PHAGE Study: Effects of Supplemental Bacteriophage Intake on Inflammation and Gut Microbiota in Healthy Adults
Abstract
1. Introduction
2. Materials and Methods
2.1. Participant Characteristics
2.2. Study Design
2.3. DNA Extraction and Sequencing
2.4. Microbiota Analysis
2.5. Short-Chain Fatty Acids
2.6. Fecal Triglycerides
2.7. Local Inflammation and Immune Responses
2.8. Systemic Inflammation
2.9. Plasma Lipids
2.10. Statistical Analysis
3. Results
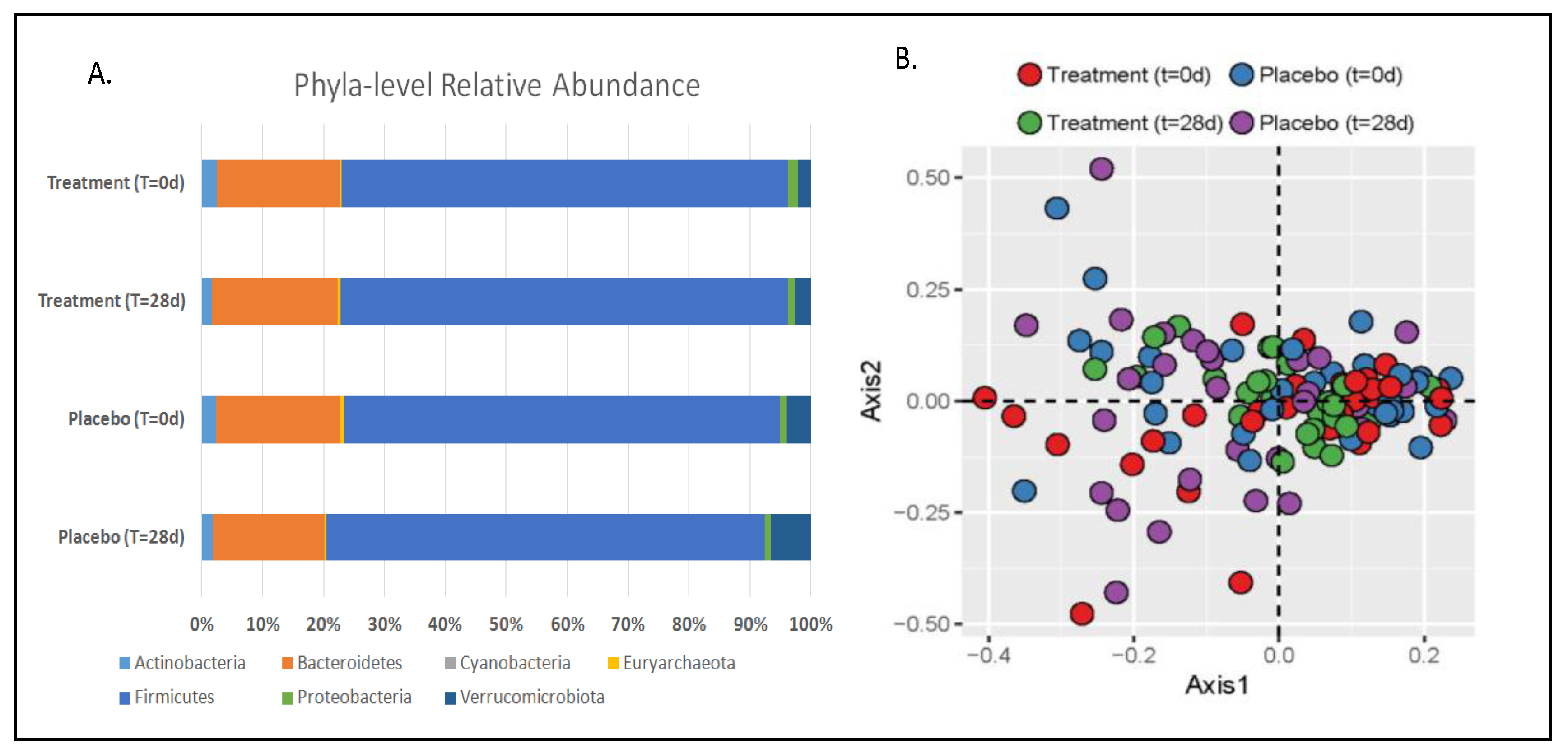
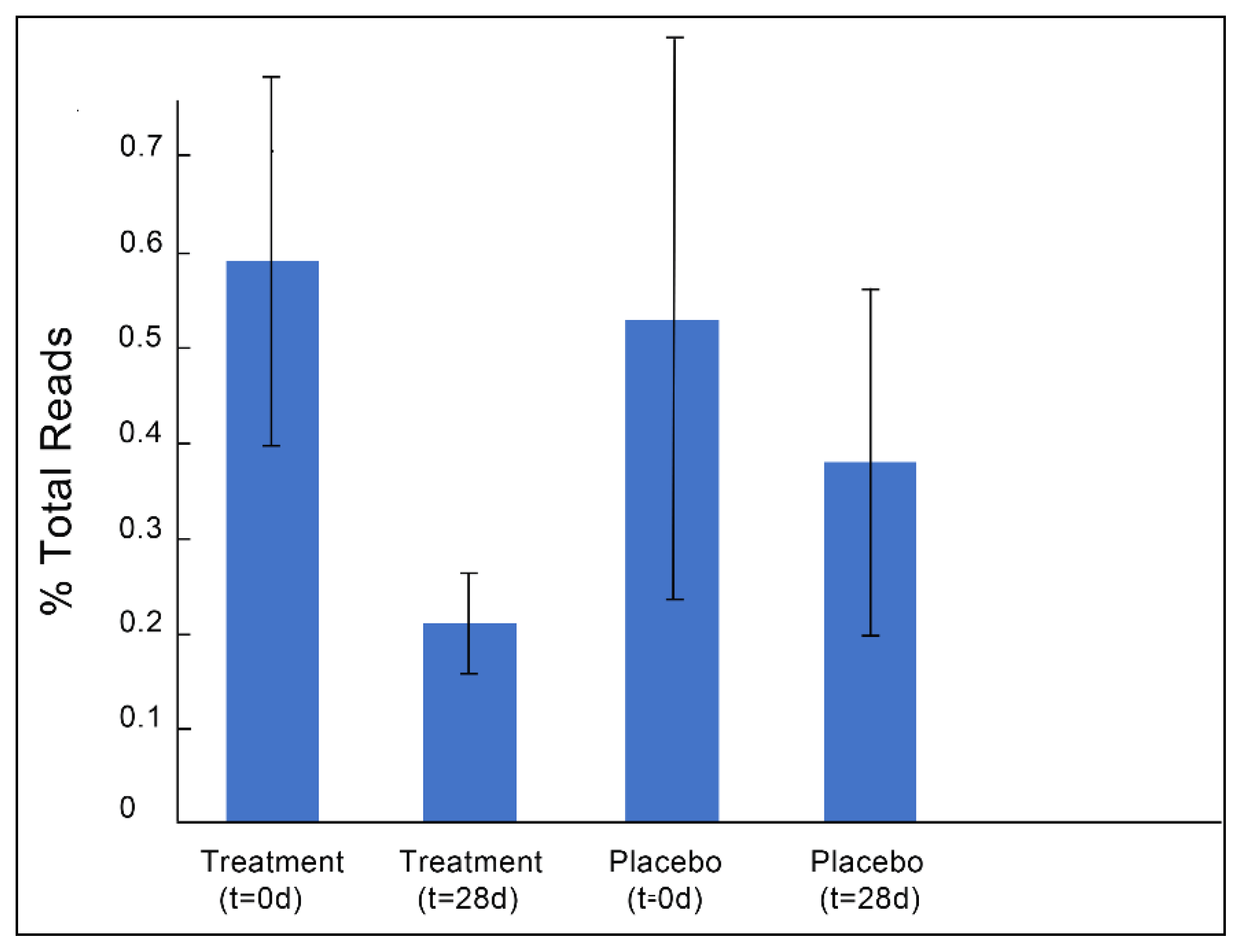
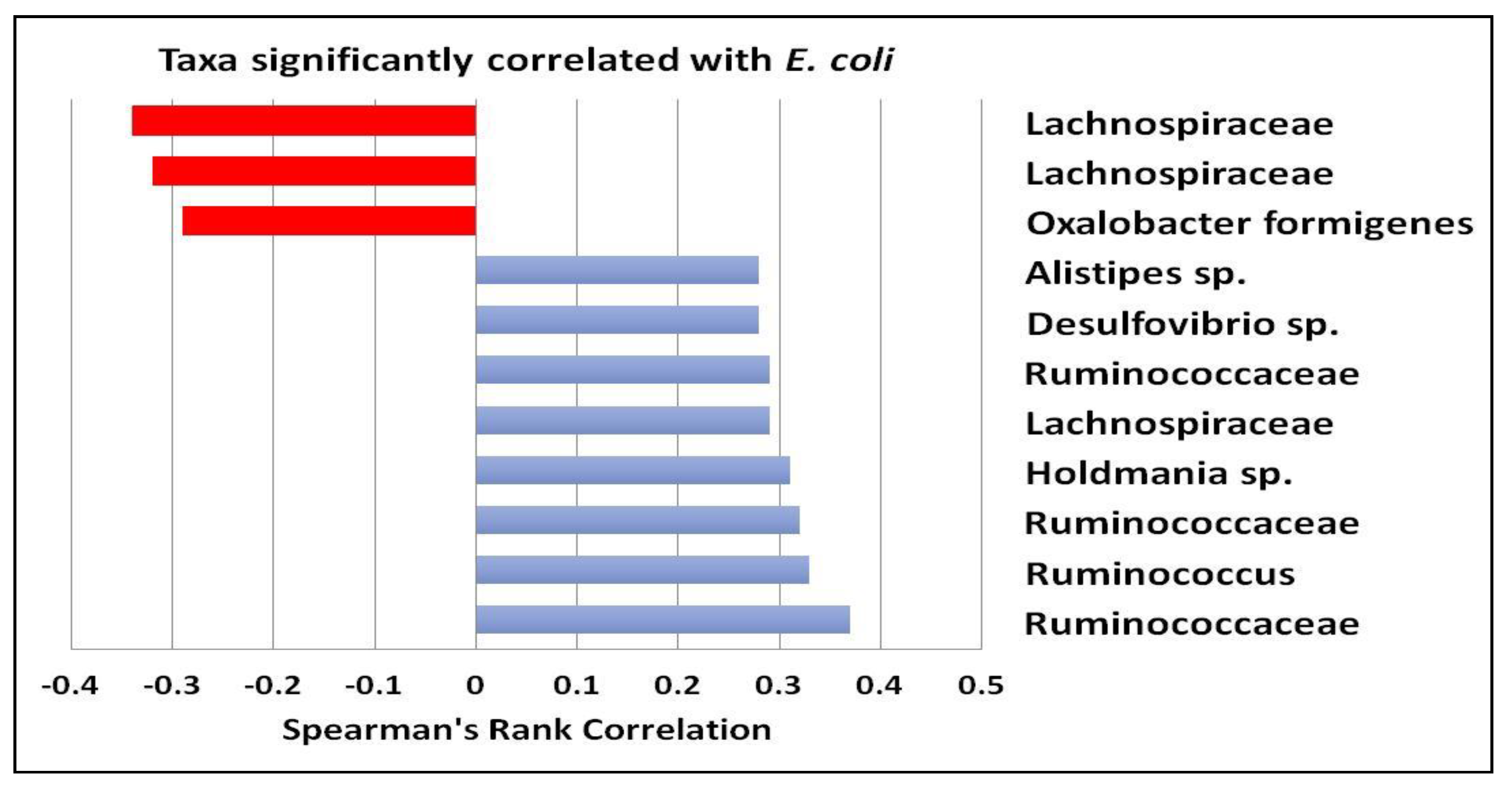
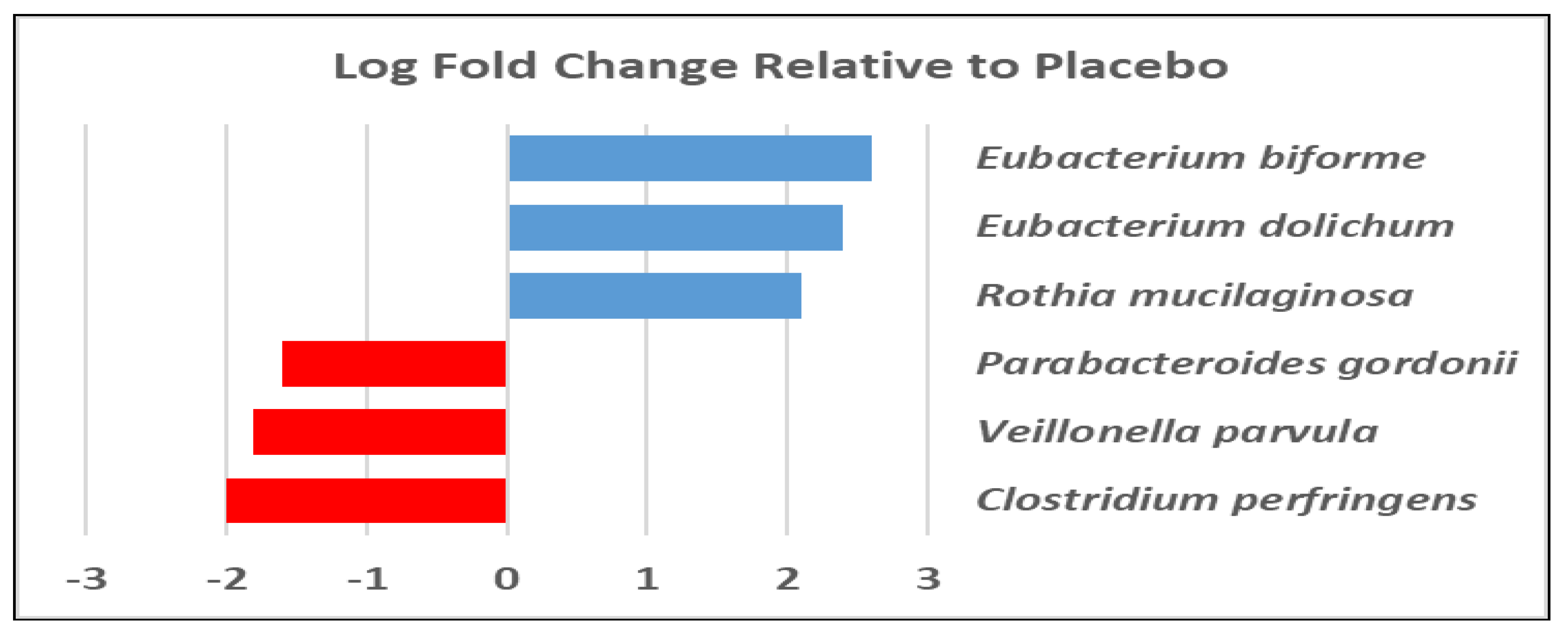
3.1. Gut Microbiota and Metabolite Analysis
3.2. Stool and Plasma Lipid Profiles
3.3. Immunological and Inflammatory Markers
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gentile, C.L.; Weir, T.L. The gut microbiota at the intersection of diet and human health. Science 2018, 362, 776–780. [Google Scholar] [CrossRef] [PubMed]
- Schippa, S.; Conte, M.P. Dysbiotic events in gut microbiota: Impact on human health. Nutrients 2014, 6, 5786–5805. [Google Scholar] [CrossRef] [PubMed]
- Nutraceutical News. Available online: http://www.nutra-insurance.com/nutraceutical-news (accessed on 25 February 2019).
- Gordillo Altamirano, F.L.; Barr, J.J. Phage Therapy in the Postantibiotic Era. Clin. Microbiol. Rev. 2019, 32, e00066-18. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T.; Thomas-Abedon, C.; Thomas, A.; Mazure, H. Bacteriophage prehistory: Is or is not Hankin, 1896, a phage reference? Bacteriophage 2011, 1, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, H.W. On an invisible microbe antagonistic to dysentery bacilli. C. R. Acad. Sci. 1917, 165, 373–375. [Google Scholar]
- Kutateladze, M.; Adamia, R. Bacteriophages as potential new therapeutics to replace or supplement antibiotics. Trends Biotechnol. 2010, 28, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Francino, M.P. Antibiotics and the Human Gut Microbiome: Dysbioses and Accumulation of Resistances. Front. Microbiol. 2016, 6, 1543. [Google Scholar] [CrossRef] [PubMed]
- Gindin, M.; Febvre, H.P.; Rao, S.; Wallace, T.C.; Weir, T.L. Bacteriophage for Gastrointestinal Health (PHAGE) Study: Evaluating the Safety and Tolerability of Supplemental Bacteriophage Consumption. J. Am. Coll. Nutr. 2018, 38, 68–75. [Google Scholar] [CrossRef]
- Finegold, S.M.; Sutter, V.L.; Mathisen, G.E. Chapter 1—Normal Indigenous Intestinal Flora. In Human Intestinal Microflora in Health and Disease; Hentges, D.J., Ed.; Academic Press: Cambridge, MA, USA, 1983; pp. 3–31. [Google Scholar]
- May, R.D.; Fung, M. Strategies targeting the IL-4/IL-13 axes in disease. Cytokine 2015, 75, 89–116. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Jansson, J.K.; Knight, R. The Earth Microbiome project: Successes and aspirations. BMC Biol. 2014, 12, 69. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Manter, D.K.; Korsa, M.; Tebbe, C.; Delgado, J.A. myPhyloDB: A local web server for the storage and analysis of metagenomic data. J. Biol. Database Curation 2016. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. J. Life Environ. Sci. 2016, 4, e2584. [Google Scholar] [CrossRef]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- Sheflin, A.M.; Borresen, E.C.; Kirkwood, J.S.; Boot, C.M.; Whitney, A.K.; Lu, S.; Brown, R.J.; Broeckling, C.D.; Ryan, E.P.; Weir, T.L. Dietary supplementation with rice bran or navy bean alters gut bacterial metabolism in colorectal cancer survivors. Mol. Nutr. Food Res. 2017, 61, 1500905. [Google Scholar] [CrossRef]
- Manning, C.D.; Schütze, H. Foundations of Statistical Natural Language Processing; The MIT Press: Cambridge, MA, USA, 1999. [Google Scholar]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Adlerberth, I.; Wold, A.E. Establishment of the gut microbiota in Western infants. Acta Paediatr. 2009, 98, 229–238. [Google Scholar] [CrossRef]
- Martinez-Guryn, K.; Hubert, N.; Frazier, K.; Urlass, S.; Musch, M.W.; Ojeda, P.; Pierre, J.F.; Miyoshi, J.; Sontag, T.J.; Cham, C.M. Small Intestine Microbiota Regulate Host Digestive and Absorptive Adaptive Responses to Dietary Lipids. Cell Host Microbe 2018, 23, 458–469. [Google Scholar] [CrossRef]
- Semova, I.; Carten, J.D.; Stombaugh, J.; Mackey, L.C.; Knight, R.; Farber, S.A.; Rawls, J.F. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe 2012, 12, 277–288. [Google Scholar] [CrossRef]
- Brown, K.A.; Khanafer, N.; Daneman, N.; Fisman, D.N. Meta-analysis of antibiotics and the risk of community-associated Clostridium difficile infection. Antimicrob. Agents Chemother. 2013, 57, 2326–2332. [Google Scholar] [CrossRef]
- Fite, A.; Macfarlane, S.; Furrie, E.; Bahrami, B.; Cummings, J.H.; Steinke, D.T.; Macfarlane, G.T. Longitudinal analyses of gut mucosal microbiotas in ulcerative colitis in relation to patient age and disease severity and duration. J. Clin. Microbiol. 2013, 51, 849–856. [Google Scholar] [CrossRef]
- Kassinen, A.; Krogius-Kurikka, L.; Mäkivuokko, H.; Rinttilä, T.; Paulin, L.; Corander, J.; Malinen, E.; Apajalahti, J.; Palva, A. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology 2007, 133, 24–33. [Google Scholar] [CrossRef]
- Malinen, E.; Krogius-Kurikka, L.; Lyra, A.; Nikkilä, J.; Jääskeläinen, A.; Rinttilä, T.; Vilpponen-Salmela, T.; von Wright, A.J.; Palva, A. Association of symptoms with gastrointestinal microbiota in irritable bowel syndrome. World J. Gastroenterol. 2010, 16, 4532–4540. [Google Scholar] [CrossRef]
- Gobert, A.P.; Sagrestani, G.; Delmas, E.; Wilson, K.T.; Verriere, T.G.; Dapoigny, M.; Del’homme, C.; Bernalier-Donadille, A. The human intestinal microbiota of constipated-predominant irritable bowel syndrome patients exhibits anti-inflammatory properties. Sci. Rep. 2016, 6, 39399. [Google Scholar] [CrossRef]
- Matsuoka, K.; Kanai, T. The gut microbiota and inflammatory bowel disease. Semin. Immunopathol. 2015, 37, 47–55. [Google Scholar] [CrossRef]
- Scanlan, P.D.; Shanahan, F.; Marchesi, J.R. Culture-independent analysis of desulfovibrios in the human distal colon of healthy, colorectal cancer and polypectomized individuals. FEMS Microbiol. Ecol. 2009, 69, 213–221. [Google Scholar] [CrossRef]
- Muir, A.B.; Benitez, A.J.; Dods, K.; Spergel, J.M.; Fillon, S.A. Microbiome and its impact on gastrointestinal atopy. Allergy 2016, 71, 1256–1263. [Google Scholar] [CrossRef]
- Sherry, C.L.; Kim, S.S.; Dilger, R.N.; Bauer, L.L.; Moon, M.L.; Tapping, R.I.; Fahey, G.C., Jr.; Tappenden, K.A.; Freund, G.G. Sickness behavior induced by endotoxin can be mitigated by the dietary soluble fiber, pectin, through up-regulation of IL-4 and Th2 polarization. Brain Behav. Immun. 2010, 24, 631–640. [Google Scholar] [CrossRef]
- Sherer, F.; Van Simaeys, G.; Kers, J.; Yuan, Q.; Doumont, G.; Laute, M.; Peleman, C.; Egrise, D.; Lahoutte, T.; Flamand, V. Dynamic molecular imaging for hepatic function assessment in mice: Evaluation in endotoxin-induced and warm ischemia-reperfusion models of acute liver failure. J. Liver 2015, 4, 1000170. [Google Scholar]
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Chabo, C. Metabolic Endotoxemia Initiates Obesity and Insulin Resistance. Diabetes 2007, 56, 1761–1772. [Google Scholar] [CrossRef]
- Deerland Enzymes. Available online: https://www.deerlandenzymes.com/preforpro/the-science/ (accessed on 18 January 2019).
| Treatment (t = 0) | Treatment (t = 28) | Placebo (t = 0) | Placebo (t = 28) | |
|---|---|---|---|---|
| Fecal Triglycerides (mg/dL) | 6.70 (±0.70) | 8.12 (±0.89) | 9.25 (±1.14) | 7.46 (±0.79) |
| Cholesterol (mg/dL) | 189.70 (±4.93) | 187.18 (±4.83) | 192.03 (±5.76) | 189.35 (±6.08) |
| LDL (mg/dL) | 103.48 (±3.56) | 100.82 (±4.23) | 106.76 (±4.85) | 103.06 (±4.67) |
| vLDL (mg/dL) | 20.21 (±1.82) | 20.52 (±1.87) | 19.62 (±1.94) | 19.85 (±1.94) |
| HDL (mg/dL) | 65.06 (±2.51) | 65.24 (±2.58) | 65.74 (±2.33) | 63.91 (±2.84) |
| nHDLc (mg/dL) | 123.61 (±4.89) | 122.85 (±4.98) | 126.21 (±5.68) | 125.62 (±5.40) |
| TC/H | 3.03 (±0.13) a | 3.01 (±0.14) a | 3.02 (±0.13) a | 3.11 (±0.14) b |
| Plasma Triglycerides (ng/dL) | 99.94 (±9.11) | 102.61 (±9.46) | 97.74 (±9.70) | 99.41 (±9.77) |
| Treatment (t = 0) | Treatment (t = 28) | Placebo (t = 0) | Placebo (t = 28) | |
|---|---|---|---|---|
| hsCRP (mg/mL) | 1.76 (±0.51) | 1.79 (±0.52) | 1.56 (±0.41) | 2.45 (±0.68) |
| GMCSF (pg/mL) | 80.69 (±9.99) | 80.54 (±10.35) | 83.17 (±9.78) | 80.59 (±10.10) |
| IFN-γ (pg/mL) | 12.67 (±1.12) | 12.29 (±1.02) | 14.21 (±1.86) | 13.81 (±1.89) |
| Il-10 (pg/mL) | 24.13 (±3.30) | 23.53 (±2.85) | 26.03 (±3.75) | 24.83 (±3.84) |
| Il-12 (pg/mL) | 3.57 (±0.33) | 3.57 (±0.32) | 3.75 (±0.35) | 3.55 (±0.37) |
| Il-13 (pg/mL) | 23.05 (±5.53) | 22.16 (±5.71) | 25.39 (±6.06) | 24.5 (±5.73) |
| Il-1β (pg/mL) | 1.76 (±0.13) | 1.71 (±0.10) | 1.85 (±0.13) | 1.73 (±0.12) |
| Il-2 (pg/mL) | 2.24 (±0.14) | 2.15 (±0.18) | 2.46 (±0.28) | 2.36 (±0.31) |
| Il-4 (pg/mL) | 69.48 (±5.75) a | 59.83 (±4.43) b | 63.79 (±4.95) a,b | 61.71 (±3.88) a,b |
| Il-5 (pg/mL) | 8.16 (±3.26) | 5.55 (±1.35) | 7.85 (±2.71) | 6.38 (±1.57) |
| Il-6 (pg/mL) | 3.37 (±0.33) | 3.43 (±0.33) | 3.76 (±0.38) | 3.82 (±0.41) |
| Il-7 (pg/mL) | 13.37 (±1.07) | 13.39 (±1.14) | 13.91 (±1.16) | 13.35 (±1.12) |
| Il-8 (pg/mL) | 4.14 (±0.70) | 4.24 (±0.77) | 4.51 (±0.83) | 4.45 (±0.81) |
| TNFα (pg/mL) | 4.45 (±0.29) | 4.18 (±0.25) | 4.23 (±0.24) | 4.09 (±0.26) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Febvre, H.P.; Rao, S.; Gindin, M.; Goodwin, N.D.M.; Finer, E.; Vivanco, J.S.; Lu, S.; Manter, D.K.; Wallace, T.C.; Weir, T.L. PHAGE Study: Effects of Supplemental Bacteriophage Intake on Inflammation and Gut Microbiota in Healthy Adults. Nutrients 2019, 11, 666. https://doi.org/10.3390/nu11030666
Febvre HP, Rao S, Gindin M, Goodwin NDM, Finer E, Vivanco JS, Lu S, Manter DK, Wallace TC, Weir TL. PHAGE Study: Effects of Supplemental Bacteriophage Intake on Inflammation and Gut Microbiota in Healthy Adults. Nutrients. 2019; 11(3):666. https://doi.org/10.3390/nu11030666
Chicago/Turabian StyleFebvre, Hallie P., Sangeeta Rao, Melinda Gindin, Natalie D. M. Goodwin, Elijah Finer, Jorge S. Vivanco, Shen Lu, Daniel K. Manter, Taylor C. Wallace, and Tiffany L. Weir. 2019. "PHAGE Study: Effects of Supplemental Bacteriophage Intake on Inflammation and Gut Microbiota in Healthy Adults" Nutrients 11, no. 3: 666. https://doi.org/10.3390/nu11030666
APA StyleFebvre, H. P., Rao, S., Gindin, M., Goodwin, N. D. M., Finer, E., Vivanco, J. S., Lu, S., Manter, D. K., Wallace, T. C., & Weir, T. L. (2019). PHAGE Study: Effects of Supplemental Bacteriophage Intake on Inflammation and Gut Microbiota in Healthy Adults. Nutrients, 11(3), 666. https://doi.org/10.3390/nu11030666







