Gluten Detection Methods and Their Critical Role in Assuring Safe Diets for Celiac Patients
Abstract
1. Introduction
2. Celiac Disease Prevalence
3. Hidden Gluten or Gluten Contamination
4. Gluten Threshold or Tolerance Level
5. Food Labeling
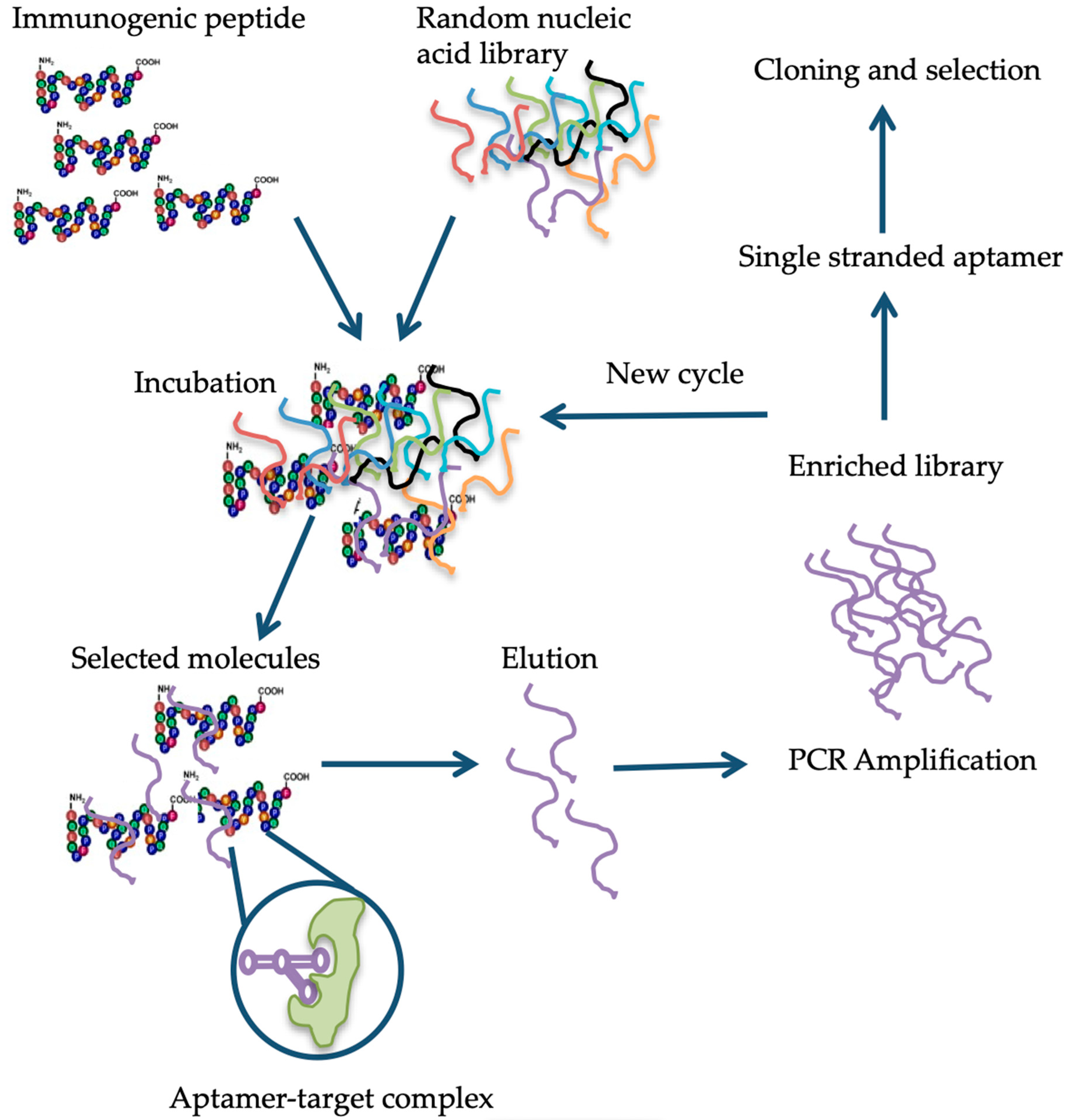
6. Available Detection Methods
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Domenico, L.; Donald, K. One and Two Dimensional (two pH) Polyacrilamide Gel Electrophoresis in a Single Gel: Separation of Wheat Proteins. Cereal Chem. 1985, 62, 314–319. [Google Scholar]
- Kasarda, D.D. Can an increase in celiac disease be attributed to an increase in the gluten content of wheat as a consequence of wheat breeding? J. Agric. Food Chem. 2013, 61, 1155–1159. [Google Scholar] [CrossRef]
- Arranz-Otaegui, A.; Carretero, L.G.; Ramsey, M.N.; Fuller, D.Q.; Richter, T. Archaeobotanical evidence reveals the origins of bread 14,400 years ago in northeastern Jordan. Proc. Natl. Acad. Sci. USA 2018, 115, 7925–7930. [Google Scholar] [CrossRef] [PubMed]
- Elli, L.; Branchi, F.; Tomba, C.; Villalta, D.; Norsa, L.; Ferretti, F.; Roncoroni, L.; Bardella, M.T. Diagnosis of gluten related disorders: Celiac disease, wheat allergy and non-celiac gluten sensitivity. World J. Gastroenterol. WJG 2015, 21, 7110. [Google Scholar] [CrossRef] [PubMed]
- Kilian, B.; Martin, W.; Salamini, F. Genetic diversity, evolution and domestication of wheat and barley in the Fertile Crescent. In Evolution in Action; Springer: Berlin, Germany, 2010; pp. 137–166. [Google Scholar]
- Turner, G.D.; Dunne, M.R.; Ryan, A.W. Celiac Disease: Background and Historical Context. In Celiac Disease; Humana Press: New York, NY, USA, 2015; pp. 3–14. [Google Scholar]
- Simpson, R. Gluten-Free Rome: Celiac disease in the bioarchaeological record. COMPASS 2017, 1, 13–24. [Google Scholar] [CrossRef]
- Van den Broeck, H.C.; de Jong, H.C.; Salentijn, E.M.; Dekking, L.; Bosch, D.; Hamer, R.J.; Gilissen, L.J.; van der Meer, I.M.; Smulders, M.J. Presence of celiac disease epitopes in modern and old hexaploid wheat varieties: Wheat breeding may have contributed to increased prevalence of celiac disease. TAG. Theor. Appl. Genet. 2010, 121, 1527–1539. [Google Scholar] [CrossRef]
- Sapone, A.; Lammers, K.M.; Casolaro, V.; Cammarota, M.; Giuliano, M.T.; De Rosa, M.; Stefanile, R.; Mazzarella, G.; Tolone, C.; Russo, M.I. Divergence of gut permeability and mucosal immune gene expression in two gluten-associated conditions: Celiac disease and gluten sensitivity. BMC Med. 2011, 9, 23. [Google Scholar] [CrossRef]
- Freeman, H.J. The Neolithic revolution and subsequent emergence of the celiac affection. Int. J. Celiac. Dis. 2013, 1, 19–22. [Google Scholar] [CrossRef]
- Sapone, A.; Bai, J.C.; Ciacci, C.; Dolinsek, J.; Green, P.H.; Hadjivassiliou, M.; Kaukinen, K.; Rostami, K.; Sanders, D.S.; Schumann, M. Spectrum of gluten-related disorders: Consensus on new nomenclature and classification. BMC Med. 2012, 10, 13. [Google Scholar] [CrossRef]
- Wen, S.; Wen, N.; Pang, J.; Langen, G.; Brew-Appiah, R.A.; Mejias, J.H.; Osorio, C.; Yang, M.; Gemini, R.; Moehs, C.P.; et al. Structural genes of wheat and barley 5-methylcytosine DNA glycosylases and their potential applications for human health. Proc. Natl. Acad. Sci. USA 2012, 109, 20543–20548. [Google Scholar] [CrossRef]
- Shewry, P.R.; Sayanova, O.; Tatham, A.S.; Tamas, L.; Turner, M.; Richard, G.; Hickman, D.; Fido, R.; Halford, N.G.; Greenfield, J.; et al. Structure, Assembly and Targeting of Wheat Storage Proteins. J. Plant Physiol. 1995, 145, 620–625. [Google Scholar] [CrossRef]
- Osorio, C.; Wen, N.; Gemini, R.; Zemetra, R.; von Wettstein, D.; Rustgi, S. Targeted modification of wheat grain protein to reduce the content of celiac causing epitopes. Funct. Integr. Genom. 2012, 12, 417–438. [Google Scholar] [CrossRef] [PubMed]
- Lebwohl, B.; Ludvigsson, J.F.; Green, P.H. Celiac disease and non-celiac gluten sensitivity. BMJ 2015, 351, h4347. [Google Scholar] [CrossRef] [PubMed]
- Biesiekierski, J.R. What is gluten? J. Gastroenterol. Hepatol. 2017, 32, 78–81. [Google Scholar] [CrossRef]
- Shan, L.; Molberg, O.; Parrot, I.; Hausch, F.; Filiz, F.; Gray, G.M.; Sollid, L.M.; Khosla, C. Structural basis for gluten intolerance in celiac sprue. Science 2002, 297, 2275–2279. [Google Scholar] [CrossRef] [PubMed]
- Shewry, P.R.; Tatham, A.S. Disulphide bonds in wheat gluten proteins. J. Cereal Sci. 1997, 25, 207–227. [Google Scholar] [CrossRef]
- Sollid, L.M. Coeliac disease: Dissecting a complex inflammatory disorder. Nat. Rev. Immunol. 2002, 2, 647–655. [Google Scholar] [CrossRef]
- Shan, L.; Qiao, S.W.; Arentz-Hansen, H.; Molberg, O.; Gray, G.M.; Sollid, L.M.; Khosla, C. Identification and analysis of multivalent proteolytically resistant peptides from gluten: Implications for Celiac Sprue. J. Proteome Res. 2005, 4, 1732–1741. [Google Scholar] [CrossRef]
- Theethira, T.G.; Dennis, M.; Leffler, D.A. Nutritional consequences of celiac disease and the gluten-free diet. Expert Rev. Gastroenterol. Hepatol. 2014, 8, 123–129. [Google Scholar] [CrossRef]
- Green, P.H.; Lebwohl, B.; Greywoode, R. Celiac disease. J. Allergy Clin. Immunol. 2015, 135, 1099–1106. [Google Scholar] [CrossRef]
- Leonard, M.M.; Sapone, A.; Catassi, C.; Fasano, A. Celiac disease and nonceliac gluten sensitivity: A review. JAMA 2017, 318, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Guandalini, S.; Discepolo, V. Celiac disease. In Textbook of Pediatric Gastroenterology, Hepatology and Nutrition; Springer: Cham, Switzerland, 2016; pp. 453–469. [Google Scholar]
- Molberg, O.; McAdam, S.N.; Korner, R.; Quarsten, H.; Kristiansen, C.; Madsen, L.; Fugger, L.; Scott, H.; Noren, O.; Roepstorff, P.; et al. Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat. Med. 1998, 4, 713–717. [Google Scholar] [CrossRef] [PubMed]
- Dieterich, W.; Ehnis, T.; Bauer, M.; Donner, P.; Volta, U.; Riecken, E.O.; Schuppan, D. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat. Med. 1997, 3, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Bethune, M.T.; Khosla, C. Parallels between pathogens and gluten peptides in celiac sprue. PLoS Pathog. 2008, 4, e34. [Google Scholar] [CrossRef]
- Siegel, M.; Strnad, P.; Watts, R.E.; Choi, K.; Jabri, B.; Adler, G.; Ornary, B.; Khosla, C. Extracellular transglutaminase 2 is catalytically inactive, but is transiently activated upon tissue injury in the small intestine. Gastroenterology 2008, 134, A151. [Google Scholar] [CrossRef]
- Sollid, L.M.; Jabri, B. Celiac disease and transglutaminase 2: A model for posttranslational modification of antigens and HLA association in the pathogenesis of autoimmune disorders. Curr. Opin. Immunol. 2011, 23, 732–738. [Google Scholar] [CrossRef]
- Sollid, L.M.; Jabri, B. Triggers and drivers of autoimmunity: Lessons from coeliac disease. Nat. Rev. Immunol. 2013, 13, 294. [Google Scholar] [CrossRef]
- Fasano, A.; Sapone, A.; Zevallos, V.; Schuppan, D. Nonceliac gluten sensitivity. Gastroenterology 2015, 148, 1195–1204. [Google Scholar] [CrossRef]
- Tatham, A.S.; Shewry, P.R. Allergens to wheat and related cereals. Clin. Exp. Allergy 2008, 38, 1712–1726. [Google Scholar] [CrossRef]
- Czaja-Bulsa, G. Non coeliac gluten sensitivity–A new disease with gluten intolerance. Clin. Nutr. 2015, 34, 189–194. [Google Scholar] [CrossRef]
- Bascunan, K.A.; Vespa, M.C.; Araya, M. Celiac disease: Understanding the gluten-free diet. Eur. J. Nutr. 2017, 56, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, S.; Gibson, P. Nutritional inadequacies of the gluten-free diet in both recently-diagnosed and long-term patients with coeliac disease. J. Hum. Nutr. Diet. 2013, 26, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Løvik, A.; Skodje, G.; Bratlie, J.; Brottveit, M.; Lundin, K. Diet adherence and gluten exposure in coeliac disease and self-reported non-coeliac gluten sensitivity. Clin. Nutr. 2017, 36, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Gatti, S.; Galeazzi, T.; Monachesi, C.; Padella, L.; Del, B.G.; Annibali, R.; Lionetti, E.; Catassi, C. Detection of gluten content in the naturally gluten free and gluten free labelled commercially available food products in Italy. Dig. Liver Dis. 2016, 48, e279. [Google Scholar] [CrossRef]
- Verma, A.; Gatti, S.; Galeazzi, T.; Monachesi, C.; Padella, L.; Baldo, G.; Annibali, R.; Lionetti, E.; Catassi, C. Gluten contamination in naturally or labeled gluten-free products marketed in Italy. Nutrients 2017, 9, 115. [Google Scholar] [CrossRef]
- Catassi, C.; Bai, J.; Bonaz, B.; Bouma, G.; Calabrò, A.; Carroccio, A.; Castillejo, G.; Ciacci, C.; Cristofori, F.; Dolinsek, J. Non-celiac gluten sensitivity: The new frontier of gluten related disorders. Nutrients 2013, 5, 3839–3853. [Google Scholar] [CrossRef]
- Allen, P.J. Primary Care Approaches. Gluten-Related Disorders: Celiac Disease, Gluten Allergy, Non-Celiac Gluten Sensitivity. Pediatric Nurs. 2015, 41, 3. [Google Scholar]
- Dominguez-Ortega, G.; Borrelli, O.; Meyer, R.; Dziubak, R.; De Koker, C.; Godwin, H.; Fleming, C.; Thapar, N.; Elawad, M.; Kiparissi, F. Extraintestinal manifestations in children with gastrointestinal food allergy. J. Pediatr. Gastroenterol. Nutr. 2014, 59, 210–214. [Google Scholar] [CrossRef]
- Volta, U.; Caio, G.; Tovoli, F.; De Giorgio, R. Non-celiac gluten sensitivity: Questions still to be answered despite increasing awareness. Cell. Mol. Immunol. 2013, 10, 383. [Google Scholar] [CrossRef]
- Codex Alimentarius Commission. Codex Standard 118-1979 (rev. 2008), Foods for special dietary use for persons intolerant to gluten. In Codex Alimentarium; FAO: Rome, Italy; WHO: Geneva, Switzerland, 2008; pp. 1–3. [Google Scholar]
- Thompson, T.; Lee, A.R.; Grace, T. Gluten contamination of grains, seeds, and flours in the United States: A pilot study. J. Am. Diet. Assoc. 2010, 110, 937–940. [Google Scholar] [CrossRef]
- Thompson, T.; Dennis, M.; Emerson, L. Gluten-Free Labeling: Are Growth Media Containing Wheat, Barley, and Rye Falling through the Cracks? J. Acad. Nutr. Diet. 2018, 118, 2025. [Google Scholar] [CrossRef] [PubMed]
- Falcomer, A.L.; Araújo, L.S.; Farage, P.; Monteiro, J.S.; Nakano, E.Y.; Zandonadi, R.P. Gluten contamination in food services and industry: A systematic review. Crit. Rev. Food Sci. Nutr. 2018, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Farage, P.; de Medeiros Nóbrega, Y.K.; Pratesi, R.; Gandolfi, L.; Assunção, P.; Zandonadi, R.P. Gluten contamination in gluten-free bakery products: A risk for coeliac disease patients. Public Health Nutr. 2017, 20, 413–416. [Google Scholar] [CrossRef] [PubMed]
- Don, C.; Halbmayr-Jech, E.; Rogers, A.; Koehler, P. AACCI Approved Methods Technical Committee report: Collaborative study on the immunochemical quantitation of intact gluten in rice flour and rice-based products using G12 sandwich ELISA. Cereal Foods World 2014, 59, 187–193. [Google Scholar] [CrossRef]
- Allred, L.K.; Kupper, C.; Iverson, G.; Perry, T.B.; Smith, S.; Stephen, R. Definition of the “Purity Protocol” for Producing Gluten-Free Oats. Cereal Chem. 2017, 94, 377–379. [Google Scholar] [CrossRef]
- Van Eckert, R.; Pfannhauser, W.; Riedl, O. Beitrag zur Qualitätssicherung bei der Herstellung von glutenfreien Lebensmitteln. Ernährung. Nutrition 1992, 16, 511–512. [Google Scholar]
- Fritschy, F.; Windemann, H.; Baumgartner, E. Determination of wheat gliadins in food with ELISA. Zeitschrift fur Lebensmittel-Untersuchung und-Forschung 1985, 181, 379–385. [Google Scholar] [CrossRef]
- Janssen, F.; Hägele, G.; de Baaij, J. Gluten-free products, the Dutch experience. In Coeliac Disease; Springer: Dordrecht, The Netherlands, 1991; pp. 95–100. [Google Scholar]
- Olexova, L.; Dovičovičová, L.; Švec, M.; Siekel, P.; Kuchta, T. Detection of gluten-containing cereals in flours and “gluten-free” bakery products by polymerase chain reaction. Food Control 2006, 17, 234–237. [Google Scholar] [CrossRef]
- Hernando, A.; Mujico, J.R.; Mena, M.C.; Lombardía, M.; Mendez, E. Measurement of wheat gluten and barley hordeins in contaminated oats from Europe, the United States and Canada by Sandwich R5 ELISA. Eur. J. Gastroenterol. Hepatol. 2008, 20, 545–554. [Google Scholar] [CrossRef]
- Geng, T.; Westphal, C.; Yeung, J. Detection of Gluten by Commercial Test Kits: Effects of Food Matrices and Extraction Procedures; American Chemical Society Symposium Series: Washington, DC, USA, 2008; Volume 1001, pp. 462–475. [Google Scholar]
- Kelly, C.P.; Bai, J.C.; Liu, E.; Leffler, D.A. Advances in diagnosis and management of celiac disease. Gastroenterology 2015, 148, 1175–1186. [Google Scholar] [CrossRef]
- Rostami, K.; Kerckhaert, J.; Tiemessen, R.; Von Blomberg, B.M.E.; Meijer, J.W.; Mulder, C.J. Sensitivity of antiendomysium and antigliadin antibodies in untreated celiac disease: Disappointing in clinical practice. Am. J. Gastroenterol. 1999, 94, 888. [Google Scholar] [CrossRef] [PubMed]
- Catassi, C.; Fasano, A. Celiac disease. In Gluten-Free Cereal Products and Beverages; Academic Press: Cambridge, MA, USA, 2008; p. 1-I. [Google Scholar]
- Singh, P.; Arora, A.; Strand, T.A.; Leffler, D.A.; Catassi, C.; Green, P.H.; Kelly, C.P.; Ahuja, V.; Makharia, G.K. Global prevalence of celiac disease: Systematic review and meta-analysis. Clin. Gastroenterol. Hepatol. 2018, 16, 823–836.e2. [Google Scholar] [CrossRef] [PubMed]
- Tanveer, M.; Ahmed, A. Non-Celiac Gluten Sensitivity: A Systematic Review. J. Coll. Physicians Surg. Pak. 2019, 29, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Tapia, A.; Ludvigsson, J.F.; Brantner, T.L.; Murray, J.A.; Everhart, J.E. The prevalence of celiac disease in the United States. Am. J. Gastroenterol. 2012, 107, 1538. [Google Scholar] [CrossRef] [PubMed]
- Barada, K.; Bitar, A.; Mokadem, M.A.-R.; Hashash, J.G.; Green, P. Celiac disease in Middle Eastern and North African countries: A new burden? World J. Gastroenterol. WJG 2010, 16, 1449. [Google Scholar] [CrossRef] [PubMed]
- Peña, A.S.; Rodrigo, L. Epidemiology of celiac disease and non-celiac gluten-related disorders. In Advances in the Understanding of Gluten Related Pathology and the Evolution of Gluten-Free Foods; OmniaScience: Barcelona, Spain, 2015; pp. 27–73. [Google Scholar]
- Malekzadeh, R.; Sachdev, A.; Ali, A.F. Coeliac disease in developing countries: Middle East, India and North Africa. Best Pract. Res. Clin. Gastroenterol. 2005, 19, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, B.; Makharia, G.K.; Chetri, K.; Dutta, S.; Mathur, P.; Ahuja, V.; Amarchand, R.; Balamurugan, R.; Chowdhury, S.D.; Daniel, D. Prevalence of adult celiac disease in India: Regional variations and associations. Am. J. Gastroenterol. 2016, 111, 115. [Google Scholar] [CrossRef]
- Makharia, G.K.; Verma, A.K.; Amarchand, R.; Bhatnagar, S.; Das, P.; Goswami, A.; Bhatia, V.; Ahuja, V.; Datta Gupta, S.; Anand, K. Prevalence of celiac disease in the northern part of India: A community based study. J. Gastroenterol. Hepatol. 2011, 26, 894–900. [Google Scholar] [CrossRef]
- Rajpoot, P.; Makharia, G. Problems and challenges to adaptation of gluten free diet by Indian patients with celiac disease. Nutrients 2013, 5, 4869–4879. [Google Scholar] [CrossRef]
- Wu, J.; Xia, B.; von Blomberg, B.; Zhao, C.; Yang, X.; Crusius, J.; Peña, A. Coeliac disease: Emerging in China? Gut 2010, 59, 418–419. [Google Scholar] [CrossRef]
- Yuan, J.; Zhou, C.; Gao, J.; Li, J.; Yu, F.; Lu, J.; Li, X.; Wang, X.; Tong, P.; Wu, Z. Prevalence of celiac disease autoimmunity among adolescents and young adults in China. Clin. Gastroenterol. Hepatol. 2017, 15, 1572–1579.e1. [Google Scholar] [CrossRef] [PubMed]
- Reese, I.; Schäfer, C.; Kleine-Tebbe, J.; Ahrens, B.; Bachmann, O.; Ballmer-Weber, B.; Beyer, K.; Bischoff, S.C.; Blümchen, K.; Dölle, S.; et al. Non-celiac gluten/wheat sensitivity (NCGS)—A currently undefined disorder without validated diagnostic criteria and of unknown prevalence. Allergo J. Int. 2018, 27, 147–151. [Google Scholar] [CrossRef] [PubMed]
- Allred, L.K.; Kupper, C.; Quinn, C. The Use of Visual Examination for Determining the Presence of Gluten-Containing Grains in Gluten Free Oats and Other Grains, Seeds, Beans, Pulses, and Legumes. J. AOAC Int. 2018, 101, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Bustamante, M.; Fernández-Gil, M.; Churruca, I.; Miranda, J.; Lasa, A.; Navarro, V.; Simón, E. Evolution of gluten content in cereal-based gluten-free products: An overview from 1998 to 2016. Nutrients 2017, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.M.; Pereira, M.; Williams, K.M. Gluten detection in foods available in the United States–A market survey. Food Chem. 2015, 169, 120–126. [Google Scholar] [CrossRef]
- Miranda, J.; Simón, E. Gluten Content Change Over the Two Last Decades. In Nutritional and Analytical Approaches of Gluten-Free Diet in Celiac Disease; Springer: Cham, Switzerland, 2017; pp. 47–57. [Google Scholar]
- Moreno, M.D.; Rodríguez-Herrera, A.; Sousa, C.; Comino, I. Biomarkers to monitor gluten-free diet compliance in celiac patients. Nutrients 2017, 9, 46. [Google Scholar] [CrossRef] [PubMed]
- Ciclitira, P.; Evans, D.; Fagg, N.; Lennox, E.; Dowling, R. Clinical testing of gliadin fractions in coeliac patients. Clin. Sci. 1984, 66, 357–364. [Google Scholar] [CrossRef]
- Ciclitira, P.; Ellis, H.; Evans, D.; Lennox, E. A radioimmunoassay for wheat gliadin to assess the suitability of gluten free foods for patients with coeliac disease. Clin. Exp. Immunol. 1985, 59, 703. [Google Scholar]
- Ejderhamn, J.; Veress, B.; Strandvik, B. The long-term effect of continual ingestion of wheat starch-containing gluten-free products in coeliac patients. In Coeliac Disease: One Hundred Years; Kumar, P.J., Ed.; University Press: Leeds, UK, 1988; pp. 294–297. [Google Scholar]
- Kaukinen, K.; Collin, P.; Holm, K.; Rantala, I.; Vuolteenaho, N.; Reunala, T.; Mäki, M. Wheat starch-containing gluten-free flour products in the treatment of coeliac disease and dermatitis herpetiformis: A long-term follow-up study. Scand. J. Gastroenterol. 1999, 34, 163–169. [Google Scholar] [CrossRef]
- Peräaho, M.; Kaukinen, K.; Paasikivi, K.; Sievänen, H.; Lohiniemi, S.; Mäki, M.; Collin, P. Wheat-starch-based gluten-free products in the treatment of newly detected coeliac disease: Prospective and randomized study. Aliment. Pharmacol. Ther. 2003, 17, 587–594. [Google Scholar] [CrossRef]
- Hischenhuber, C.; Crevel, R.; Jarry, B.; Mäki, M.; Moneret-Vautrin, D.; Romano, A.; Troncone, R.; Ward, R. Safe amounts of gluten for patients with wheat allergy or coeliac disease. Aliment. Pharmacol. Ther. 2006, 23, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Catassi, C.; Rossini, M.; Rätsch, I.; Bearzi, I.; Santinelli, A.; Castagnani, R.; Pisani, E.; Coppa, G.; Giorgi, P. Dose dependent effects of protracted ingestion of small amounts of gliadin in coeliac disease children: A clinical and jejunal morphometric study. Gut 1993, 34, 1515–1519. [Google Scholar] [CrossRef] [PubMed]
- Catassi, C.; Fabiani, E.; Iacono, G.; D’Agate, C.; Francavilla, R.; Biagi, F.; Volta, U.; Accomando, S.; Picarelli, A.; De Vitis, I. A prospective, double-blind, placebo-controlled trial to establish a safe gluten threshold for patients with celiac disease. Am. J. Clin. Nutr. 2007, 85, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Brouns, F.; Van Rooy, G.; Shewry, P.; Rustgi, S.; Jonkers, D. Adverse reactions to wheat or wheat components. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1437–1452. [Google Scholar] [CrossRef]
- Gibert, A.; Espadaler, M.; Canela, M.A.; Sanchez, A.; Vaque, C.; Rafecas, M. Consumption of gluten-free products: Should the threshold value for trace amounts of gluten be at 20, 100 or 200 ppm? Eur. J. Gastroenterol. Hepatol. 2006, 18, 1187–1195. [Google Scholar] [CrossRef]
- Gendel, S.M. Comparison of international food allergen labeling regulations. Regul. Toxicol. Pharmacol. 2012, 63, 279–285. [Google Scholar] [CrossRef]
- EC Regulation. No 1169/2011 of the European Parliament and of the Council of 25 October 2011 on the provision of food information to consumers. Off. J. Eur. Union 2011, 1–45.
- Thompson, T.; Simpson, S. A comparison of gluten levels in labeled gluten-free and certified gluten-free foods sold in the United States. Eur. J. Clin. Nutr. 2015, 69, 143. [Google Scholar] [CrossRef]
- Scherf, K.A.; Poms, R.E. Recent developments in analytical methods for tracing gluten. J. Cereal Sci. 2016, 67, 112–122. [Google Scholar] [CrossRef]
- Allmann, M.; Candrian, U.; Höfelein, C.; Lüthy, J. Polymerase chain reaction (PCR): A possible alternative to immunochemical methods assuring safety and quality of food Detection of wheat contamination in non-wheat food products. Z Lebensm Unters Forch 1993, 196, 248–251. [Google Scholar] [CrossRef]
- Köppel, E.; Stadler, M.; Lüthy, J.; HuÈbner, P. Detection of wheat contamination in oats by polymerase chain reaction (PCR) and enzyme-linked immunosorbent assay (ELISA). Zeitschrift für Lebensmitteluntersuchung und-Forschung A 1998, 206, 399–403. [Google Scholar]
- Dahinden, I.; von Büren, M.; Lüthy, J. A quantitative competitive PCR system to detect contamination of wheat, barley or rye in gluten-free food for coeliac patients. Eur. Food Res. Technol. 2001, 212, 228–233. [Google Scholar] [CrossRef]
- Sandberg, M.; Lundberg, L.; Ferm, M.; Yman, I.M. Real time PCR for the detection and discrimination of cereal contamination in gluten free foods. Eur. Food Res. Technol. 2003, 217, 344–349. [Google Scholar] [CrossRef]
- Henterich, N.; Osman, A.A.; Méndez, E.; Mothes, T. Assay of gliadin by real-time immunopolymerase chain reaction. Food Nahr. 2003, 47, 345–348. [Google Scholar] [CrossRef] [PubMed]
- Mujico, J.R.; Lombardía, M.; Mena, M.C.; Méndez, E.; Albar, J.P. A highly sensitive real-time PCR system for quantification of wheat contamination in gluten-free food for celiac patients. Food Chem. 2011, 128, 795–801. [Google Scholar] [CrossRef]
- Scharf, A.; Kasel, U.; Wichmann, G.; Besler, M. Performance of ELISA and PCR methods for the determination of allergens in food: An evaluation of six years of proficiency testing for soy (Glycine max L.) and wheat gluten (Triticum aestivum L.). J. Agric. Food Chem. 2013, 61, 10261–10272. [Google Scholar] [PubMed]
- Mejías, J.H.; Lu, X.; Osorio, C.; Ullman, J.L.; von Wettstein, D.; Rustgi, S. Analysis of wheat prolamins, the causative agents of celiac sprue, using reversed phase high performance liquid chromatography (RP-HPLC) and matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF-MS). Nutrients 2014, 6, 1578–1597. [Google Scholar] [CrossRef]
- Camafeita, E.; Alfonso, P.; Mothes, T.; Méndez, E. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometric micro-analysis: The first non-immunological alternative attempt to quantify gluten gliadins in food samples. J. Mass Spectrom. 1997, 32, 940–947. [Google Scholar] [CrossRef]
- Camafeita, E.; Solís, J.; Alfonso, P.; López, J.A.; Sorell, L.; Méndez, E. Selective identification by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry of different types of gluten in foods made with cereal mixtures. J. Chromatogr. A 1998, 823, 299–306. [Google Scholar] [CrossRef]
- Ferranti, P.; Mamone, G.; Picariello, G.; Addeo, F. Mass spectrometry analysis of gliadins in celiac disease. J. Mass Spectrom. 2007, 42, 1531–1548. [Google Scholar] [CrossRef]
- Weber, D.; Cléroux, C.; Godefroy, S.B. Emerging analytical methods to determine gluten markers in processed foods—method development in support of standard setting. Anal. Bioanal. Chem. 2009, 395, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Sealey-Voyksner, J.A.; Khosla, C.; Voyksner, R.D.; Jorgenson, J.W. Novel aspects of quantitation of immunogenic wheat gluten peptides by liquid chromatography–mass spectrometry/mass spectrometry. J. Chromatogr. A 2010, 1217, 4167–4183. [Google Scholar] [CrossRef] [PubMed]
- Tanner, G.J.; Colgrave, M.L.; Blundell, M.J.; Goswami, H.P.; Howitt, C.A. Measuring hordein (gluten) in beer—A comparison of ELISA and mass spectrometry. PLoS ONE 2013, 8, e56452. [Google Scholar] [CrossRef] [PubMed]
- Tanner, G.J.; Blundell, M.J.; Colgrave, M.L.; Howitt, C.A. Quantification of hordeins by ELISA: The correct standard makes a magnitude of difference. PLoS ONE 2013, 8, e56456. [Google Scholar] [CrossRef]
- Colgrave, M.L.; Goswami, H.; Blundell, M.; Howitt, C.A.; Tanner, G.J. Using mass spectrometry to detect hydrolysed gluten in beer that is responsible for false negatives by ELISA. J. Chromatogr. A 2014, 1370, 105–114. [Google Scholar] [CrossRef]
- Fiedler, K.L.; McGrath, S.C.; Callahan, J.H.; Ross, M.M. Characterization of grain-specific peptide markers for the detection of gluten by mass spectrometry. J. Agric. Food Chem. 2014, 62, 5835–5844. [Google Scholar] [CrossRef]
- Colgrave, M.L.; Goswami, H.; Byrne, K.; Blundell, M.; Howitt, C.A.; Tanner, G.J. Proteomic profiling of 16 cereal grains and the application of targeted proteomics to detect wheat contamination. J. Proteome Res. 2015, 14, 2659–2668. [Google Scholar] [CrossRef]
- Scherf, K.A.; Wieser, H.; Koehler, P. Improved quantitation of gluten in wheat starch for celiac disease patients by gel-permeation high-performance liquid chromatography with fluorescence detection (GP-HPLC-FLD). J. Agric. Food Chem. 2016, 64, 7622–7631. [Google Scholar] [CrossRef]
- Seilmeier, W.; Wieser, H. Comparative investigations of gluten proteins from different wheat species. Eur. Food Res. Technol. 2003, 217, 360–364. [Google Scholar] [CrossRef]
- Radman, M.; Jurina, T.; Benković, M.; Tušek, A.J.; Valinger, D.; Kljusurić, J.G. Application of NIR spectroscopy in gluten detection as a cross-contaminant in food. Croat. J. Food Technol. Biotechnol. Nutr. 2018, 13, 120–127. [Google Scholar] [CrossRef][Green Version]
- Codex Alimentarius Commission. Codex Standard 234-1999 (amended 2011), Recommended methods of analysis and sampling. Gluten-free foods: Enzyme-linked immunoassay R5 Mendez (ELISA) method. In Codex Alimentarious; FAO: Rome, Italy; WHO: Geneva, Switzerland, 2019; pp. 1–80. [Google Scholar]
- Ellis, H.; Doyle, A.; Wieser, H.; Sturgess, R.; Day, P.; Ciclitira, P. Measurement of gluten using a monoclonal antibody to a sequenced peptide of α-gliadin from the coeliac-activating domain I. J. Biochem. Biophys. Methods 1994, 28, 77–82. [Google Scholar] [CrossRef]
- Rumbo, M.; Chirdo, F.G.; Fossati, C.A.; Añón, M.C. Analysis of the effects of heat treatment on gliadin immunochemical quantification using a panel of anti-prolamin antibodies. J. Agric. Food Chem. 2001, 49, 5719–5726. [Google Scholar] [CrossRef] [PubMed]
- Vinci, G.; Rapa, M. Noble Metal Nanoparticles Applications: Recent Trends in Food Control. Bioengineering 2019, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Khot, L.R.; Sankaran, S.; Maja, J.M.; Ehsani, R.; Schuster, E.W. Applications of nanomaterials in agricultural production and crop protection: A review. Crop Prot. 2012, 35, 64–70. [Google Scholar] [CrossRef]
- Thaxton, C.S.; Rosi, N.L.; Mirkin, C.A. Optically and chemically encoded nanoparticle materials for DNA and protein detection. MRS Bull. 2005, 30, 376–380. [Google Scholar] [CrossRef]
- Nam, J.-M.; Thaxton, C.S.; Mirkin, C.A. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science 2003, 301, 1884–1886. [Google Scholar] [CrossRef]
- Manfredi, A.; Giannetto, M.; Mattarozzi, M.; Costantini, M.; Mucchino, C.; Careri, M. Competitive immunosensor based on gliadin immobilization on disposable carbon-nanogold screen-printed electrodes for rapid determination of celiotoxic prolamins. Anal. Bioanal. Chem. 2016, 408, 7289–7298. [Google Scholar] [CrossRef]
- Chu, P.-T.; Lin, C.-S.; Chen, W.-J.; Chen, C.-F.; Wen, H.-W. Detection of gliadin in foods using a quartz crystal microbalance biosensor that incorporates gold nanoparticles. J. Agric. Food Chem. 2012, 60, 6483–6492. [Google Scholar] [CrossRef]
- Yin, H.-Y.; Chu, P.-T.; Tsai, W.-C.; Wen, H.-W. Development of a barcode-style lateral flow immunoassay for the rapid semi-quantification of gliadin in foods. Food Chem. 2016, 192, 934–942. [Google Scholar] [CrossRef]
- Mercadal, P.A.; Fraire, J.C.; Motrich, R.D.; Coronado, E.A. Enzyme-Free Immunoassay Using Silver Nanoparticles for Detection of Gliadin at Ultralow Concentrations. ACS Omega 2018, 3, 2340–2350. [Google Scholar] [CrossRef]
- Doña, V.; Fossati, C.; Chirdo, F. Interference of denaturing and reducing agents on the antigen/antibody interaction. Impact on the performance of quantitative immunoassays in gliadin analysis. Eur. Food Res. Technol. 2008, 226, 591–602. [Google Scholar] [CrossRef]
- Song, K.-M.; Lee, S.; Ban, C. Aptamers and their biological applications. Sensors 2012, 12, 612–631. [Google Scholar] [CrossRef] [PubMed]
- Berezovski, M.V.; Musheev, M.U.; Drabovich, A.P.; Jitkova, J.V.; Krylov, S.N. Non-SELEX: Selection of aptamers without intermediate amplification of candidate oligonucleotides. Nat. Protoc. 2006, 1, 1359. [Google Scholar] [CrossRef] [PubMed]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. SELEX—a (r) evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007, 24, 381–403. [Google Scholar] [CrossRef]
- Zhou, J.; Battig, M.R.; Wang, Y. Aptamer-based molecular recognition for biosensor development. Anal. Bioanal. Chem. 2010, 398, 2471–2480. [Google Scholar] [CrossRef]
- Van Dorst, B.; Mehta, J.; Bekaert, K.; Rouah-Martin, E.; De Coen, W.; Dubruel, P.; Blust, R.; Robbens, J. Recent advances in recognition elements of food and environmental biosensors: A review. Biosens. Bioelectron. 2010, 26, 1178–1194. [Google Scholar] [CrossRef]
- Nimjee, S.M.; Rusconi, C.P.; Sullenger, B.A. Aptamers: An emerging class of therapeutics. Annu. Rev. Med. 2005, 56, 555–583. [Google Scholar] [CrossRef]
- Robertson, D.L.; Joyce, G.F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 1990, 344, 467. [Google Scholar] [CrossRef]
- Tombelli, S.; Mascini, M. Aptamers as molecular tools for bioanalytical methods. Curr. Opin. Mol. Ther. 2009, 11, 179–188. [Google Scholar]
- Jenison, R.D.; Gill, S.C.; Pardi, A.; Polisky, B. High-resolution molecular discrimination by RNA. Science 1994, 263, 1425–1429. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818. [Google Scholar] [CrossRef] [PubMed]
- Strehlitz, B.; Nikolaus, N.; Stoltenburg, R. Protein detection with aptamer biosensors. Sensors 2008, 8, 4296–4307. [Google Scholar] [CrossRef] [PubMed]
- Jayasena, S.D. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar] [PubMed]
- Wang, J.; Meng, W.; Zheng, X.; Liu, S.; Li, G. Combination of aptamer with gold nanoparticles for electrochemical signal amplification: Application to sensitive detection of platelet-derived growth factor. Biosens. Bioelectron. 2009, 24, 1598–1602. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, J.; Nilsen-Hamilton, M. Aptamers: Multifunctional molecules for biomedical research. J. Mol. Med. 2013, 91, 1333–1342. [Google Scholar] [CrossRef]
- Tombelli, S.; Minunni, M.; Mascini, M. Analytical applications of aptamers. Biosens. Bioelectron. 2005, 20, 2424–2434. [Google Scholar] [CrossRef]
- Amaya-González, S.; de-los-Santos-Álvarez, N.; Miranda-Ordieres, A.J.; Lobo-Castañón, M.J.s. Aptamer binding to celiac disease-triggering hydrophobic proteins: A sensitive gluten detection approach. Anal. Chem. 2014, 86, 2733–2739. [Google Scholar] [CrossRef]
- Pinto, A.; Polo, P.N.; Henry, O.; Redondo, M.C.B.; Svobodova, M.; O’Sullivan, C.K. Label-free detection of gliadin food allergen mediated by real-time apta-PCR. Anal. Bioanal. Chem. 2014, 406, 515–524. [Google Scholar] [CrossRef]
- Amaya-González, S.; López-López, L.; Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Miranda-Ordieres, A.J.; Lobo-Castañón, M.J. Affinity of aptamers binding 33-mer gliadin peptide and gluten proteins: Influence of immobilization and labeling tags. Anal. Chim. Acta 2015, 873, 63–70. [Google Scholar] [CrossRef]
- López-López, L.; Miranda-Castro, R.; de-Los-Santos-Alvarez, N.; Miranda-Ordieres, A.J.; Lobo-Castañón, M.J. Disposable electrochemical aptasensor for gluten determination in food. Sens. Actuators B Chem. 2017, 241, 522–527. [Google Scholar] [CrossRef]
- Schopf, M.; Scherf, K.A. Wheat cultivar and species influence variability of gluten ELISA analyses based on polyclonal and monoclonal antibodies R5 and G12. Cereal Sci. 2018, 83, 32–41. [Google Scholar] [CrossRef]
- Slot, I.D.B.; Bremer, M.G.; van der Fels-Klerx, I.; Hamer, R.J. Evaluating the performance of gluten ELISA test kits: The numbers do not tell the tale. Cereal Chem. 2015, 92, 513–521. [Google Scholar] [CrossRef]
- Schalk, K.; Lexhaller, B.; Koehler, P.; Scherf, K.A. Isolation and characterization of gluten protein types from wheat, rye, barley and oats for use as reference materials. PLoS ONE 2017, 12, e0172819. [Google Scholar] [CrossRef] [PubMed]
| Company | Neogen Corp. | R-Biopharm AG | R-Biopharm AG | Inmunología y Genética Aplicada SA | Romer Labs | Tepnel Biosystem | Morinaga Inc. |
| Product | Veratox | RIDA- SCREEN | Ridascreen® Gliadin Competitive | INgezim Gluten | AgraQuant® Gluten G12 | Gluten assay | Wheat protein |
| Antibody | 2 mAb | R5 mAb | R5 mAb | R5 mAb | G12 mAb | Skerritt mAb | Wheat pAb |
| ELISA type | Sandwich | Sandwich | Competitive | Sandwich | Sandwich | Sandwich | Sandwich |
| Time | 30 min | 1.5 h | 40 min | 60 min | 60 min | 30 min | 2.5 h |
| Target | gliadin | ω, α/β- & γ-gliadins and LMWg | ω, α/β- & γ-gliadins and LMWg | ω, α/β- & γ-gliadins and LMWg | α gliadins | ω gliadins and HMWg | Wheat proteins |
| Antigen | |||||||
| LOD (mg/kg) | n/a | 3 | 1.36 | 3 | 2 | 1 | 0.3 |
| LOQ (mg/kg) | 10 | 5 | 5 | 10 | 4 | 10 | 3.12 |
| Properties | Aptamers | Antibodies | Reference |
|---|---|---|---|
| Affinity | Very high target affinity, dissociation constants from micro to picomolar range. | Lower target affinity except for some monoclonal antibodies. | [129] |
| Immunogenic effect | Independent of immunogenic effect, due to their in vitro production. | Immune response can fail when the target molecule, has a structure similar to an endogenous protein. | [130] |
| Specificity | High binding specificity, e.g., the Anti-theophyllin aptamer displayed 10,000-fold discrimination against caffeine (Theophyllin differs from caffeine by a single methyl group). | Depends on target type. | [131] |
| Production | In vitro. | In vivo. Use of animals or cell lines. | [132] |
| Consistency | Chemical synthesis, extreme accuracy, and reproducibility. Little or no batch-to-batch variation. | May have in vivo variations. Restricted to environmental conditions. | [130] |
| Properties | Can be optimized on demand for increasing binding affinity and specificity. | Properties cannot be changed on demand. | [126,133] |
| Stability | Undergo denaturation, but reversible within minutes. | Irreversible denaturation. Stable under physiological conditions | [130] |
| Range of targets | Combinatorial library can be produced against any type of target, even toxic targets. | Restricted to molecules that produce immunogenic effect. | [134] |
| Shelf-life | Stable to long-term storage at ambient temperature. | Limited shelf-life. | [132] |
| Functionalization | Labeling does not affect affinity. | Attachment of molecules can cause loss in affinity. | [126,133,135] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Osorio, C.E.; Mejías, J.H.; Rustgi, S. Gluten Detection Methods and Their Critical Role in Assuring Safe Diets for Celiac Patients. Nutrients 2019, 11, 2920. https://doi.org/10.3390/nu11122920
Osorio CE, Mejías JH, Rustgi S. Gluten Detection Methods and Their Critical Role in Assuring Safe Diets for Celiac Patients. Nutrients. 2019; 11(12):2920. https://doi.org/10.3390/nu11122920
Chicago/Turabian StyleOsorio, Claudia E., Jaime H. Mejías, and Sachin Rustgi. 2019. "Gluten Detection Methods and Their Critical Role in Assuring Safe Diets for Celiac Patients" Nutrients 11, no. 12: 2920. https://doi.org/10.3390/nu11122920
APA StyleOsorio, C. E., Mejías, J. H., & Rustgi, S. (2019). Gluten Detection Methods and Their Critical Role in Assuring Safe Diets for Celiac Patients. Nutrients, 11(12), 2920. https://doi.org/10.3390/nu11122920






