Combined Omics Reveals That Disruption of the Selenocysteine Lyase Gene Affects Amino Acid Pathways in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Diets
2.2. Reagents and Antibodies
2.3. RNA Extraction and Sequencing
2.4. Quantitative PCR (qPCR)
2.5. Metabolomics Analysis
2.6. Bioinformatics Analysis
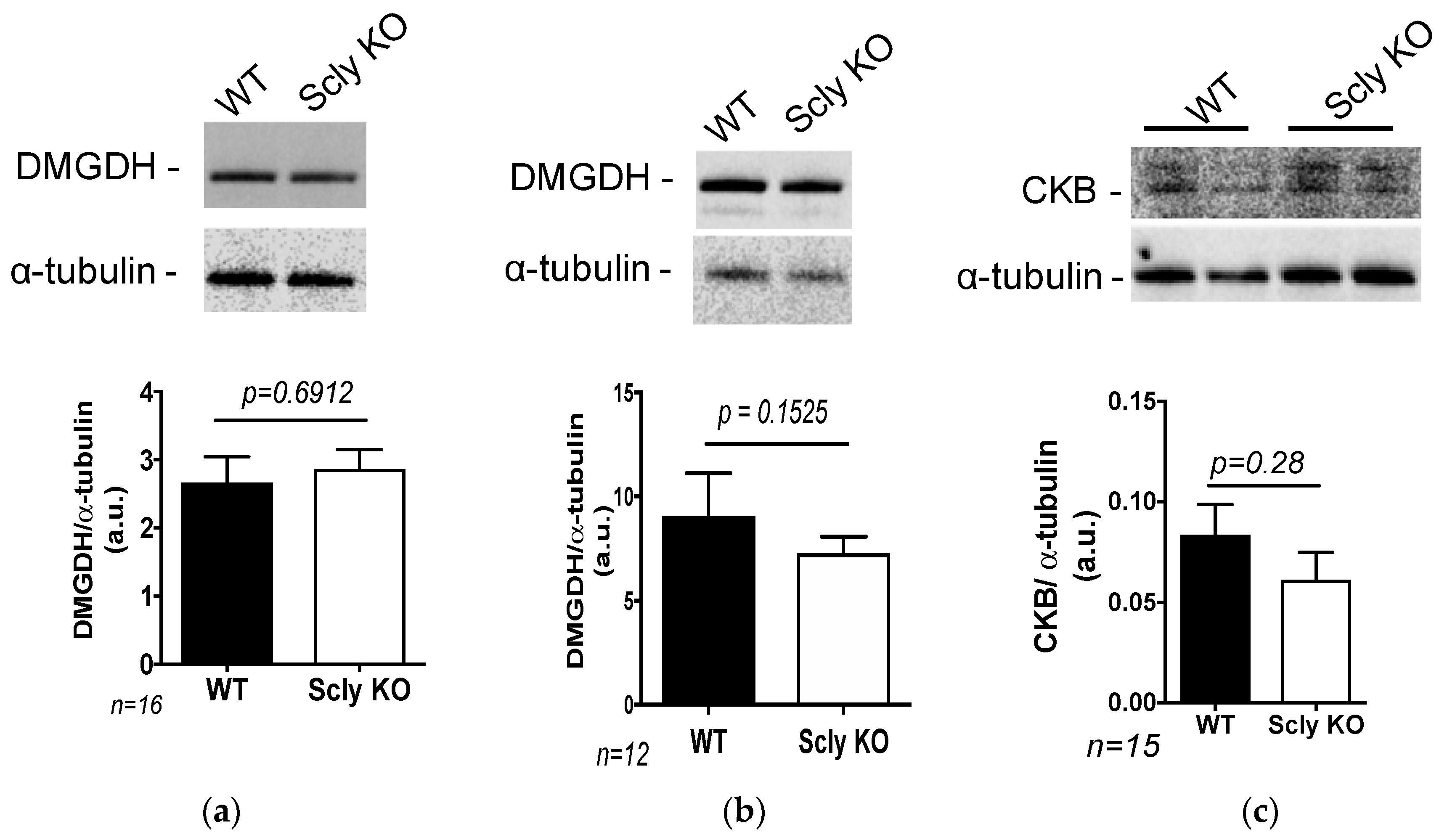
2.7. Western Blot
2.8. Amino Acid Assays
2.9. Statistical Analysis
3. Results
3.1. Effect of Scly Disruption on the Hepatic Transcriptome
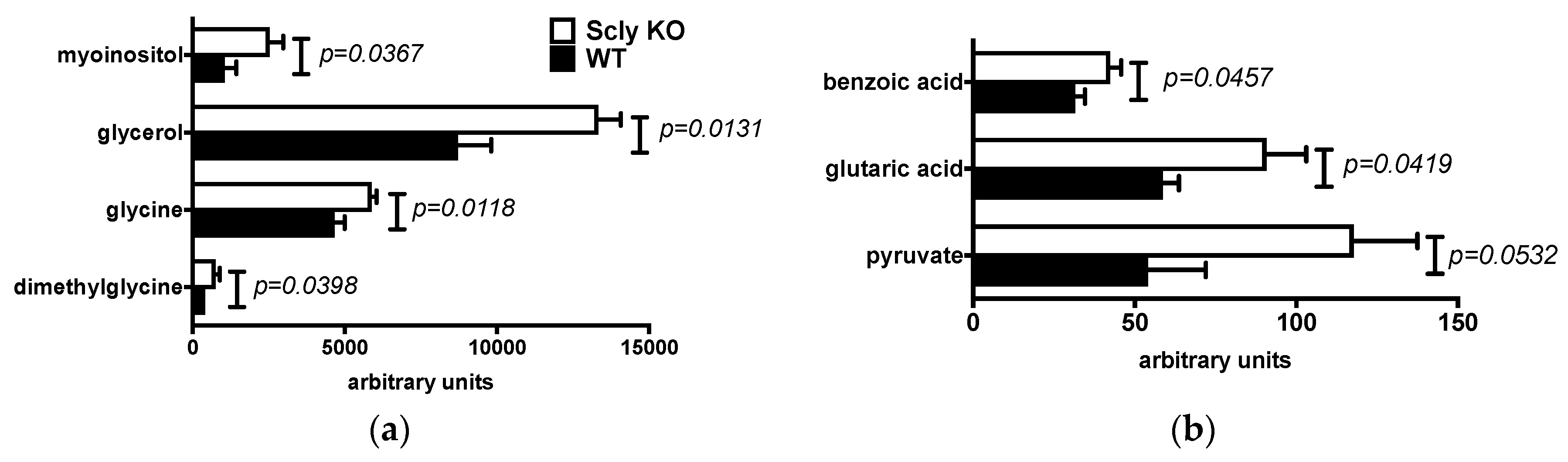
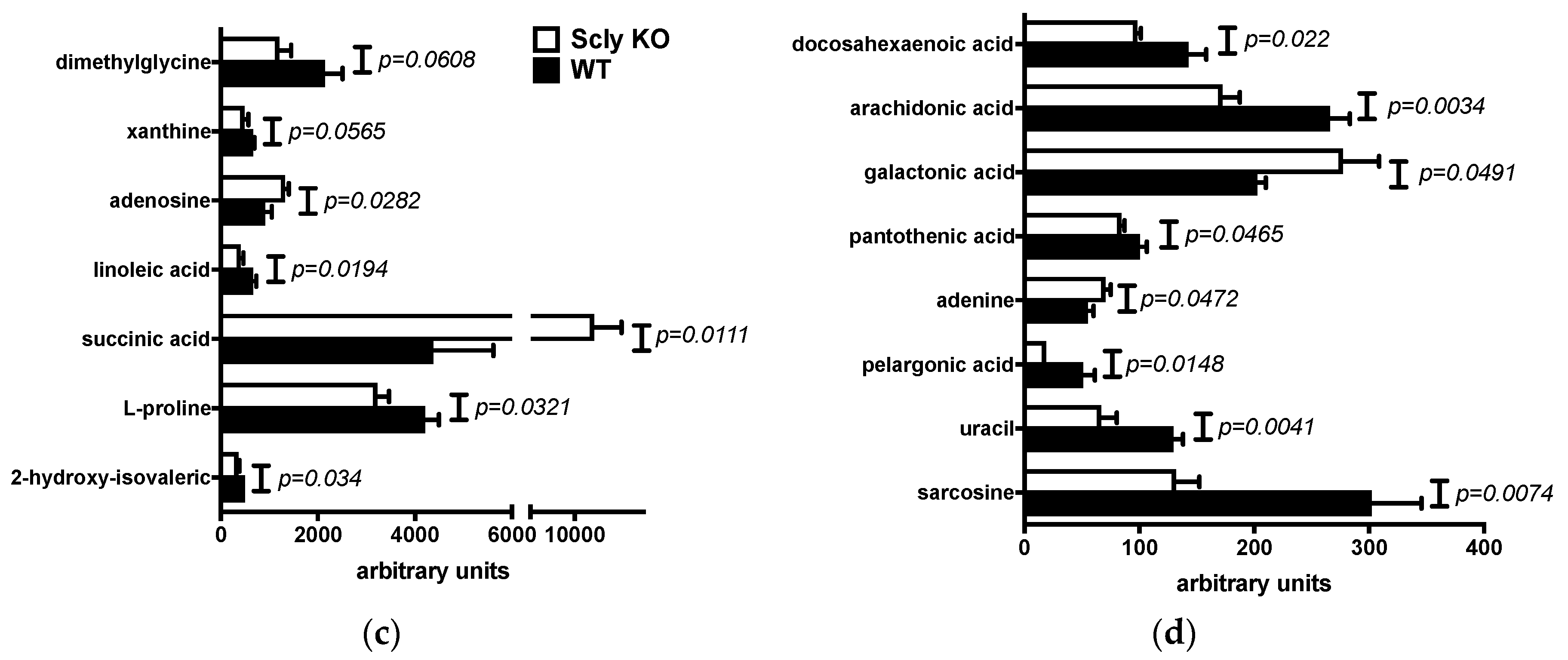
3.2. Sex Differences in Hepatic Metabolites of Scly KO Mice
3.3. Pathway Enrichment Analysis of the Livers of Scly KO Mice
3.4. Glycine Metabolism in the Scly KO Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Labunskyy, V.M.; Hatfield, D.L.; Gladyshev, V.N. Selenoproteins: Molecular pathways and physiological roles. Physiol. Rev. 2014, 94, 739–777. [Google Scholar] [CrossRef] [PubMed]
- Ha, H.Y.; Alfulaij, N.; Berry, M.J.; Seale, L.A. From Selenium Absorption to Selenoprotein Degradation. Biol. Trace Elem. Res. 2019, 192, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Esaki, N.; Karai, N.; Nakamura, T.; Tanaka, H.; Soda, K. Mechanism of reactions catalyzed by selenocysteine beta-lyase. Arch. Biochem. Biophys. 1985, 238, 418–423. [Google Scholar] [CrossRef]
- Kurokawa, S.; Takehashi, M.; Tanaka, H.; Mihara, H.; Kurihara, T.; Tanaka, S.; Hill, K.; Burk, R.; Esaki, N. Mammalian selenocysteine lyase is involved in selenoprotein biosynthesis. J. Nutr. Sci. Vitam. 2011, 57, 298–305. [Google Scholar] [CrossRef]
- Seale, L.A.; Ha, H.Y.; Hashimoto, A.C.; Berry, M.J. Relationship between selenoprotein P and selenocysteine lyase: Insights into selenium metabolism. Free Radic. Biol. Med. 2018, 127, 182–189. [Google Scholar] [CrossRef]
- Seale, L.A.; Gilman, C.L.; Hashimoto, A.C.; Ogawa-Wong, A.N.; Berry, M.J. Diet.-Induced Obesity in the Selenocysteine Lyase Knockout Mouse. Antioxid. Redox Signal. 2015, 23, 761–774. [Google Scholar] [CrossRef]
- Seale, L.A.; Hashimoto, A.C.; Kurokawa, S.; Gilman, C.L.; Seyedali, A.; Bellinger, F.P.; Raman, A.V.; Berry, M.J. Disruption of the selenocysteine lyase-mediated selenium recycling pathway leads to metabolic syndrome in mice. Mol. Cell. Biol. 2012, 32, 4141–4154. [Google Scholar] [CrossRef]
- Pan, L.; Qiu, Y.P.; Chen, T.L.; Lin, J.C.; Chi, Y.; Su, M.M.; Zhao, A.H.; Jia, W. An optimized procedure for metabonomic analysis of rat liver tissue using gas chromatography/time-of-flight mass spectrometry. J. Pharm. Biomed. Anal. 2010, 52, 589–596. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Kir, J.; Liu, D.; Bryant, D.; Guo, Y.J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; et al. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007, 35, 169–175. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.N.; Wang, Z.C.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.N.; Wang, Z.C.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Li, C.I.; Guo, Y.; Sheng, Q.; Shyr, Y. RnaSeqSampleSize: Real data based sample size estimation for RNA sequencing. BMC Bioinform. 2018, 19, 191. [Google Scholar] [CrossRef] [PubMed]
- Ogawa-Wong, A.N.; Hashimoto, A.C.; Ha, H.; Pitts, M.W.; Seale, L.A.; Berry, M.J. Sexual Dimorphism in the Selenocysteine Lyase Knockout Mouse. Nutrients 2018, 10, 159. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wu, Z.; Dai, Z.; Yang, Y.; Wang, J.; Wu, G. Glycine metabolism in animals and humans: Implications for nutrition and health. Amino Acids 2013, 45, 463–477. [Google Scholar] [CrossRef]
- Tobe, R.; Mihara, H. Delivery of selenium to selenophosphate synthetase for selenoprotein biosynthesis. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2433–2440. [Google Scholar] [CrossRef]
- Mihara, H.; Esaki, N. Selenocysteine lyase: Mechanism, structure, and biological role. In Selenium—Its Molecular Biology and Role in Human Health, 1st ed.; Hatfield, D.L., Gladyshev, V.N., Berry, M.J., Eds.; Springer: New York, NY, USA, 2012; pp. 95–105. [Google Scholar]
- Kwak, M.S.; Mihara, H.; Esaki, N. A novel regulatory function of selenocysteine lyase, a unique catalyst to modulate major urinary protein. J. Mol. Catal. B Enzym. 2003, 23, 367–372. [Google Scholar] [CrossRef]
- Cave, M.C.; Clair, H.B.; Hardesty, J.E.; Falkner, K.C.; Feng, W.; Clark, B.J.; Sidey, J.; Shi, H.; Aqel, B.A.; McClain, C.J.; et al. Nuclear receptors and nonalcoholic fatty liver disease. Biochim. Biophys. Acta Gene Regul. Mech. 2016, 1859, 1083–1099. [Google Scholar] [CrossRef] [Green Version]
- Semple, R.K.; Chatterjee, V.K.; O‘Rahilly, S. PPAR gamma and human metabolic disease. J. Clin. Investig. 2006, 116, 581–589. [Google Scholar] [CrossRef]
- Liss, K.H.; Finck, B.N. PPARs and nonalcoholic fatty liver disease. Biochimie 2017, 136, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Viscarra, J.; Kim, S.J.; Sul, H.S. Transcriptional regulation of hepatic lipogenesis. Nat. Rev. Mol. Cell Biol. 2015, 16, 678–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yim, S.H.; Clish, C.B.; Gladyshev, V.N. Selenium Deficiency Is Associated with Pro-Longevity Mechanisms. Cell Rep. 2019, 27, 2785–2797. [Google Scholar] [CrossRef] [PubMed]
- Mickiewicz, B.; Villemaire, M.L.; Sandercock, L.E.; Jirik, F.R.; Vogel, H.J. Metabolic changes associated with selenium deficiency in mice. Biometals Int. J. Role Met. Ions Biol. Biochem. Med. 2014, 27, 1137–1147. [Google Scholar] [CrossRef]
- Gar, C.; Rottenkolber, M.; Prehn, C.; Adamski, J.; Seissler, J.; Lechner, A. Serum and plasma amino acids as markers of prediabetes, insulin resistance, and incident diabetes. Crit. Rev. Clin. Lab. Sci. 2018, 55, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Alves, A.; Bassot, A.; Bulteau, A.L.; Pirola, L.; Morio, B. Glycine Metabolism and Its Alterations in Obesity and Metabolic Diseases. Nutrients 2019, 11, 1356. [Google Scholar] [CrossRef] [PubMed]
- Sunde, R.A.; Raines, A.M.; Barnes, K.M.; Evenson, J.K. Selenium status highly regulates selenoprotein mRNA levels for only a subset of the selenoproteins in the selenoproteome. Biosci. Rep. 2009, 29, 329–338. [Google Scholar] [CrossRef] [Green Version]
- Labunskyy, V.M.; Lee, B.C.; Handy, D.E.; Loscalzo, J.; Hatfield, D.L.; Gladyshev, V.N. Both maximal expression of selenoproteins and selenoprotein deficiency can promote development of type 2 diabetes-like phenotype in mice. Antioxid. Redox Signal. 2011, 14, 2327–2336. [Google Scholar] [CrossRef]
- Speckmann, B.; Schulz, S.; Hiller, F.; Hesse, D.; Schumacher, F.; Kleuser, B.; Geisel, J.; Obeid, R.; Grune, T.; Kipp, A.P. Selenium increases hepatic DNA methylation and modulates one-carbon metabolism in the liver of mice. J. Nutr. Biochem. 2017, 48, 112–119. [Google Scholar] [CrossRef]
- Mao, J.; Vanderlelie, J.J.; Perkins, A.V.; Redman, C.W.; Ahmadi, K.R.; Rayman, M.P. Genetic polymorphisms that affect selenium status and response to selenium supplementation in United Kingdom pregnant women. Am. J. Clin. Nutr. 2016, 103, 100–106. [Google Scholar] [CrossRef]
- Lennicke, C.; Rahn, J.; Kipp, A.P.; Dojcinovic, B.P.; Muller, A.S.; Wessjohann, L.A.; Lichtenfels, R.; Seliger, B. Individual effects of different selenocompounds on the hepatic proteome and energy metabolism of mice. Biochim. Biophys. Acta 2017, 1861, 3323–3334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schomburg, L. Sex-specific differences in biological effects and metabolism of selenium. In Selenium—Its Molecular Biology and Role in Human Health, 4th ed.; Hatfield, D.L., Schweizer, U., Tsuji, P.A., Gladyshev, V.N., Eds.; Springer: New York, NY, USA, 2016. [Google Scholar] [CrossRef]
- Seale, L.A.; Ogawa-Wong, A.N.; Berry, M.J. Sexual Dimorphism in Selenium Metabolism and Selenoproteins. Free Radic. Biol. Med. 2018, 127, 198–205. [Google Scholar] [CrossRef] [PubMed]


| Gene | 0.08 g/g Se | 0.25 g/g Se | 2-Way ANOVA | ||||
|---|---|---|---|---|---|---|---|
| WT (n = 5) | Scly KO (n = 5) | WT (n = 4) | Scly KO (n = 4) | pinteraction | pSe | pgenotype | |
| Abcb1a | 0.403 ± 0.016 | 0.302 ± 0.052 | 0.43 ± 0.038 | 0.571 ± 0.06 | 0.0193 | 0.0063 | 0.6606 |
| Abcc3 | 0.556 ± 0.034 | 0.498 ± 0.077 | 0.476 ± 0.235 | 1.205 ± 0.143 | 0.0003 | 0.0019 | 0.0012 |
| Acot3 | 0.402 ± 0.177 | 0.278 ± 0.205 | 0.526 ± 0.384 | 0.638 ± 0.190 | 0.3654 | 0.0783 | 0.9596 |
| Angptl6 | 0.56 ± 0.049 | 0.501 ± 0.127 | 0.622 ± 0.172 | 0.584 ± 0.2 | 0.8866 | 0.3468 | 0.5221 |
| Arl4d | 0.86 ± 0.287 | 0.858 ± 0.092 | 0.408 ± 0.135 | 1.275 ± 0.604 | 0.0321 | 0.9247 | 0.0329 |
| Cesg1 | 5.394 ± 0.903 | 3.382 ± 0.158 | 2.399 ± 0.873 | 4.382 ± 1.778 | 0.0043 | 0.1055 | 0.9801 |
| Eif4ebp3 | 1.118 ± 0.234 | 0.436 ± 0.108 | 1.482 ± 0.4 | 0.518 ± 0.198 | 0.316 | 0.123 | <0.0001 |
| Inhbe | 3.332 ± 0.484 | 1.187 ± 0.241 | 3.815 ± 0.739 | 2.297 ± 0.454 | 0.2633 | 0.0114 | <0.0001 |
| Lcn2 | 0.428 ± 0.297 | 0.348 ± 0.099 | 0.642 ± 0.357 | 0.426 ± 0.187 | 0.6491 | 0.3353 | 0.3292 |
| Mt1 | 0.709 ± 0.322 | 0.133 ± 0.044 | 0.201 ± 0.096 | 0.304 ± 0.207 | 0.01 | 0.1562 | 0.0551 |
| Mt2 | 0.688 ± 0.292 | 0.177 ± 0.054 | 0.25 ± 0.112 | 0.344 ± 0.13 | 0.0088 | 0.1869 | 0.0524 |
| Pparg | 0.166 ± 0.052 | 0.692 ± 0.103 | 0.193 ± 0.082 | 0.683 ± 0.087 | 0.8335 | 0.9209 | <0.0001 |
| Ppl | 0.089 ± 0.022 | 0.112 ± 0.059 | 0.151 ± 0.03 | 0.196 ± 0.028 | 0.5362 | 0.0011 | 0.0754 |
| Selenbp2 | 13.92 ± 7.954 | 6.691 ± 1.122 | 20.88 ± 10.12 | 6.224 ± 1.178 | 0.3281 | 0.3504 | 0.008 |
| Slc25a25 | 6.697 ± 0.888 | 3.163 ± 0.958 | 6.147 ± 3.191 | 4.869 ± 0.781 | 0.1732 | 0.5319 | 0.0357 |
| Gene | 0.08 g/g Se | 0.25 g/g Se | 2-Way ANOVA | ||||
|---|---|---|---|---|---|---|---|
| WT (n = 4) | Scly KO (n = 5) | WT (n = 4) | Scly KO (n = 5) | pinteraction | pSe | pgenotype | |
| Abcb1a | 0.579 ± 0.203 | 1.065 ± 0.503 | 0.727 ± 0.265 | 1.77 ± 0.707 | 0.2541 | 0.0914 | 0.0065 |
| Abcc3 | 1.075 ± 0.578 | 1.496 ± 0.741 | 1.401 ± 0.605 | 1.673 ± 0.556 | 0.8063 | 0.4129 | 0.2647 |
| Acot3 | 0.251 ± 0.043 | 0.114 ± 0.066 | 0.124 ± 0.029 | 0.106 ± 0.057 | 0.0323 | 0.0163 | 0.0074 |
| Angptl6 | 0.012 ± 0.002 | 0.014 ± 0.007 | 0.009 ± 0.001 | 0.016 ± 0.006 | 0.3646 | 0.7616 | 0.0713 |
| Arl4d | 0.561 ± 0.235 | 0.382 ± 0.171 | 1.147 ± 0.662 | 0.852 ± 0.541 | 0.7996 | 0.0345 | 0.3081 |
| Cesg1 | 4.006 ± 1.288 | 4.223 ± 1.249 | 4.339 ± 0.751 | 5.236 ± 1.125 | 0.537 | 0.231 | 0.3175 |
| Eif4ebp3 | 0.016 ± 0.008 | 0.017 ± 0.0146 | 0.011 ± 0.003 | 0.016 ± 0.007 | 0.6982 | 0.5723 | 0.5054 |
| Inhbe | 1.899 ± 0.719 | 3.481 ± 0.845 | 2.338 ± 0.949 | 4.11 ± 0.961 | 0.8286 | 0.2351 | 0.0018 |
| Lcn2 | 0.026 ± 0.01 | 0.017 ± 0.006 | 0.025 ± 0.009 | 0.016 ± 0.005 | 0.9079 | 0.7187 | 0.024 |
| Mt1 | 0.314 ± 0.092 | 0.154 ± 0.12 | 0.205 ± 0.124 | 0.073 ± 0.062 | 0.7741 | 0.0696 | 0.0092 |
| Mt2 | 0.397 ± 0.135 | 0.186 ± 0.121 | 0.259 ± 0.18 | 0.1 ± 0.063 | 0.6717 | 0.0844 | 0.0082 |
| Pparg | 0.524 ± 0.246 | 0.465 ± 0.234 | 0.753 ± 0.165 | 0.6 ± 0.208 | 0.6549 | 0.098 | 0.319 |
| Ppl | 0.004 ± 0.003 | 0.003 ± 0.002 | 0.003 ± 0.002 | 0.003 ± 0.001 | 0.5335 | 0.6621 | 0.4059 |
| Selenbp2 | 0.122 ± 0.048 | 0.069 ± 0.038 | 0.077 ± 0.022 | 0.057 ± 0.017 | 0.3098 | 0.0889 | 0.0338 |
| Slc25a25 | 0.209 ± 0.064 | 0.043 ± 0.013 | 0.087 ± 0.054 | 0.111 ± 0.06 | 0.0015 | 0.27 | 0.0109 |
| Ingenuity Canonical Pathways | −log (p-Value) | Ratio |
|---|---|---|
| PXR/RXR Activation | 4.11 | 0.0462 |
| LPS/IL-1 Mediated Inhibition of RXR Function | 3.76 | 0.018 |
| Xenobiotic Metabolism Signaling | 3.32 | 0.0138 |
| Nicotine Degradation III | 2.65 | 0.0364 |
| Melatonin Degradation I | 2.52 | 0.0312 |
| Nicotine Degradation II | 2.52 | 0.0312 |
| Superpathway of Melatonin Degradation | 2.45 | 0.029 |
| Acyl-CoA Hydrolysis | 1.82 | 0.0833 |
| Hepatic Cholestasis | 1.75 | 0.0125 |
| Ubiquinol-10 Biosynthesis (Eukaryotic) | 1.62 | 0.0526 |
| Biological Pathways | Hits | p-Value |
|---|---|---|
| Alanine, aspartate and glutamate metabolism | 10 | 0.000285 |
| D-glutamine and D-glutamate metabolism | 4 | 0.000299 |
| Galactose metabolism | 9 | 0.000886 |
| Aminoacyl-tRNA biosynthesis | 12 | 0.000907 |
| Biosynthesis of unsaturated fatty acids | 6 | 0.002 |
| Pantothenate and CoA biosynthesis | 5 | 0.021 |
| Valine, leucine and isoleucine biosynthesis | 3 | 0.022 |
| Glyoxylate and dicarboxylate metabolism | 6 | 0.045 |
| Taurine and hypotaurine metabolism | 3 | 0.051 |
| Glycine, serine and threonine metabolism | 7 | 0.053 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seale, L.A.; Khadka, V.S.; Menor, M.; Xie, G.; Watanabe, L.M.; Sasuclark, A.; Guirguis, K.; Ha, H.Y.; Hashimoto, A.C.; Peplowska, K.; et al. Combined Omics Reveals That Disruption of the Selenocysteine Lyase Gene Affects Amino Acid Pathways in Mice. Nutrients 2019, 11, 2584. https://doi.org/10.3390/nu11112584
Seale LA, Khadka VS, Menor M, Xie G, Watanabe LM, Sasuclark A, Guirguis K, Ha HY, Hashimoto AC, Peplowska K, et al. Combined Omics Reveals That Disruption of the Selenocysteine Lyase Gene Affects Amino Acid Pathways in Mice. Nutrients. 2019; 11(11):2584. https://doi.org/10.3390/nu11112584
Chicago/Turabian StyleSeale, Lucia A., Vedbar S. Khadka, Mark Menor, Guoxiang Xie, Ligia M. Watanabe, Alexandru Sasuclark, Kyrillos Guirguis, Herena Y. Ha, Ann C. Hashimoto, Karolina Peplowska, and et al. 2019. "Combined Omics Reveals That Disruption of the Selenocysteine Lyase Gene Affects Amino Acid Pathways in Mice" Nutrients 11, no. 11: 2584. https://doi.org/10.3390/nu11112584
APA StyleSeale, L. A., Khadka, V. S., Menor, M., Xie, G., Watanabe, L. M., Sasuclark, A., Guirguis, K., Ha, H. Y., Hashimoto, A. C., Peplowska, K., Tiirikainen, M., Jia, W., Berry, M. J., & Deng, Y. (2019). Combined Omics Reveals That Disruption of the Selenocysteine Lyase Gene Affects Amino Acid Pathways in Mice. Nutrients, 11(11), 2584. https://doi.org/10.3390/nu11112584






