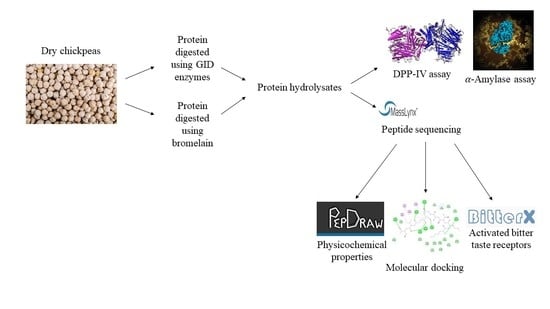
Identification and Comparison of Peptides from Chickpea Protein Hydrolysates Using Either Bromelain or Gastrointestinal Enzymes and Their Relationship with Markers of Type 2 Diabetes and Bitterness
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Preparation of Chickpea Protein Isolate
2.3. Preparation of Chickpea Protein Hydrolysates
2.4. Peptide Sequencing
2.5. Molecular Docking
2.6. Dipeptidyl Peptidase-IV Activity and α-Amylase Activity Assays
2.7. Properties of Peptides
2.8. Statistical Analysis
3. Results
3.1. Peptide Sequencing of Chickpea Protein Hydrolysates
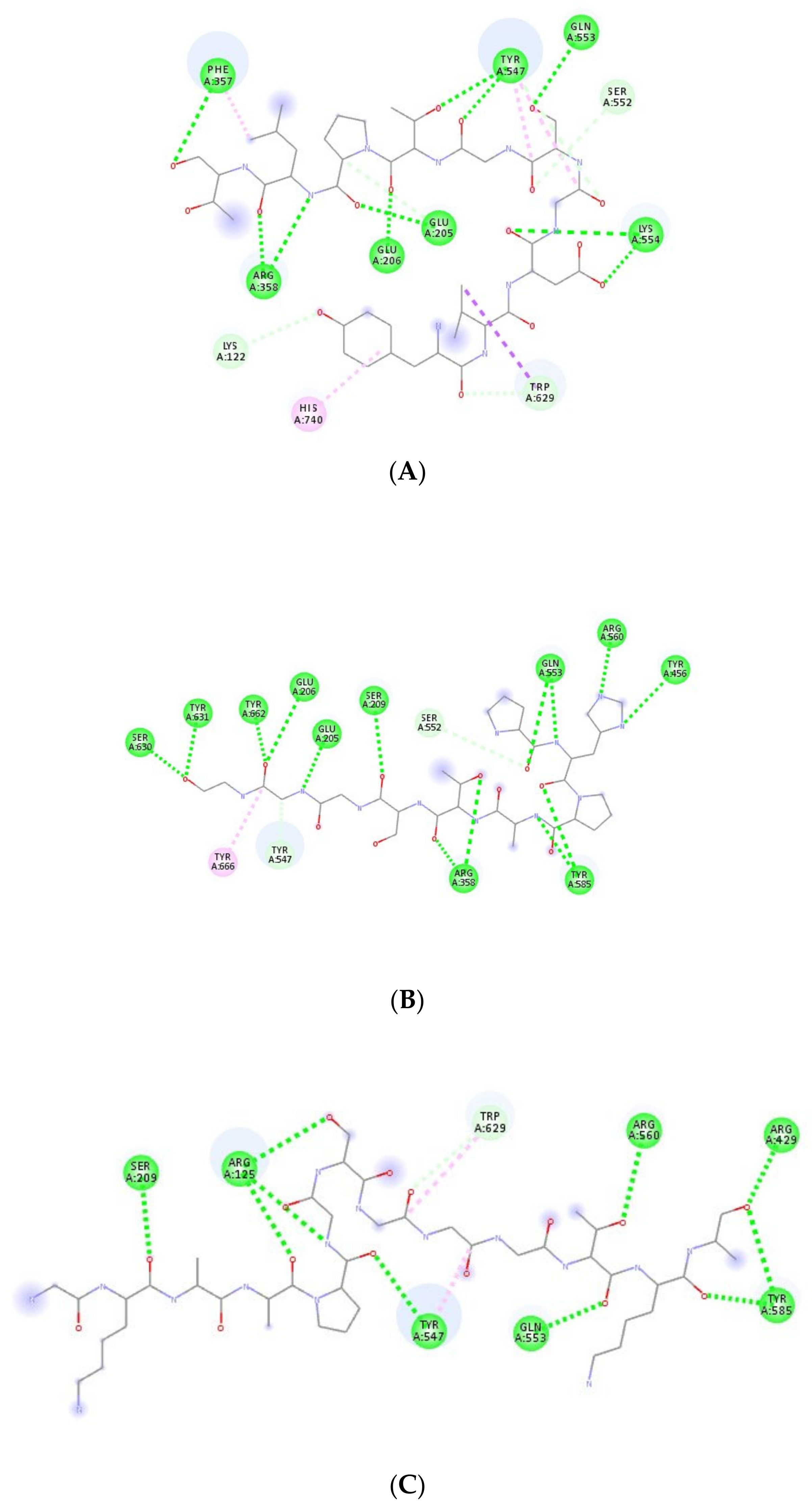
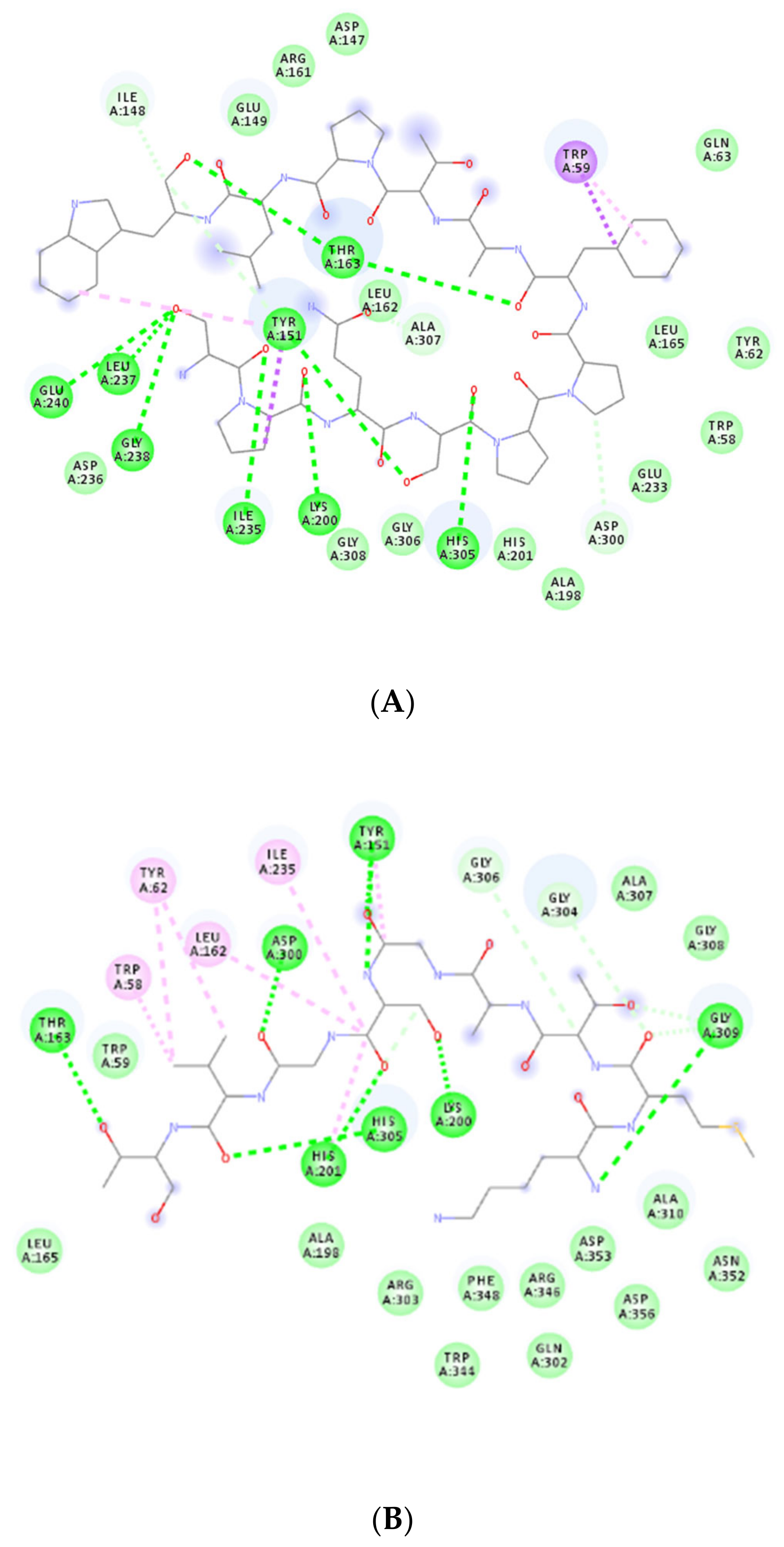
3.2. Molecular Docking
3.3. DPP-IV Inhibitory Activities
3.4. Bitterness-Related Properties of Sequences
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Aisa, H.A.; Gao, Y.; Yili, A.; Ma, Q.; Cheng, Z. Beneficial role of chickpea (cicer arietinum L.) functional factors in the intervention of metabolic syndrome and diabetes mellitus. In Bioactive Food as Dietary Interventions for Diabetes, 2nd ed.; Watson, R.R., Preedy, V.R., Eds.; Academic Press: Cambridge, MA, USA, 2019; Volume 2, pp. 615–627. [Google Scholar] [CrossRef]
- Cid-Gallegos, M.S.; Sánchez-Chino, X.M.; Álvarez-González, I.; Madrigal-Bujaidar, E.; Vásquez-Garzón, V.R.; Baltiérrez-Hoyos, R.; Villa-Treviño, S.; Dávila-Ortiz, G.; Jiménez-Martínez, C. Modification of in vitro and in vivo antioxidant activity by consumption of cooked chickpea in a colon cancer model. Nutrients 2020, 12, 2572. [Google Scholar] [CrossRef]
- Daliri, E.B.-M.; Oh, D.-H.; Lee, B.H. Bioactive peptides—Review. Foods 2017, 6, 32. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Galanopoulos, M.; Sismour, E.; Ren, S.; Mersha, Z.; Lynch, P.; Almutaimi, A. Effect of enzymatic hydrolysis using endo- and exo-proteases on secondary structure, functional, and antioxidant properties of chickpea protein hydrolysates. J. Food Meas. Charact. 2020, 14, 343–352. [Google Scholar] [CrossRef]
- Li-Chan, E.C. Bioactive peptides and protein hydrolysates: Research trends and challenges for application as nutraceuticals and functional food ingredients. Curr. Opin. Food Sci. 2015, 1, 28–37. [Google Scholar] [CrossRef]
- Atanes, P.; Persaud, S.J. GPCR targets in type 2 diabetes. In GPCRs: Elstevieructure BV, Function, and Drug Discovery, 1st ed.; Jastrzebska, B., Park, P.S.-H., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 367–391. [Google Scholar]
- McMacken, M.; Shah, S. A plant-based diet for the prevention and treatment of type 2 diabetes. J. Geriatr. Cardiol. 2017, 14, 342–354. [Google Scholar] [CrossRef]
- Bommer, C.; Sagalova, V.; Heesemann, E.; Manne-Goehler, J.; Atun, R.; Bärnighausen, T.; Davies, J.; Vollmer, S. Global economic burden of diabetes in adults: Projections from 2015 to 2030. Diabetes Care 2018, 41, 963–970. [Google Scholar] [CrossRef]
- Artasensi, A.; Pedretti, A.; Vistoli, G.; Fumagalli, L. Type 2 diabetes mellitus: A review of multi-target drugs. Molecules 2020, 25, 1987. [Google Scholar] [CrossRef]
- Bashary, R.; Vyas, M.; Nayak, S.K.; Suttee, A.; Verma, S.; Narang, R.; Khatik, G.L. An insight of alpha-amylase inhibitors as a valuable tool in the management of type 2 diabetes mellitus. Curr. Diabetes Rev. 2019, 16, 117–136. [Google Scholar] [CrossRef]
- Usman, B.; Sharma, N.; Satija, S.; Mehta, M.; Vyas, M.; Khatik, G.L.; Khurana, N.; Hansbro, P.M.; Williams, K.A.; Dua, K. Recent developments in alpha-glucosidase inhibitors for management of type-2 diabetes: An update. Curr. Pharm. Des. 2019, 25, 2510–2525. [Google Scholar] [CrossRef]
- Barrea, L.; Annunziata, G.; Muscogiuri, G.; Arnone, A.; Tenore, G.C.; Colao, A.; Savastano, S. Could hop-derived bitter compounds improve glucose homeostasis by stimulating the secretion of GLP-1? Crit. Rev. Food Sci. Nutr. 2019, 59, 528–535. [Google Scholar] [CrossRef]
- Zhang, C.; Alashi, A.M.; Singh, N.; Chelikani, P.; Aluko, R.E. Glycated beef protein hydrolysates as sources of bitter taste modifiers. Nutrients 2019, 11, 2166. [Google Scholar] [CrossRef] [PubMed]
- Shaabani, S.; Yarmand, M.S.; Kiani, H.; Emam-Djomeh, Z. The effect of chickpea protein isolate in combination with transglutaminase and xanthan on the physical and rheological characteristics of gluten free muffins and batter based on millet flour. LWT Food Sci. Technol. 2018, 90, 362–372. [Google Scholar] [CrossRef]
- Torres-Fuentes, C.; Alaiz, M.; Vioque, J. Affinity purification and characterisation of chelating peptides from chickpea protein hydrolysates. Food Chem. 2011, 129, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Corzo, C.A.; Waliszewski, K.N.; Welti-Chanes, J. Pineapple fruit bromelain affinity to different protein substrates. Food Chem. 2012, 133, 631–635. [Google Scholar] [CrossRef]
- Grancieri, M.; Martino, H.S.D.; De Mejia, E.G. Digested total protein and protein fractions from chia seed (Salvia hispanica L.) had high scavenging capacity and inhibited 5-LOX, COX-1-2, and iNOS enzymes. Food Chem. 2019, 289, 204–214. [Google Scholar] [CrossRef]
- Burley, S.K.; Berman, H.M.; Bhikadiya, C.; Bi, C.; Chen, L.; Di Costanzo, L.; Christie, C.; Dalenberg, K.; Duarte, J.M.; Dutta, S.; et al. RCSB protein data bank: Biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019, 47, D464–D474. [Google Scholar] [CrossRef]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef]
- Mojica, L.; De Mejia, E.G.; Granados-Silvestre, M.Á.; Menjivar, M. Evaluation of the hypoglycemic potential of a black bean hydrolyzed protein isolate and its pure peptides using in silico, in vitro and in vivo approaches. J. Funct. Foods 2017, 31, 274–286. [Google Scholar] [CrossRef]
- Wang, W.; Zhou, X.; Liu, Y. Characterization and evaluation of umami taste: A review. TrAC Trends Anal. Chem. 2020, 127, 115876. [Google Scholar] [CrossRef]
- Huang, W.; Shen, Q.; Su, X.; Ji, M.; Liu, X.; Chen, Y.; Lu, S.; Zhuang, H.; Zhang, J. BitterX: A tool for understanding bitter taste in humans. Sci. Rep. 2016, 6, 23450. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, I.; Leclercq, C.C.; Simões, N.; Toureiro, A.; Duarte, I.; Freire, J.; Chaves, M.; Renaut, J.; Pinheiro, C.C. Identification of chickpea seed proteins resistant to simulated in vitro human digestion. J. Proteom. 2017, 169, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Torres-Fuentes, C.; Contreras, M.D.M.; Recio, I.; Alaiz, M.; Vioque, J. Identification and characterization of antioxidant peptides from chickpea protein hydrolysates. Food Chem. 2015, 180, 194–202. [Google Scholar] [CrossRef]
- Hernandez, L.M.R.; De Mejia, E.G. Enzymatic production, bioactivity, and bitterness of chickpea (Cicer arietinum) peptides. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1913–1946. [Google Scholar] [CrossRef]
- Cai, J.; Li, C.; Liu, Z.; Du, J.; Ye, J.; Gu, Q.; Xu, J. Predicting DPP-IV inhibitors with machine learning approaches. J. Comput. Mol. Des. 2017, 31, 393–402. [Google Scholar] [CrossRef]
- Jhong, C.-H.; Riyaphan, J.; Lin, S.-H.; Chia, Y.-C.; Weng, C.-F. Screening alpha-glucosidase and alpha-amylase inhibitors from natural compounds by molecular dockingin silico. BioFactors 2015, 41, 242–251. [Google Scholar] [CrossRef]
- Siow, H.-L.; Gan, C.-Y. Extraction, identification, and structure–activity relationship of antioxidative and α-amylase inhibitory peptides from cumin seeds (Cuminum cyminum). J. Funct. Foods 2016, 22, 1–12. [Google Scholar] [CrossRef]
- Ngoh, Y.-Y.; Lim, T.S.; Gan, C.-Y. Screening and identification of five peptides from pinto bean with inhibitory activities against α-amylase using phage display technique. Enzym. Microb. Technol. 2016, 89, 76–84. [Google Scholar] [CrossRef]
- Awosika, T.O.; Aluko, R.E. Inhibition of the in vitro activities of α-amylase, α-glucosidase and pancreatic lipase by yellow field pea (Pisum sativum L.) protein hydrolysates. Int. J. Food Sci. Technol. 2019, 54, 2021–2034. [Google Scholar] [CrossRef]
- Teng, H.; Chen, L. α-Glucosidase and α-amylase inhibitors from seed oil: A review of liposoluble substance to treat diabetes. Crit. Rev. Food Sci. Nutr. 2017, 57, 3438–3448. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.M.M.; Freitas, M.; Fernandes, E. A comprehensive review on xanthone derivatives as α-glucosidase inhibitors. Eur. J. Med. Chem. 2018, 157, 1460–1479. [Google Scholar] [CrossRef] [PubMed]
- Tundis, R.; Loizzo, M.; Menichini, F. Natural products as α-amylase and α-glucosidase inhibitors and their hypoglycaemic potential in the treatment of diabetes: An update. Mini Rev. Med. Chem. 2010, 10, 315–331. [Google Scholar] [CrossRef] [PubMed]
- Udani, J.; Tan, O.; Molina, J. Systematic review and meta-analysis of a proprietary alpha-amylase inhibitor from white bean (Phaseolus vulgaris L.) on weight and fat loss in humans. Foods 2018, 7, 63. [Google Scholar] [CrossRef] [PubMed]
- Aluko, R.E. Structural characteristics of food protein-derived bitter peptides. In Bitterness: Perception, Chemistry and Food Processing; Aliani, M., Eskin, N.A., Eds.; IFT Press: Chicago, IL, USA, 2017; pp. 105–129. [Google Scholar]
- Fujimaki, M.; Yamashita, M.; Okazawa, Y.; Arai, S. Diffusable bitter peptides in peptic hydrolyzate of soybean protein. Agric. Biol. Chem. 1968, 32, 794–795. [Google Scholar] [CrossRef][Green Version]
- Wang, Q.; Liszt, K.I.; Depoortere, I. Extra-oral bitter taste receptors: New targets against obesity? Peptides 2020, 127, 170284. [Google Scholar] [CrossRef]
- Ning, X.; He, J.; Shi, X.; Yang, G. Regulation of adipogenesis by quinine through the ERK/S6 pathway. Int. J. Mol. Sci. 2016, 17, 504. [Google Scholar] [CrossRef]
- Guilherme, A.; Virbasius, J.V.; Puri, V.; Czech, M.P. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat. Rev. Mol. Cell Biol. 2008, 9, 367–377. [Google Scholar] [CrossRef]
- Glusac, J.; Isaschar-Ovdat, S.; Fishman, A. Transglutaminase modifies the physical stability and digestibility of chickpea protein-stabilized oil-in-water emulsions. Food Chem. 2020, 315, 126301. [Google Scholar] [CrossRef]
- Lopes, M.; Pierrepont, C.; Duarte, C.M.; Filipe, A.; Medronho, B.; Sousa, I. Legume beverages from chickpea and lupin, as new milk alternatives. Foods 2020, 9, 1458. [Google Scholar] [CrossRef]

| Sequences | Molecular Mass (g/mol) | Isoelectric Point | Net Charge | Hydrophobicity (kcal/mol) | Length | Potential Bitterness Sequences | Potential Umami Taste Sequences | Potential Biological Activity |
|---|---|---|---|---|---|---|---|---|
| LR | 287.2 | 10.7 | 1 | 8.5 | 2 | L, R | - | Inhibits ACE, DPP-III, renin |
| PLLVE | 563.3 | 3.0 | −1 | 8.7 | 5 | P, V, L, VE, LL, LV, PL | E, VE | Inhibits ACE, alpha-glucosidase, DPP-IV |
| SPKAGAGK | 714.4 | 10.5 | 2 | 17.4 | 8 | P, K, PK | - | Inhibits ACE, DPP-IV, DPP-III |
| HATGGGSGR | 798.4 | 10.7 | 1 | 17.9 | 9 | R, GR | - | Inhibits ACE, DPP-IV |
| PHPATSGGGL | 892.4 | 7.8 | 0 | 13.9 | 10 | P, L, GL, PA, GGL, GGGL | - | Inhibits ACE, DPP-IV, DPP-III |
| TPKASATAAL | 929.5 | 10.1 | 1 | 12.6 | 10 | P, K, L, PK | - | Inhibits ACE, DPP-IV, DPP-III |
| TLTTGTGGLL | 932.5 | 5.4 | 0 | 8.6 | 10 | L, LL, GL, GLL, GGL | - | Inhibits ACE, DPP-IV |
| YVDGSGTPLT | 1008.5 | 3.1 | −1 | 12.5 | 10 | P, V, L, VD, PL, DG | D, VD, DG | Inhibits ACE, DPP-IV |
| TKTPGAGTSAGL | 1059.5 | 10.1 | 1 | 15.3 | 12 | P, K, L, GL, PG | - | Antiamnesic, antithrombotic, inhibits ACE, DPP-IV |
| KEGGGTGTGAAR | 1060.5 | 9.8 | 1 | 23.4 | 12 | R, K, EG, EGG | E, EG | Antioxidative, inhibits ACE, DPP-IV |
| STGPNAGGGAGGY | 1064.5 | 5.4 | 0 | 16.8 | 13 | P, GP, GY, GGY | - | Antiamnesic, antithrombotic, inhibits ACE DPP-IV |
| TLLFTELLF | 1095.6 | 3.1 | −1 | 3.6 | 9 | F, L, LF, LL, EL, ELL | E, EL, TE | Antioxidative, inhibits ACE, DPP-IV, renin |
| KNGAAGPSTVAR | 1127.6 | 11.5 | 2 | 17.6 | 12 | R, P, K, V, GP, VA | - | Antiamnestic, antithrombotic, inhibits ACE, DPP-IV |
| LASEGASAATGAF | 1151.5 | 3.2 | −1 | 14.5 | 13 | F, L, LA, EG, AF | E, EG | Inhibits ACE, DPP-IV |
| VLTSGAGSGAAALT | 1174.6 | 5.5 | 0 | 11.8 | 14 | V, L, VL | - | Antioxidative, inhibits ACE, DPP-IV |
| KNGLGAGAGAGSAR | 1185.6 | 11.5 | 2 | 20.3 | 14 | R, K, L, LG, GL, GLG | - | Inhibits ACE, DPP-IV |
| LSAHAGGTGATLW | 1240.6 | 7.7 | 0 | 11.6 | 13 | L, W, LW | - | Antioxidative, inhibits ACE, DPP-IV, renin |
| LDLARAGGCPTKN | 1314.6 | 8.5 | 1 | 18.2 | 13 | R, P, L, L, LD, DL, LA | D | Inhibits ACE, DPP-IV, DPP-III |
| SPQSPPFATPLW | 1326.6 | 5.4 | 0 | 5.9 | 12 | P, F, L, W, PP, PF, LW, PL, PPF | - | Antioxidative, inhibits ACE, alpha-glucosidase, DPP-IV, DPP-III, renin |
| LLSASMGSQLLSF | 1352.7 | 5.5 | 0 | 4.8 | 13 | F, L, LL | - | Inhibits ACE, DPP-IV, DPP-III, renin |
| Sequences | Molecular Mass (g/mol) | Isoelectric Point | Net Charge | Hydrophobicity (kcal/mol) | Length | Potential Causes of Bitterness | Potential Causes of Umami Taste | Potential Biological Activity |
|---|---|---|---|---|---|---|---|---|
| GKGSGAF | 622.3 | 9.9 | 1 | 13.4 | 7 | F, K, AF, KG | KG | Antioxidative, inhibit: ACE, DPP-IV |
| TRGTGGR | 703.4 | 12.5 | 2 | 15.5 | 7 | R, RG, GR | - | Inhibit: ACE, DPP-IV |
| KMTAGSGVT | 850.4 | 9.8 | 1 | 13.3 | 9 | V, K, GV | - | Inhibit: ACE, DPP-IV |
| KSGGGGGGTAVT | 947.5 | 9.8 | 1 | 18.6 | 12 | V, K | - | Inhibit: ACE DPP-IV |
| GKAAPGSGGGTKA | 1057.6 | 10.7 | 2 | 21.6 | 13 | P, K, PG | - | Antiamnestic, antithrombotic, inhibit: ACE, DPP-IV, DPP-III inhibitor |
| RASAAGGGGGGVSSR | 1245.6 | 12.5 | 2 | 20.8 | 15 | R, V, GV, GGV | - | HMG-CoA reductase, Inhibit: ACE, DPP-IV |
| GKGSSGTGAGGASVSGVT | 1435.7 | 9.9 | 1 | 21.2 | 18 | V, K, GV, KG | KG | Inhibit: ACE, DPP-IV |
| NKKSGAGGGSGAGKGGVA | 1458.8 | 10.9 | 3 | 28.3 | 18 | V, K, GV, KG, VA, GGV | KG | HMG-CoA reductase, inhibit: ACE, DPP-IV |
| LLGELCGSGNTVVEL | 1502.8 | 3.0 | −2 | 14.2 | 15 | V, L, VE, GE, LL, LG, EL, VV, LLG | R, VV, VE, EL | Antioxidative, inhibit: ACE, alpha-glucosidase, DPP-IV, DPP-III |
| QNPLSSAAPTGAGKPY | 1557.8 | 9.5 | 1 | 15.8 | 16 | P, L, K, KP, PL | - | Antioxidative, inhibit: ACE, DPP-IV |
| GLTQGASLAGSGAPSPLF | 1629.8 | 5.5 | 0 | 11.2 | 18 | P, F, L, LF, GL, PL, LA, SLA | - | Inhibit: ACE, DPP-IV, DPP-III |
| AMMELGWSTSGEFLL | 1670.8 | 3.0 | −2 | 10.2 | 15 | F, L, W, FL, GE, LL, LG, EL, EF, FLL | EL | Antioxidative, hypolipidemic, inhibit: ACE, DPP-IV, DPP-III |
| Sequences | Energy of Affinity with DPP-IV (kcal/mol) | Energy of Affinity with α-Amylase (kcal/mol) | Energy of Affinity with α-Glucosidase (kcal/mol) |
|---|---|---|---|
| LR | −5.0 | −5.4 | −4.5 |
| PLLVE | −7.9 | −7.6 | −5.2 |
| SPKAGAGK | −6.7 | −7.1 | −5.3 |
| HATGGGSGR | −7.2 | −6.7 | −5.2 |
| PHPATSGGGL | −8.2 | −7.4 | −7.1 |
| TPKASATAAL | −7.7 | −6.6 | −6.8 |
| TLTTGTGGLL | −7.8 | −6.8 | −5.6 |
| YVDGSGTPLT | −8.2 | −7.3 | −7.3 |
| TKTPGAGTSAGL | −7.1 | −7.3 | −5.8 |
| KEGGGTGTGAAR | −7.2 | −6.4 | −5.6 |
| STGPNAGGGAGGY | −7.6 | −7.3 | −6.3 |
| TLLFTELLF | −7.3 | −7.4 | −6.0 |
| KNGAAGPSTVAR | −6.9 | −6.5 | −5.7 |
| LASEGASAATGAF | −7.3 | −6.9 | −6.1 |
| VLTSGAGSGAAALT | −7.0 | −6.7 | −5.1 |
| KNGLGAGAGAGSAR | −6.7 | −6.2 | −6.1 |
| LSAHAGGTGATLW | −6.7 | −7.9 | −6.0 |
| LDLARAGGCPTKN | −7.0 | −6.3 | −6.1 |
| SPQSPPFATPLW | −7.0 | −8.4 | −7.2 |
| LLSASMGSQLLSF | −6.6 | −6.3 | −5.9 |
| Sequences | Energy of Affinity with DPP-IV (kcal/mol) | Energy of Affinity with α-Amylase | Energy of Affinity with α-Glucosidase |
|---|---|---|---|
| GKGSGAF | −6.8 | −6.5 | −5.5 |
| TRGTGGR | −6.5 | −6.7 | −5.6 |
| KMTAGSGVT | −6.8 | −7.1 | −6.2 |
| KSGGGGGGTAVT | −6.9 | −6.1 | −5.5 |
| GKAAPGSGGGTKA | −7.3 | −6.5 | −6.1 |
| RASAAGGGGGGVSSR | −6.2 | −6.5 | −6.1 |
| GKGSSGTGAGGASVSGVT | −6.5 | −6.2 | −5.6 |
| NKKSGAGGGSGAGKGGVA | −5.8 | −5.9 | −6.1 |
| LLGELCGSGNTVVEL | −6.8 | −6.2 | −5.2 |
| QNPLSSAAPTGAGKPY | −6.8 | −6.1 | −6.4 |
| GLTQGASLAGSGAPSPLF | −6.2 | −6.4 | −6.5 |
| AMMELGWSTSGEFLL | −5.1 | −5.9 | −5.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chandrasekaran, S.; Luna-Vital, D.; de Mejia, E.G. Identification and Comparison of Peptides from Chickpea Protein Hydrolysates Using Either Bromelain or Gastrointestinal Enzymes and Their Relationship with Markers of Type 2 Diabetes and Bitterness. Nutrients 2020, 12, 3843. https://doi.org/10.3390/nu12123843
Chandrasekaran S, Luna-Vital D, de Mejia EG. Identification and Comparison of Peptides from Chickpea Protein Hydrolysates Using Either Bromelain or Gastrointestinal Enzymes and Their Relationship with Markers of Type 2 Diabetes and Bitterness. Nutrients. 2020; 12(12):3843. https://doi.org/10.3390/nu12123843
Chicago/Turabian StyleChandrasekaran, Subhiksha, Diego Luna-Vital, and Elvira Gonzalez de Mejia. 2020. "Identification and Comparison of Peptides from Chickpea Protein Hydrolysates Using Either Bromelain or Gastrointestinal Enzymes and Their Relationship with Markers of Type 2 Diabetes and Bitterness" Nutrients 12, no. 12: 3843. https://doi.org/10.3390/nu12123843
APA StyleChandrasekaran, S., Luna-Vital, D., & de Mejia, E. G. (2020). Identification and Comparison of Peptides from Chickpea Protein Hydrolysates Using Either Bromelain or Gastrointestinal Enzymes and Their Relationship with Markers of Type 2 Diabetes and Bitterness. Nutrients, 12(12), 3843. https://doi.org/10.3390/nu12123843