Characteristics and Glucose Uptake Promoting Effect of Chrysin-Loaded Phytosomes Prepared with Different Phospholipid Matrices
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of CP
2.3. Characterization of CP
2.3.1. Particle Size, PDI, and Zeta Potential
2.3.2. Entrapment Efficiency of CP
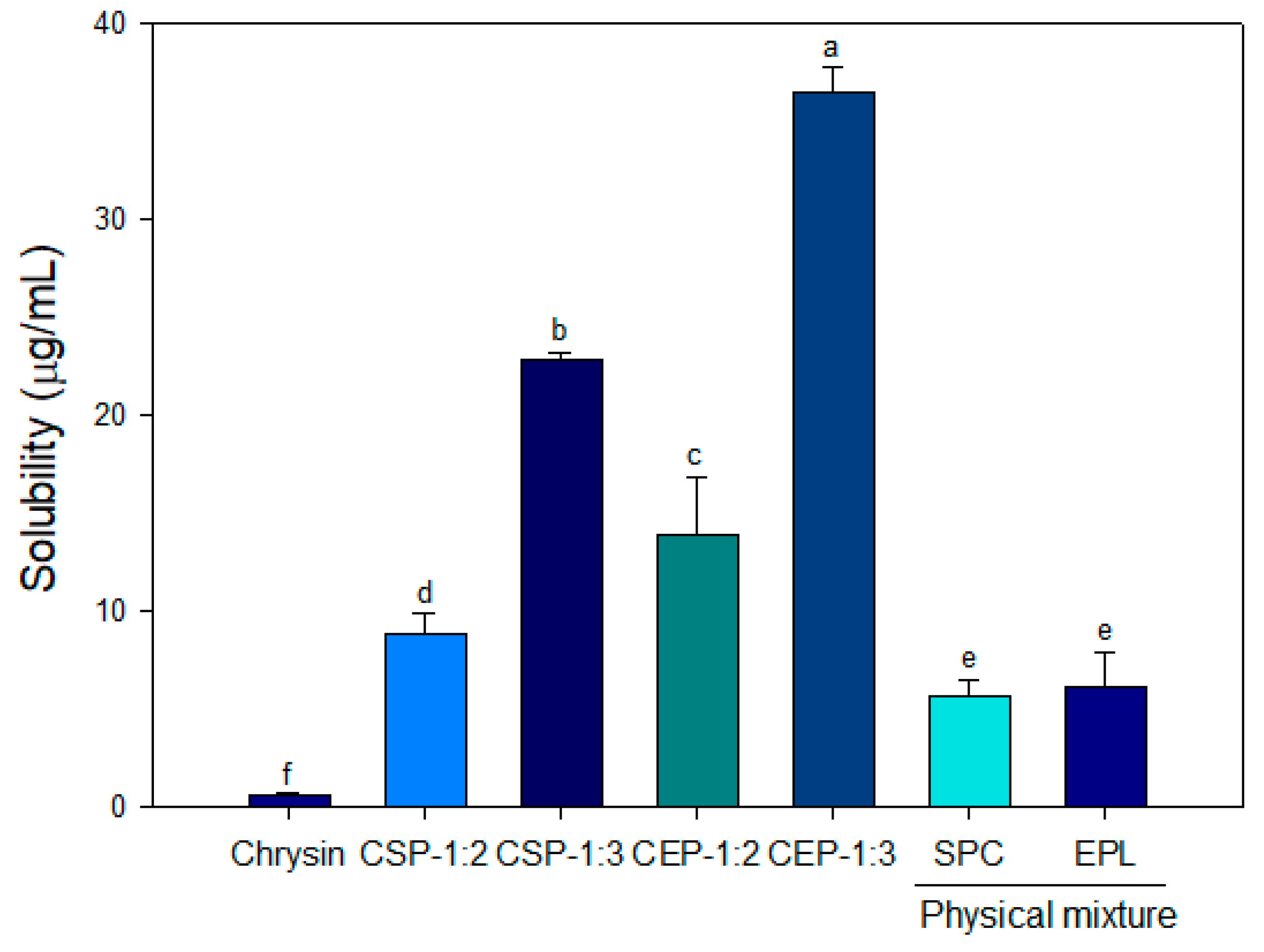
2.3.3. Solubility
2.3.4. Topographical Analysis and Measurement of Elastic Modulus
2.3.5. FTIR
2.3.6. XRD
2.3.7. SEM
2.3.8. In Vitro Release
2.4. Bioassays
2.4.1. Cell Culture
2.4.2. Glucose Uptake Assay Using C2C12 Skeletal Muscle Cells
2.4.3. RNA Extraction and Quantitative Real Time PCR (qRT-PCR)
2.5. Statistical Analysis
3. Results and Discussion
3.1. Formation of Chrysin-Loaded Phytosomes (CPs)
3.2. Size, Polydispersity Index (PDI), and Zeta Potential of CPs
3.3. Solubility of CPs
3.4. Topographical Analysis of CPs Using AFM
3.5. Fourier Transformation Infrared Spectroscopy (FTIR)
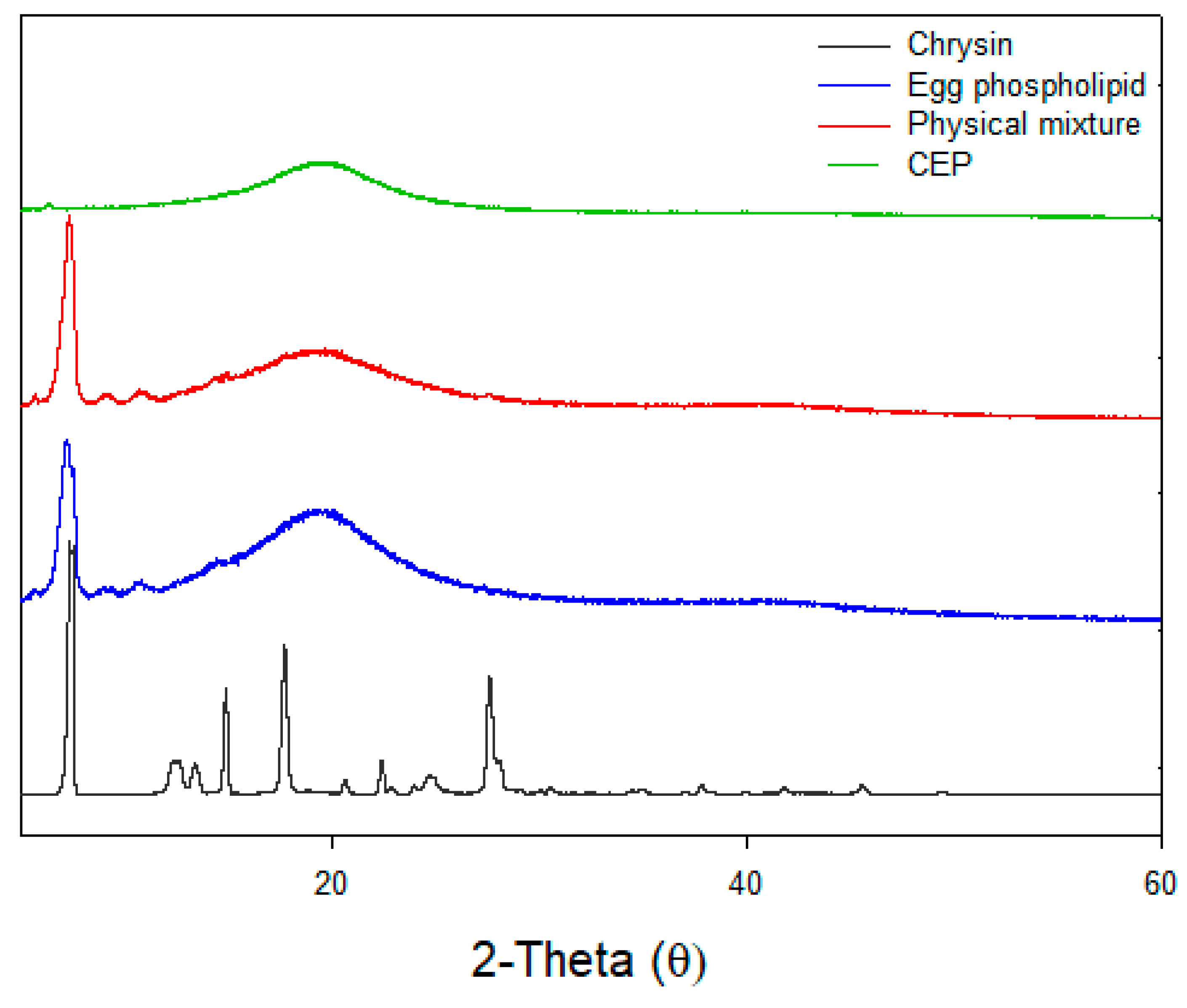
3.6. X-ray Diffraction (XRD)
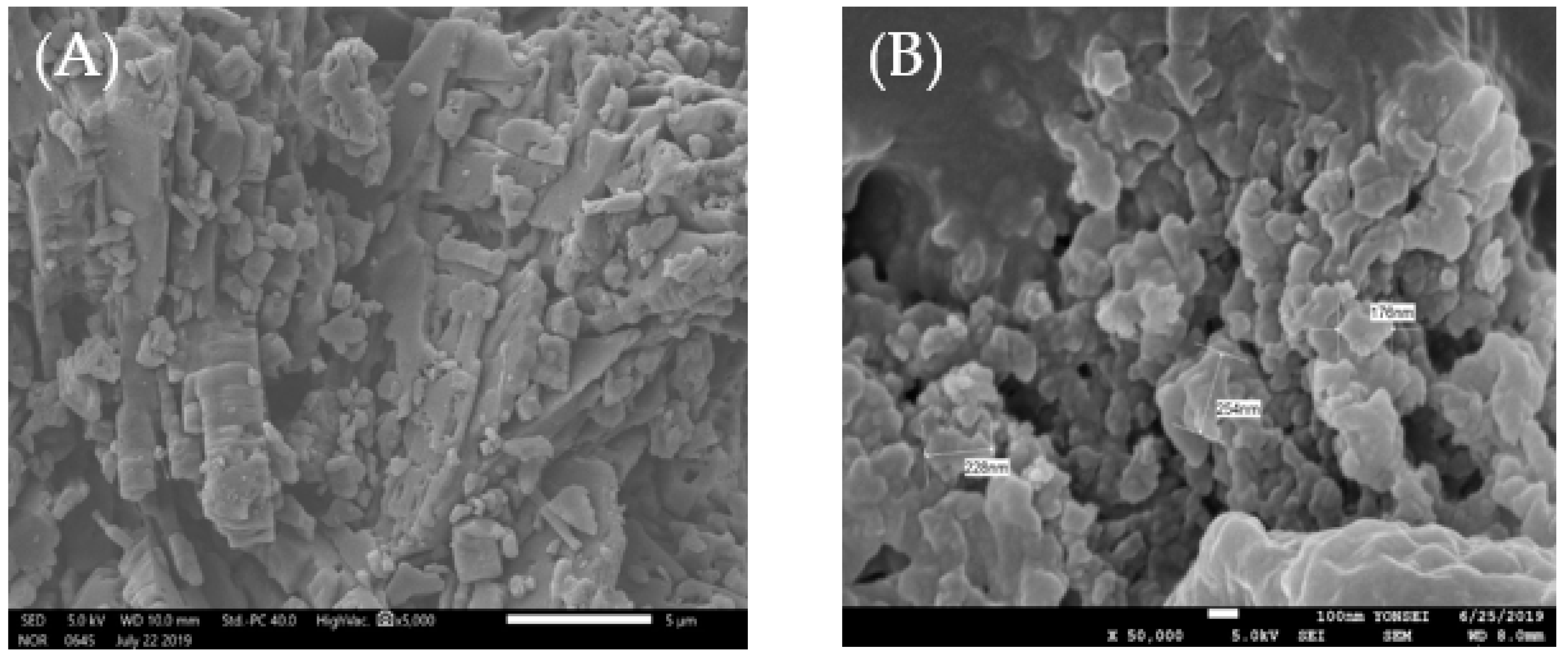
3.7. Scanning Electron Microscopy (SEM)
3.8. In Vitro Release of CEP
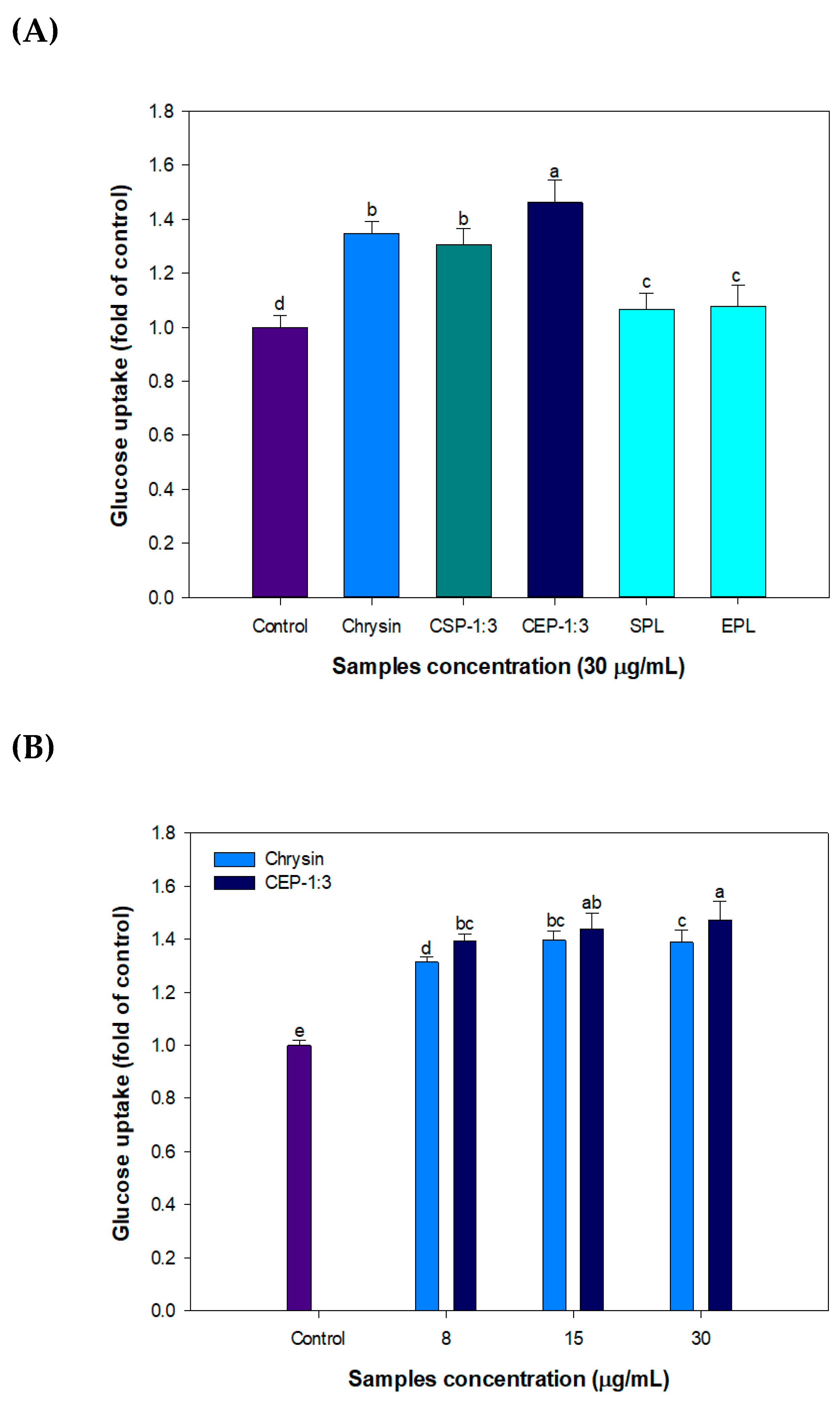
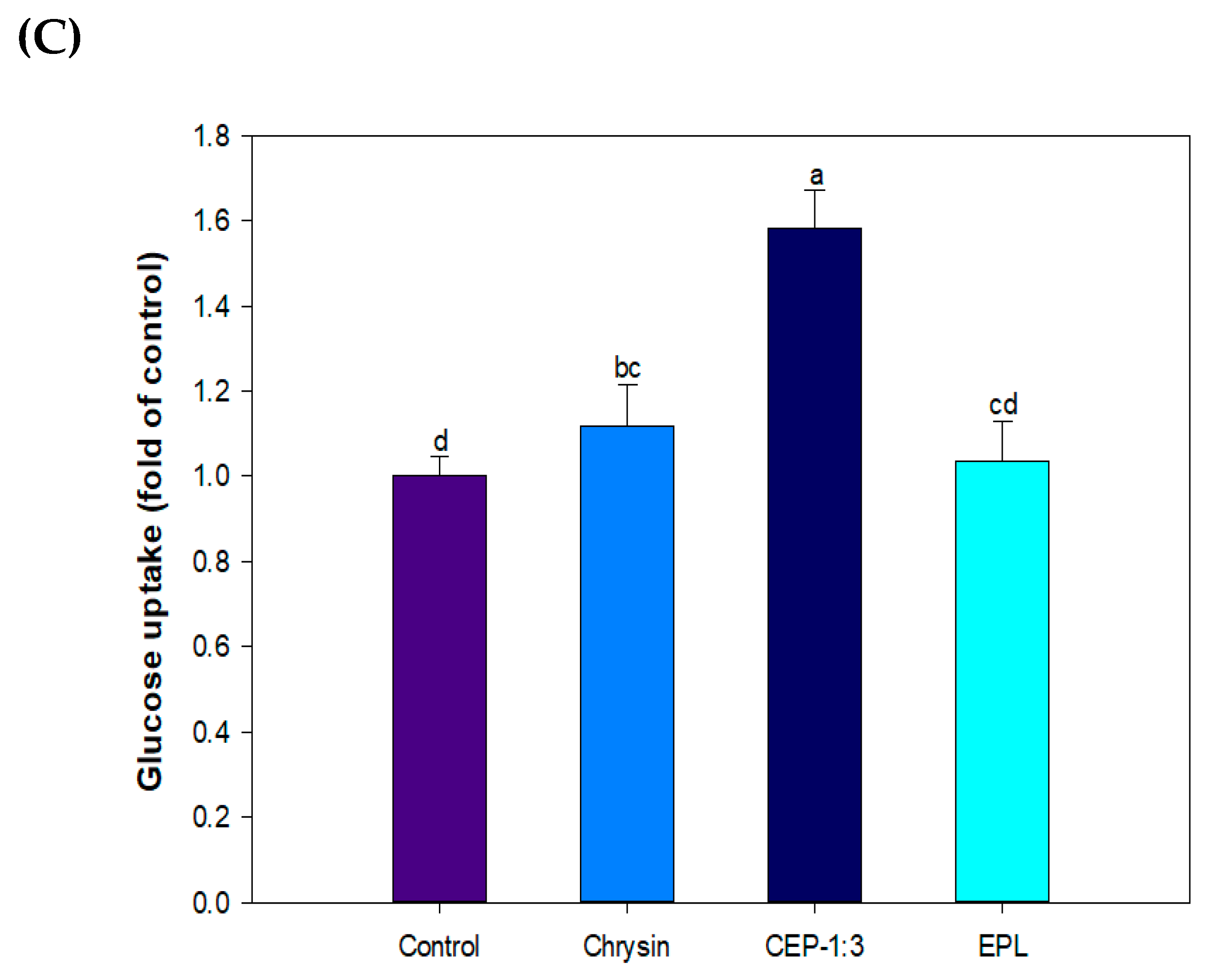
3.9. Effect of CP on Glucose Uptake in C2C12 Skeletal Muscle Cells
3.10. Effect of CEP-1:3 on the Expression of Genes Related to Glucose Uptake in C2C12 Skeletal Muscle Cells
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kahn, S.E.; Cooper, M.E.; Del Prato, S. Pathophysiology and treatment of type 2 diabetes: Perspectives on the past, present, and future. Lancet 2014, 383, 1068–1083. [Google Scholar] [CrossRef]
- Babu, P.V.A.; Liu, D.; Gilbert, E.R. Recent advances in understanding the antidiabetic actions of dietary flavonoids. J. Nutr. Biochem. 2013, 24, 1777–1789. [Google Scholar] [CrossRef] [PubMed]
- Smith, V.T.; Muscat, G.E. Skeletal muscle and nuclear hormone receptors: Implications for cardiovascular and metabolic disease. Int. J. Biochem. Cell Biol. 2005, 37, 2047–2063. [Google Scholar] [CrossRef] [PubMed]
- Farkhondeh, T.; Samarghandian, S.; Roshanravan, B. Impact of chrysin on the molecular mechanisms underlying diabetic complications. J. Cell Physiol. 2019, 234, 17144–17158. [Google Scholar] [CrossRef]
- Ramírez-Espinosa, J.J.; Saldaña-Ríos, J.; García-Jiménez, S.; Villalobos-Molina, R.; Ávila-Villarreal, G.; Rodríguez-Ocampo, A.N.; Bernal-Fernández, G.; Estrada-Soto, S. Chrysin induces antidiabetic, antidyslipidemic and anti-inflammatory effects in athymic nude diabetic mice. Molecules 2018, 23, 67. [Google Scholar] [CrossRef]
- Kandhare, A.D.; Shivakumar, V.; Rajmane, A.; Ghosh, P.; Bodhankar, S.L. Evaluation of the neuroprotective effect of chrysin via modulation of endogenous biomarkers in a rat model of spinal cord injury. J. Nat. Med. 2014, 68, 586–603. [Google Scholar] [CrossRef]
- Nabavi, S.F.; Braidy, N.; Habtemariam, S.; Orhan, I.E.; Daglia, M.; Manayi, A.; Gortzi, O.; Nabavi, S.M. Neuroprotective effects of chrysin: From chemistry to medicine. Neurochem. Int. 2015, 90, 224–231. [Google Scholar] [CrossRef]
- Song, J.H.; Kim, Y.H.; Lee, S.C.; Kim, M.H.; Lee, J.H. Inhibitory effect of chrysin (5, 7-dihydroxyflavone) on experimental choroidal neovascularization in rats. Ophthal. Res. 2016, 56, 49–55. [Google Scholar] [CrossRef]
- Walle, T.; Otake, Y.; Brubaker, J.A.; Walle, U.K.; Halushka, P.V. Disposition and metabolism of the flavonoid chrysin in normal volunteers. Br. J. Clin. Pharmacol. 2001, 51, 143–146. [Google Scholar] [CrossRef]
- Mohammadian, F.; Abhari, A.; Dariushnejad, H.; Nikanfar, A.; Pilehvar-Soltanahmadi, Y.; Zarghami, N. Effects of chrysin-PLGA-PEG nanoparticles on proliferation and gene expression of miRNAs in gastric cancer cell line. Iran J. Caner Prev. 2016, 9, e4190. [Google Scholar] [CrossRef]
- Sabzichi, M.; Mohammadian, J.; Bazzaz, R.; Pirouzpanah, M.B.; Shaaker, M.; Hamishekar, H.; Chavoshi, H.; Salehi, R.; Samadi, N. Chrysin loaded nanostructured lipid carriers (NLCs) triggers apoptosis in MCF-7 cancer cells by inhibiting the Nrf2 pathway. Process Biochem. 2017, 60, 84–91. [Google Scholar] [CrossRef]
- Li, J.; Wang, X.; Zhang, T.; Wang, C.; Huang, Z.; Luo, X.; Deng, Y. A review on phospholipids and their main applications in drug delivery systems. Asian J. Pharm. Sci. 2015, 10, 81–98. [Google Scholar] [CrossRef]
- Semalty, A.; Semalty, M.; Rawat, M.S.; Franceschi, F. Supramolecular phospholipids-polyphenolics interactions: The phytosome strategy to improve the bioavailability of phytochemicals. Fitoterapia 2010, 81, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Gangadharappa, H.V.; Mruthunjaya, K. Phospholipids: Unique carriers for drug delivery systems. J. Drug Delivery Sci. Technol. 2017, 39, 166–179. [Google Scholar] [CrossRef]
- Babazadeh, A.; Ghanbarzadeh, B.; Hamishehkar, H. Phosphatidylcholine-rutin complex as a potential nanocarrier for food applications. J. Func. Foods 2017, 33, 134–141. [Google Scholar] [CrossRef]
- Telange, D.R.; Patil, A.T.; Pethe, A.M.; Fegade, H.; Anand, S.; Dave, V.S. Formulation and characterization of an apigenin-phospholipid phytosome (APLC) for improved solubility, in vivo bioavailability, and antioxidant potential. Eur. J. Pharma. Sci. 2017, 108, 36–49. [Google Scholar] [CrossRef]
- Brochu, H.; Vermette, P. Young’s moduli of surface-bound liposomes by atomic force microscopy force measurements. Langmuir 2008, 24, 2009–2014. [Google Scholar] [CrossRef]
- Derjaguin, B.V.; Muller, V.M.; Toporov, Y.P. Effect of contact deformations on the adhesion of particles. J. Colloid Interf. Sci. 1975, 53, 314–326. [Google Scholar] [CrossRef]
- Hou, Z.; Li, Y.; Huang, Y.; Zhou, C.; Lin, J.; Wang, Y.; Cui, F.; Zhou, S.; Jia, M.; Ye, S.; et al. Phytosomes loaded with mitomycin C-soybean phosphatidylcholine complex developed for drug delivery. Mol. Pharm. 2013, 10, 90–101. [Google Scholar] [CrossRef]
- Kim, S.; Go, G.-W.; Imm, J.-Y. Promotion of glucose uptake in C2C12 myotubes by cereal flavone tricin and its underlying molecular mechanism. J. Agric. Food Chem. 2017, 65, 3819–3826. [Google Scholar] [CrossRef]
- Kim, S.; Choi, S.-I.; Kim, G.-H.; Imm, J.-Y. Anti-inflammatory effect of Ecklonia cava extract on Porphyromonas gingivalis lipopolysaccharide-stimulated macrophages and a periodontitis rat model. Nutrients 2019, 11, 1143. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, C.; Driessen, A.J.M.; Recourt, K. The uncoupling efficiency and affinity of flavonoids for vesicles. Biochem. Pharmacol. 2000, 60, 1593–1600. [Google Scholar] [CrossRef]
- El-Fattah, A.I.; Fathy, M.M.; Ali, Z.Y.; El-Garawany, A.E.A.; Mohamed, E.K. Enhanced therapeutic benefit of quercetin-loaded phytosome nanoparticles in ovariectomized rats. Chem. Biol. Interact. 2017, 271, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Mazumder, A.; Dwivedi, A.; Preez, J.L.D.; Plessis, J.D. In vitro wound healing and cytotoxic effects of sinigrin-phytosome complex. Int. J. Pharm. 2015, 498, 283–293. [Google Scholar] [CrossRef]
- Vilamarin, R.; Bernardo, J.; Videira, R.A.; Valentao, P.; Veiga, F.; Andrade, P.B. An egg yolk’s phospholipid-pennyroyal nootropic nanoformulation modulates monoamino oxidase-A (MAO-A) activity in SH-SY5Y neuronal model. J. Func. Foods 2018, 46, 335–344. [Google Scholar] [CrossRef]
- Freag, M.S.; Saleh, W.M.; Abdallah, O.Y. Self-assembled phospholipid-based phytosomal nanocarriers as promising platforms for improving oral bioavailability of the anticancer celastrol. Int. J. Pharm. 2018, 535, 18–26. [Google Scholar] [CrossRef]
- Chaudhary, S.; Garg, T.; Murthy, R.S.R.; Rath, G.; Goyal, A.K. Development, optimization and evaluation of long chain nanolipid carrier for hepatic delivery of silymarin through lymphatic transport pathway. Int. J. Pharm. 2015, 485, 108–121. [Google Scholar] [CrossRef]
- Nara, E.; Miyashita, K.; Ota, T. Oxidative stability of liposomes prepared from soybean PC, chicken egg PC, and salmon egg PC. Biosci. Biotechnol. Biochem. 1997, 61, 1736–1738. [Google Scholar] [CrossRef]
- Pathan, R.A.; Bhandari, U. Preparation & characterization of embelin—phospholipid complex as effective drug delivery tool. J. Incl. Phenom. Macrocycl. Chem. 2011, 69, 139–147. [Google Scholar]
- Lichtenberger, L.M.; Ulloa, C.; Romero, J.J.; Vanous, A.L.; Romero, J.J.; Dial, E.J.; Illich, P.A.; Walters, E.T. Zwitterionic phospholipids enhance aspirin’s therapeutic activity, as demonstrated in rodent model systems. J. Pharmacol. Exp. Ther. 1996, 277, 1221–1227. [Google Scholar]
- Liu, A.; Lou, H.; Zhao, L.; Fan, P. Validated LC/MS/MS assay for curcumin and tetrahydrocurcumin in rat plasma and application to pharmacokinetic study of phospholipid complex of curcumin. J. Pharm. Biomed. Anal. 2006, 40, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Sahebkar, A. The promise of curcumin-phosphatidylcholine complex for cardiometabolic diseases: More than just ‘more curcumin’. Nat. Prod. Res. 2015, 29, 392–393. [Google Scholar] [CrossRef] [PubMed]
- Kucˇerka, N.; Tristram-Nagle, S.; Nagle, J.F. Closer look at structure of fully hydrated fluid phase DPPC bilayers. Biophys. J. 2006, L83–L85. [Google Scholar] [CrossRef] [PubMed]
- Nzai, J.M.; Proctor, A. Determination of phospholipids in vegetable oil by Fourier transform infrared spectroscopy. J. Am. Oil Chem. Soc. 1998, 75, 1281–1289. [Google Scholar] [CrossRef]
- Uekusa, Y.; Takeshita, Y.; Ishii, T.; Nakayama, T. Partition coefficients of polyphenols for phosphatidylcholine investigated by HPLC with an immobilized artificial membrane column. Biosci. Biotechnol. Biochem. 2008, 72, 3289–3292. [Google Scholar] [CrossRef]
- Lu, M.; Qiu, Q.; Luo, X.; Liu, X.; Sun, J.; Wang, C.; Lin, X.; Deng, Y.; Song, Y. Phyto-phospholipid complexes (phytosome): A novel strategy to improve the bioavailability of active constituents. Asian J. Pharm. Sci. 2019, 14, 265–274. [Google Scholar] [CrossRef]
- Mancini, S.; Nardo, L.; Gregiri, M.; Ribeiro, I.; Mantegazza, F.; Delerue-Matos, C.; Masserini, M.; Grosso, G. Functionalized liposomes and phytosomes loading Annona murcata L. aqueous extract: Potential nanoshuttles for brain-delivery of phenolic compounds. Phytomedicine 2018, 42, 233–244. [Google Scholar] [CrossRef]
- Ishikado, A.; Nishio, Y.; Yamane, K.; Mukosea, A.; Morinoa, K.; Murakami, Y.; Sekinea, O.; Makino, T.; Maegawa, H.; Kashiwagi, A. Soy phosphatidylcholine inhibited TLR4-mediated MCP-1 expression in vascular cells. Atherosclerosis 2009, 205, 404–412. [Google Scholar] [CrossRef]
- Feng, X.; Qin, H.; Shi, Q.; Zhang, Y.; Zhou, F.; Wu, H.; Ding, S.; Niu, Z.; Lu, Y.; Shen, P. Chrysin attenuates inflammation by regulating M1/M2 status via activating PPARγ. Biochem. Pharmacol. 2014, 89, 503–514. [Google Scholar] [CrossRef]
- Dhaunsi, G.S.; Yousif, M.H.; Akhtar, S.; Chappell, M.C.; Diz, D.I.; Benter, I.F. Angiotensin-(1–7) prevents diabetes-induced attenuation in PPAR-gamma and catalase activities. Eur. J. Pharmacol. 2010, 638, 108–114. [Google Scholar] [CrossRef]
- Verma, N.K.; Singh, J.; Dey, C.S. PPAR-γ expression modulates insulin sensitivity in C2C12 skeletal muscle cells. Brit. J. Pharmacol. 2004, 143, 1006–1013. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.J.; Bang, M.-H.; Kim, H.; Imm, J.-Y. Improvement of palmitate-induced insulin resistance in C2C12 skeletal muscle cells using Platycodon grandiflorum seed extract. Food Biosci. 2018, 25, 61–67. [Google Scholar] [CrossRef]
- Yang, M.; Wei, D.; Mo, C.; Zhang, J.; Wang, X.; Han, X.; Wang, Z.; Xiao, H. Saturated fatty acid palmitate-induced insulin resistance is accompanied with myotube loss and the impaired expression of health benefit myokine genes in C2C12 myotubes. Lipids Health Dis. 2013, 12, 104. [Google Scholar] [CrossRef] [PubMed]
- Satyanarayana, K.; Sravanthi, K.; Shaker, I.V.; Ponnulakshmi, R.; Selvaraj, J. Role of chrysin on expression of insulin signaling molecules. J. Ayurveda Intrgr. Med. 2015, 6, 248–258. [Google Scholar] [CrossRef]
- Prabhakar, P.K.; Doble, M. Synergistic effect of phytochemicals in combination with hypoglycemic drugs on glucose uptake in myotubes. Phytomedicine 2009, 16, 1119–1126. [Google Scholar] [CrossRef]




| Average Particle Size (nm) | PDI | Zeta Potential (mV) | Encapsulation Efficiency (%) | |
|---|---|---|---|---|
| CSP-1:2 | 129 ± 2.0 | 0.45 ± 0.02 | −28 ± 0.2 | 98.8 ± 0.1 |
| CSP-1:3 | 134 ± 0.1 | 0.49 ± 0.01 | −33 ± 4.1 | 98.6 ± 0.1 |
| CEP-1:2 | 89 ± 0.5 | 0.25 ± 0.01 | −21 ± 0.3 | 98.8 ± 0.1 |
| CEP-1:3 | 117 ± 0.7 | 0.29 ± 0.02 | −31 ± 0.8 | 98.7 ± 0.1 |
| Storage for 21 days at 4 °C | ||||
| CEP-1:3 | 118 ± 1 | 0.30 ± 0.02 | −32 ± 0.3 | |
| CSP-1:3 | CEP-1:3 | |
|---|---|---|
| Elastic modulus (kPa) | 12.4 ± 0.52 | 33.2 ± 0.90 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.-M.; Jung, J.-I.; Chai, C.; Imm, J.-Y. Characteristics and Glucose Uptake Promoting Effect of Chrysin-Loaded Phytosomes Prepared with Different Phospholipid Matrices. Nutrients 2019, 11, 2549. https://doi.org/10.3390/nu11102549
Kim S-M, Jung J-I, Chai C, Imm J-Y. Characteristics and Glucose Uptake Promoting Effect of Chrysin-Loaded Phytosomes Prepared with Different Phospholipid Matrices. Nutrients. 2019; 11(10):2549. https://doi.org/10.3390/nu11102549
Chicago/Turabian StyleKim, Seong-Min, Jae-In Jung, Changhoon Chai, and Jee-Young Imm. 2019. "Characteristics and Glucose Uptake Promoting Effect of Chrysin-Loaded Phytosomes Prepared with Different Phospholipid Matrices" Nutrients 11, no. 10: 2549. https://doi.org/10.3390/nu11102549
APA StyleKim, S.-M., Jung, J.-I., Chai, C., & Imm, J.-Y. (2019). Characteristics and Glucose Uptake Promoting Effect of Chrysin-Loaded Phytosomes Prepared with Different Phospholipid Matrices. Nutrients, 11(10), 2549. https://doi.org/10.3390/nu11102549






