scholar]
- Liu, Y.; McNamara, R.K. Elevated Delta-6 desaturase (FADS2) gene expression in the prefrontal cortex of patients with bipolar disorder. J. Psychiatr. Res. 2011, 45, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jandacek, R.; Rider, T.; Tso, P.; McNamara, R.K. Elevated delta-6 desaturase (FADS2) expression in the postmortem prefrontal cortex of schizophrenic patients: Relationship with fatty acid composition. Schizophr. Res. 2009, 109, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.K.; Leonard, S.; Reddy, R.D. Membrane phospholipid abnormalities in postmortem brains from schizophrenic patients. Schizophr. Res. 2000, 42, 7–17. [Google Scholar] [CrossRef]
- Maekawa, M.; Watanabe, A.; Iwayama, Y.; Kimura, T.; Hamazaki, K.; Balan, S.; Ohba, H.; Hisano, Y.; Nozaki, Y.; Ohnishi, T.; et al. Polyunsaturated fatty acid deficiency during neurodevelopment in mice models the prodromal state of schizophrenia through epigenetic changes in nuclear receptor genes. Transl. Psychiatry 2017, 7. [Google Scholar] [CrossRef]
- Mercuri, O.; de Tomás, M.E.; Itarte, H. Prenatal protein depletion and Δ9, Δ6 and Δ5 desaturases in the rat. Lipids 1979, 14, 822–825. [Google Scholar] [CrossRef]
- de Tomás, M.E.; Mercuri, O.; Rodrigo, A. Effects of Dietary Protein and EFA Deficiency on Liver Δ5, Δ6 and Δ9 Desaturase Activities in the Early Developing Rat. J. Nutr. 1980, 110, 595–599. [Google Scholar] [CrossRef]
- Pawełczyk, T.; Grancow-Grabka, M.; Kotlicka-Antczak, M.; Trafalska, E.; Pawełczyk, A. A randomized controlled study of the efficacy of six-month supplementation with concentrated fish oil rich in omega-3 polyunsaturated fatty acids in first episode schizophrenia. J. Psychiatr. Res. 2016, 73, 34–44. [Google Scholar] [CrossRef]
- Bošković, M.; Vovk, T.; Koprivšek, J.; Plesničar, B.K.; Grabnar, I. Vitamin E and essential polyunsaturated fatty acids supplementation in schizophrenia patients treated with haloperidol. Nutr. Neurosci. 2016, 19, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Bentsen, H.; Osnes, K.; Refsum, H.; Solberg, D.K.; Bøhmer, T. A randomized placebo-controlled trial of an omega-3 fatty acid and vitamins E+C in schizophrenia. Transl. Psychiatry 2013, 3. [Google Scholar]
| Type of Desaturases | SNP Variants | Associated Trait | Nature of The Change | Tested Population | Reference |
|---|---|---|---|---|---|
| FADS1 | rs174551-? | Alanine transaminase levels | Higher serum ALT level | East Asian | [105] |
| rs174541-C | Bipolar disorder | Specific variant more common in subjects with the disease | East Asian, European | [106,107] | |
| rs174556-T | Breast milk fatty acid composition | Higher breast milk AA level | South Asian | [108] | |
| rs174549-A | Cancer (Laryngeal squamous cell carcinoma) | Specific variant more common in subjects with the disease | East Asian | [109] | |
| rs174564-G rs174546-T rs174549-A rs174547-C | Cardiology traits | Higher pulse Shorter QT interval Increased heart rate Lower resting heart rate | European, African, Hispanic/Latin American, Asian unspecified, Native American | [110,111,112,113] | |
| rs174546-C rs174550-T rs174551T rs174547-C rs174546-T | Cholesterol | Higher level of HDL Higher total cholesterol Higher level of LDL Lover level of HDL Lower total cholesterol Lower level of LDL | European, East Asian, South Asian, Hispanic/Latin American, African American/Afro- Caribbean, Oceanian, Native American | [114,115,116,117,118,119,120,121,122,123,124,125,126,127] | |
| rs174550-T | Fasting blood glucose | Increased fasting blood glucose | Hispanic/ Latin American, African American/Afro- Caribbean, East Asian, Oceanian, Native American, European | [123,128,129,130] | |
| rs174548-G rs174549-A | Fatty acid desaturase activity | Not specified Lower activity | East Asian, European | [91,131,132] | |
| rs174549-A rs174548-Grs174555-C rs174548-G | Hematology traits | Higher monocytes % Lower white blood cell count Higher platelet count, Sum eosinophil basophil counts Lower granulocytes count Lower red cell distribution width | European, East Asian | [133,134,135,136,137] | |
| rs174547-T | Height | Unspecified | East Asian, European | [133,138] | |
| rs174550-T rs174547-T | Plasman-3 polyunsaturated fatty acid level | Higher EPA level Lower ALA level Higher DPA level | European | [139] | |
| rs174550-T rs174547-C rs174546-T | Plasman-6 polyunsaturated fatty acid levels | Higher adrenic acid Lower ARA level Lower DGLA level | European, East Asian | [131,140] | |
| rs174546-T | Triglyceride levels | Higher TG level | European, Hispanic/ Latin American, African American or Afro-Caribbean, South Asian, East Asian, Oceanian, Native American | [105,115,116,117,118,119,120,121,123,124] | |
| FADS2 | rs174566-A rs174621-G | Asthma | Specific variant more common in subjects with the disease | European | [141] |
| rs174592-G rs174581-A | Balding (type 1, male-pattern baldness) | Specific variant more common in subjects with the disease | European | [133,142,143] | |
| rs12226877-A rs28456-G rs174576-A | Bipolar disorder | Specific variant more common in subjects with the disease | East Asian, European | [106,107] | |
| rs174594-A rs1535-A rs2072113-C | Cancer (Laryngeal squamous cell carcinoma, Colorectal cancer, lung cancer) | Specific variant more common in subjects with the disease | East Asian, European, African American/Afro- Caribbean | [109,144,145,146,147] | |
| rs174577-A rs174564-G rs174583-T rs174577-? | Cardiology traits | Shorter P-wave duration Higher pulse pressure Shorter QT interval Shorter QRS duration | South Asian, European | [111,112,113,148,149,150] | |
| rs174570-G rs174577-C rs174566-G rs174570-T | Cholesterol levels | Higher total cholesterol, HDL, LDL levels Higher HDL level Higher LDL level Lower LDL level | European, Hispanic/Latin American, African American or Afro- Caribbean, East Asian | [114,115,116,118,119,120,121,122,125,151,152] | |
| rs174583-T rs174577-A | Comprehensive strength and appendicular lean mass | Higher comprehensive strength and appendicular lean mass | East Asian | [153] | |
| rs174566-G rs2072113-T | Fatty acid desaturase activity | Decrease activity | European, East Asian | [91,131,132] | |
| rs174601-T | Gondoic acid (20:1 n-9) levels | Higher FA level | East Asian, European | [154] | |
| rs968567-T rs2727271-? rs174570-C rs174577-A rs61897795-G rs2727271-T | Hematology traits | Higher IgA level Lower albumin-globulin ratio Higher glycated hemoglobin level Higher transferrin level Lower neutrophil count Higher non-albumin protein levels | East Asian, South Asian, European | [105,133,134,136,155,156,157] | |
| rs174574-A | Heel bone mineral density | Higher bone mineral density | European | [158,159] | |
| rs174599-? | Hypothyroidism | Lower thyroid hormones | European | [133] | |
| rs4246215-T | Inflammatory bowel disease | Specific variant more common in subjects with the disease | European | [160,161] | |
| rs174574-A rs1535-A | Plasma n-3 PUFA levels | Lower level of EPA, Lower ALA level Higher DPA | European | [139] | |
| rs174577-C rs2727270-T rs174578-T | Plasma n-6 PUFA levels | Higher ARA level Higher LA level Lower LA level | European, East Asian | [131,140] | |
| rs968567-C | Rheumatoid arthritis | Specific variant more common in subjects with the disease | European, East Asian, African American Afro-Caribbean | [162,163] | |
| rs174560-C | Sleep duration | Longer habitual sleep duration | European | [164] | |
| rs174564-G rs174577-C | Triglyceride levels | Lower TG level | European, Hispanic/ Latin American, African American/ Afro-Caribbean, South Asian, East Asian | [105,115,116,117,118,119,120,121,123,124] | |
| FADS3 | rs1000778-A | Sphingolipid levels | Lower sphingolipids level | European | [165] |
| rs174468-A rs174448-A | Plasma n-3 PUFA levels | Higher level of ALA Lower level of DPA, EPA | European | [139] | |
| rs174449-A rs174448-A | Trans fatty acid levels | Higher concentrations of cis/trans-18:2 | European, African American/Afro- Caribbean, East Asian, Hispanic/Latin American | [139,166] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czumaj, A.; Śledziński, T. Biological Role of Unsaturated Fatty Acid Desaturases in Health and Disease. Nutrients 2020, 12, 356. https://doi.org/10.3390/nu12020356
Czumaj A, Śledziński T. Biological Role of Unsaturated Fatty Acid Desaturases in Health and Disease. Nutrients. 2020; 12(2):356. https://doi.org/10.3390/nu12020356
Chicago/Turabian StyleCzumaj, Aleksandra, and Tomasz Śledziński. 2020. "Biological Role of Unsaturated Fatty Acid Desaturases in Health and Disease" Nutrients 12, no. 2: 356. https://doi.org/10.3390/nu12020356
APA StyleCzumaj, A., & Śledziński, T. (2020). Biological Role of Unsaturated Fatty Acid Desaturases in Health and Disease. Nutrients, 12(2), 356. https://doi.org/10.3390/nu12020356





