Nobiletin Enhances Chemosensitivity to Adriamycin through Modulation of the Akt/GSK3β/β–Catenin/MYCN/MRP1 Signaling Pathway in A549 Human Non-Small-Cell Lung Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Cell Viability Assay
2.3. Transcriptome Analysis
2.4. Functional Annotation of Differentially Expressed Genes (DEGs)
2.5. Analysis of the Effects of Drug Combinations
2.6. Intracellular Accumulation of ADR
2.7. Cell Cycle Analysis
2.8. Western Blot Analysis
2.9. In Vivo Animal Studies
2.10. Statistical Analysis
3. Results
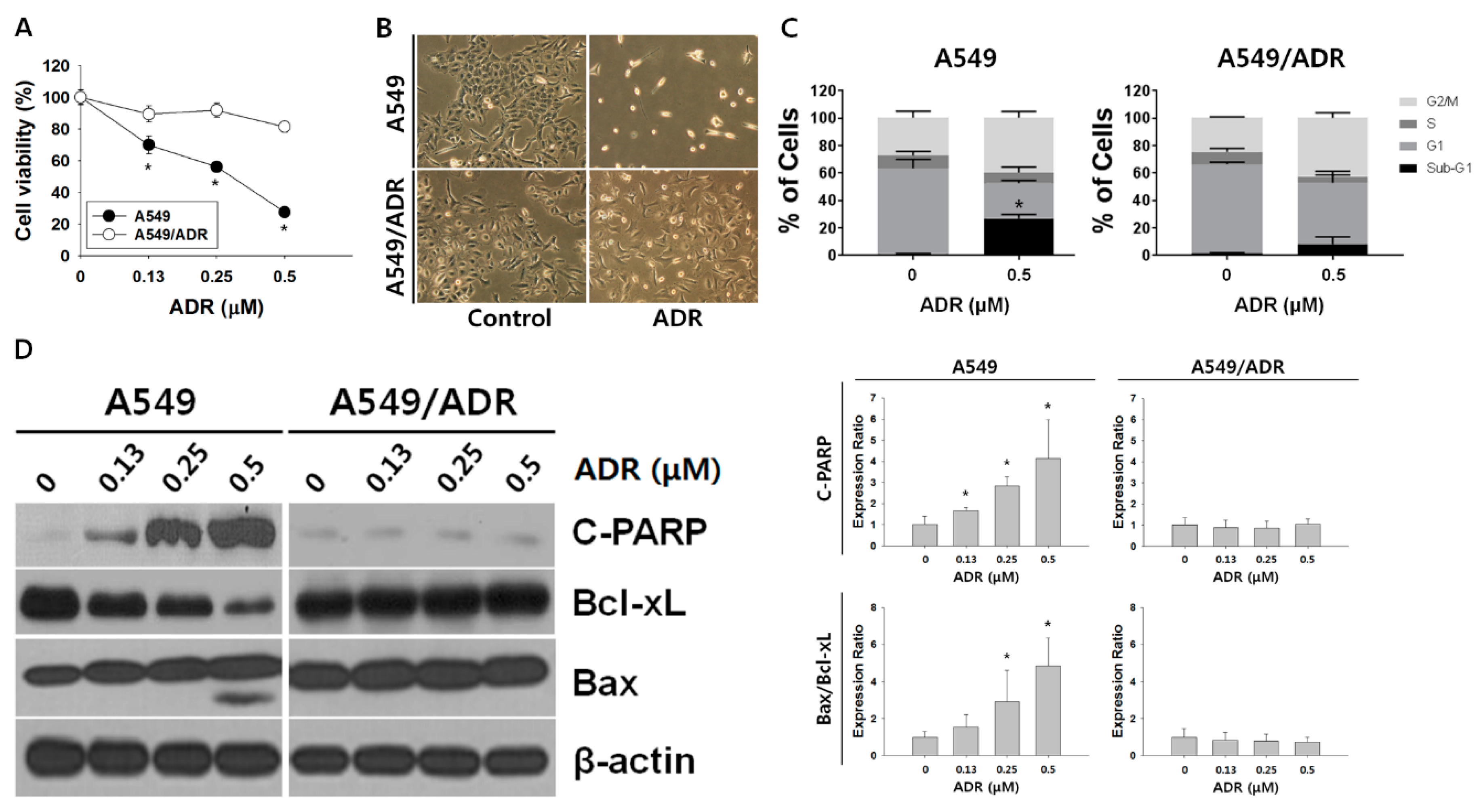
3.1. Characteristics of A549/ADR Cells
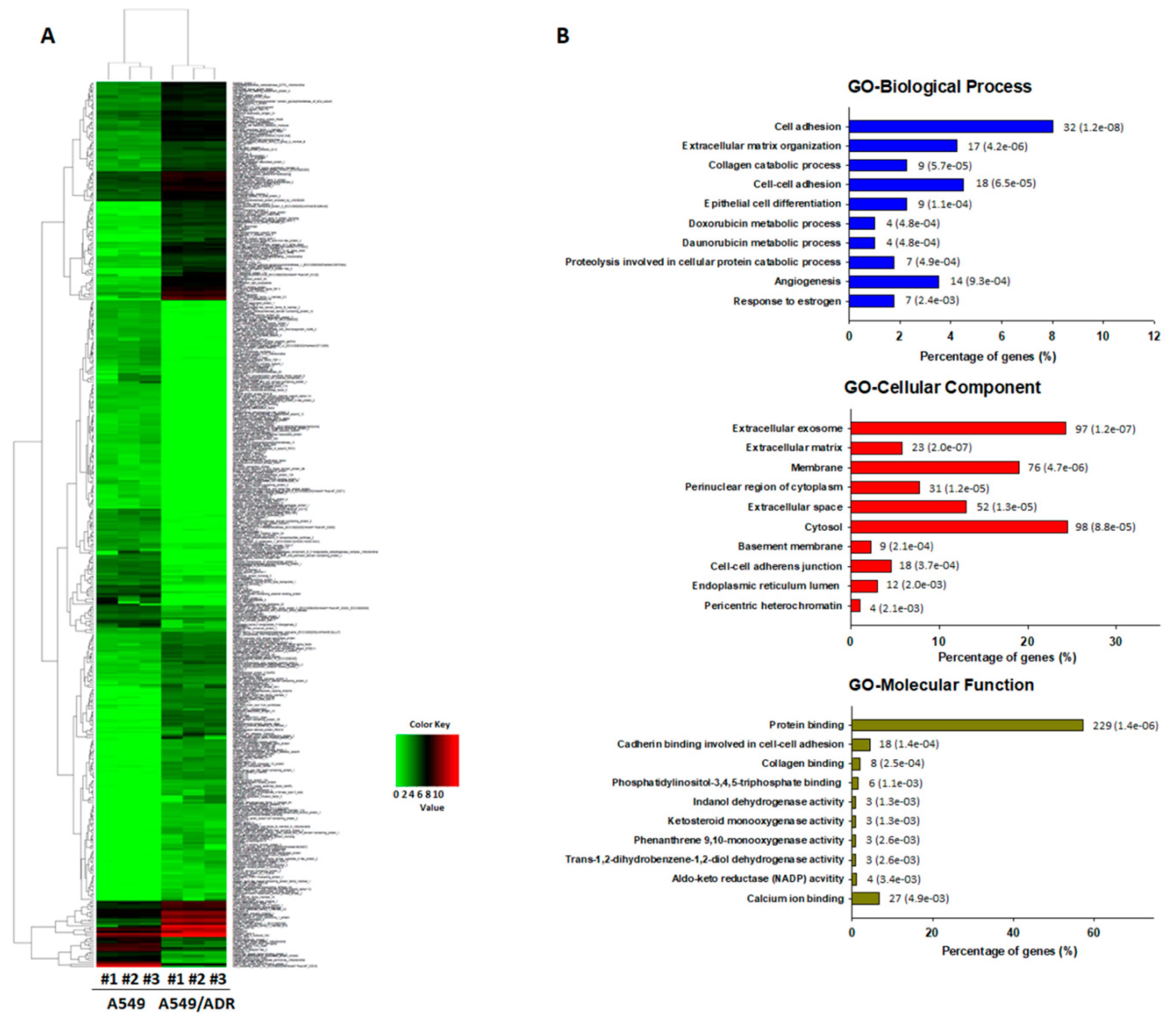
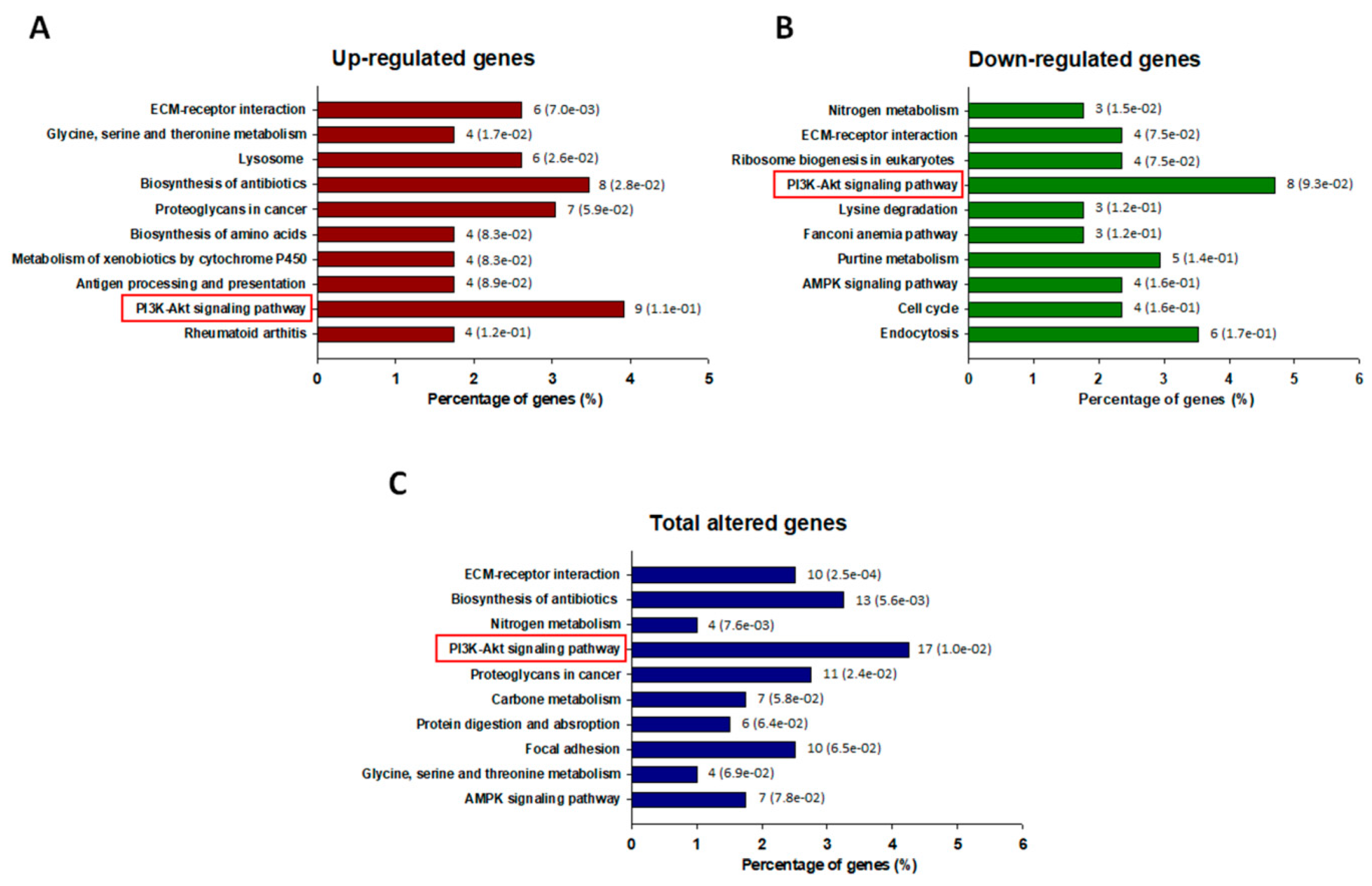
3.2. Transcriptomic Analysis of ADR-Resistant Non-Small Cell Lung Cancer (A549/ADR) Cells
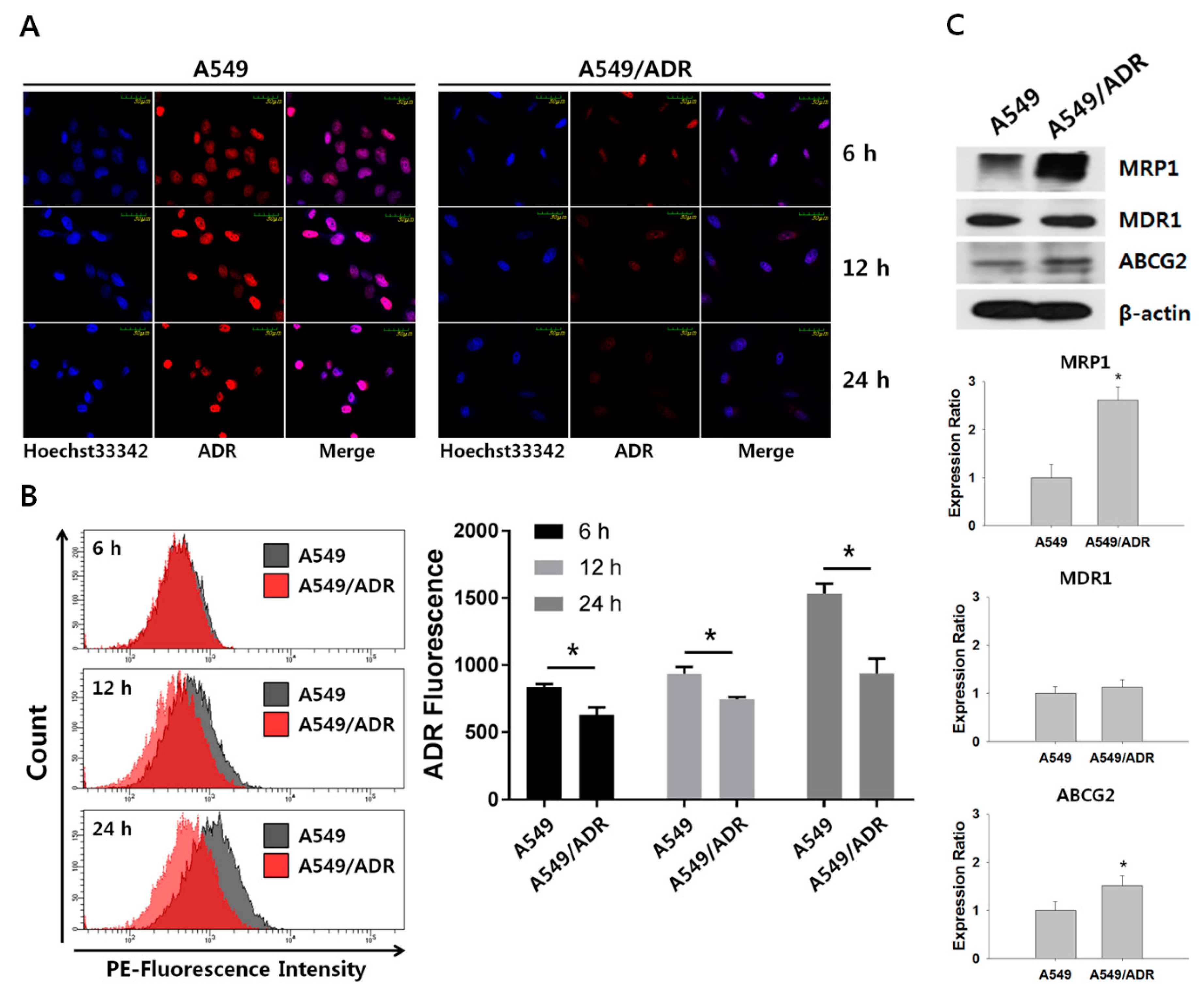
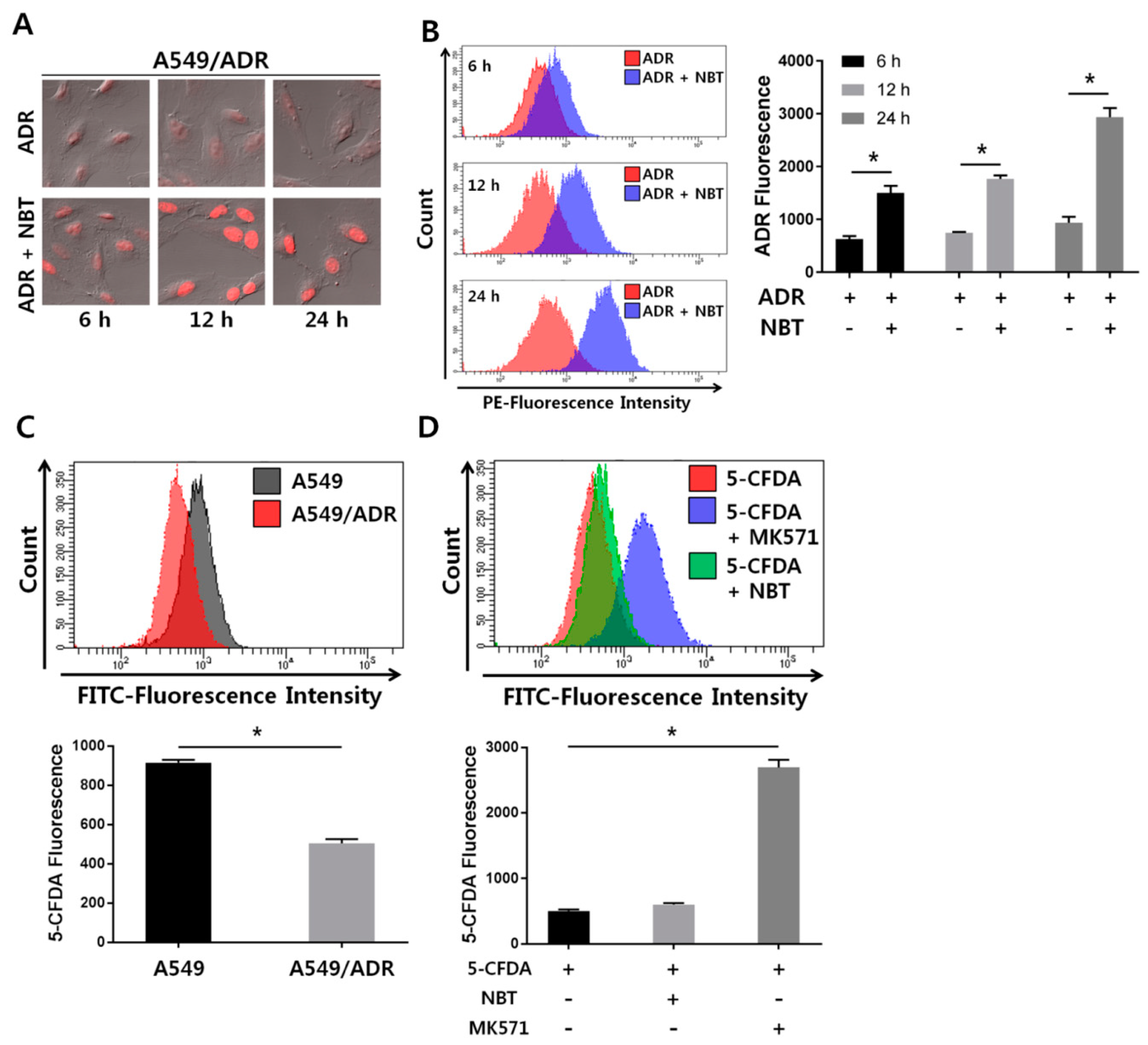
3.3. Decrease in Intracellular Accumulation of ADR in A549/ADR Cells Compared to That in the Parental A549 Cells
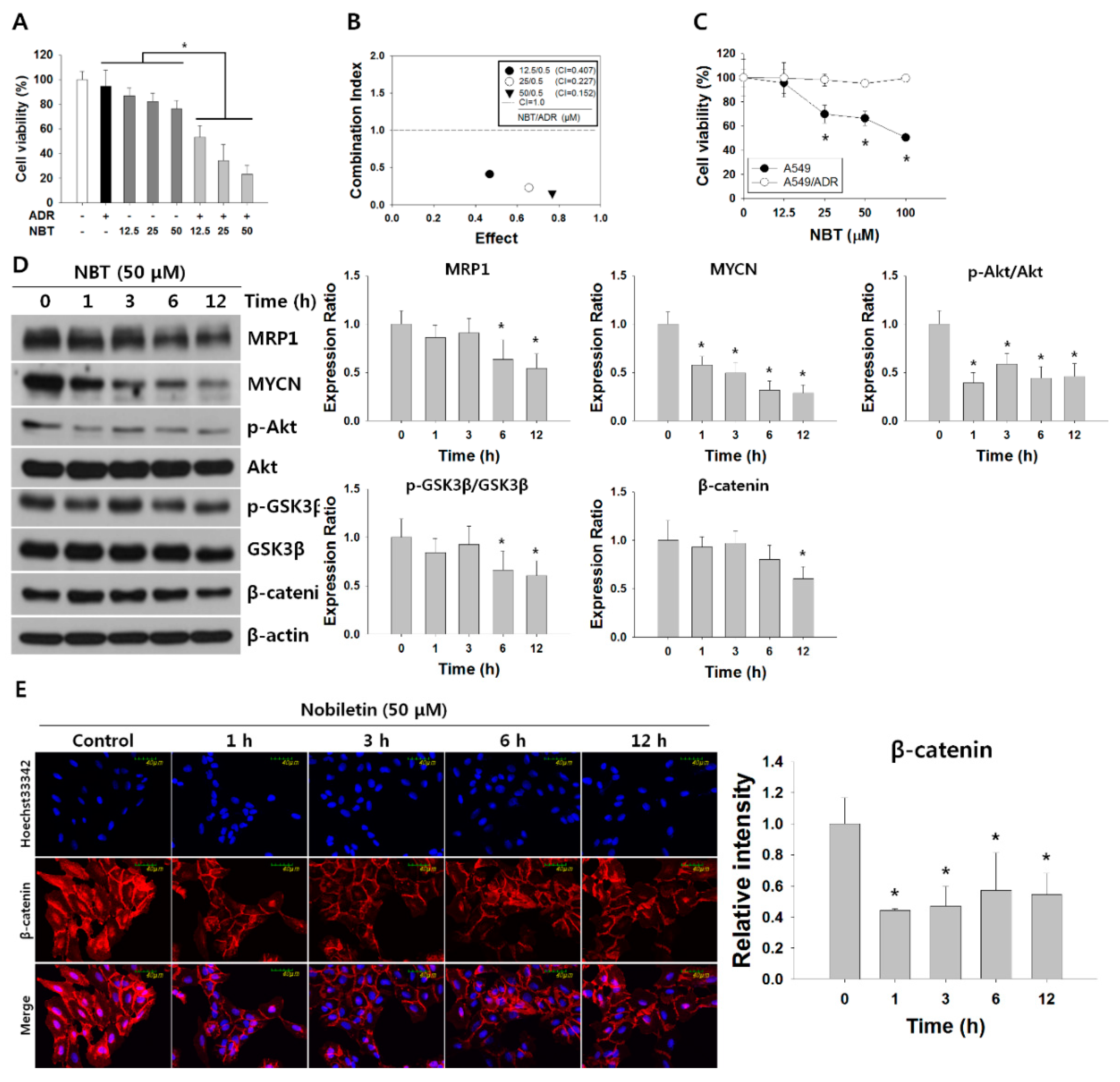
3.4. NBT Enhances Chemosensitivity of A549/ADR Cells to ADR and Suppresses the Akt/GSK3β/β-Catenin/MYCN/MRP1 Signaling Pathway
3.5. NBT Enhances Intracellular Accumulation of ADR without Changing the Function of the Efflux Pump
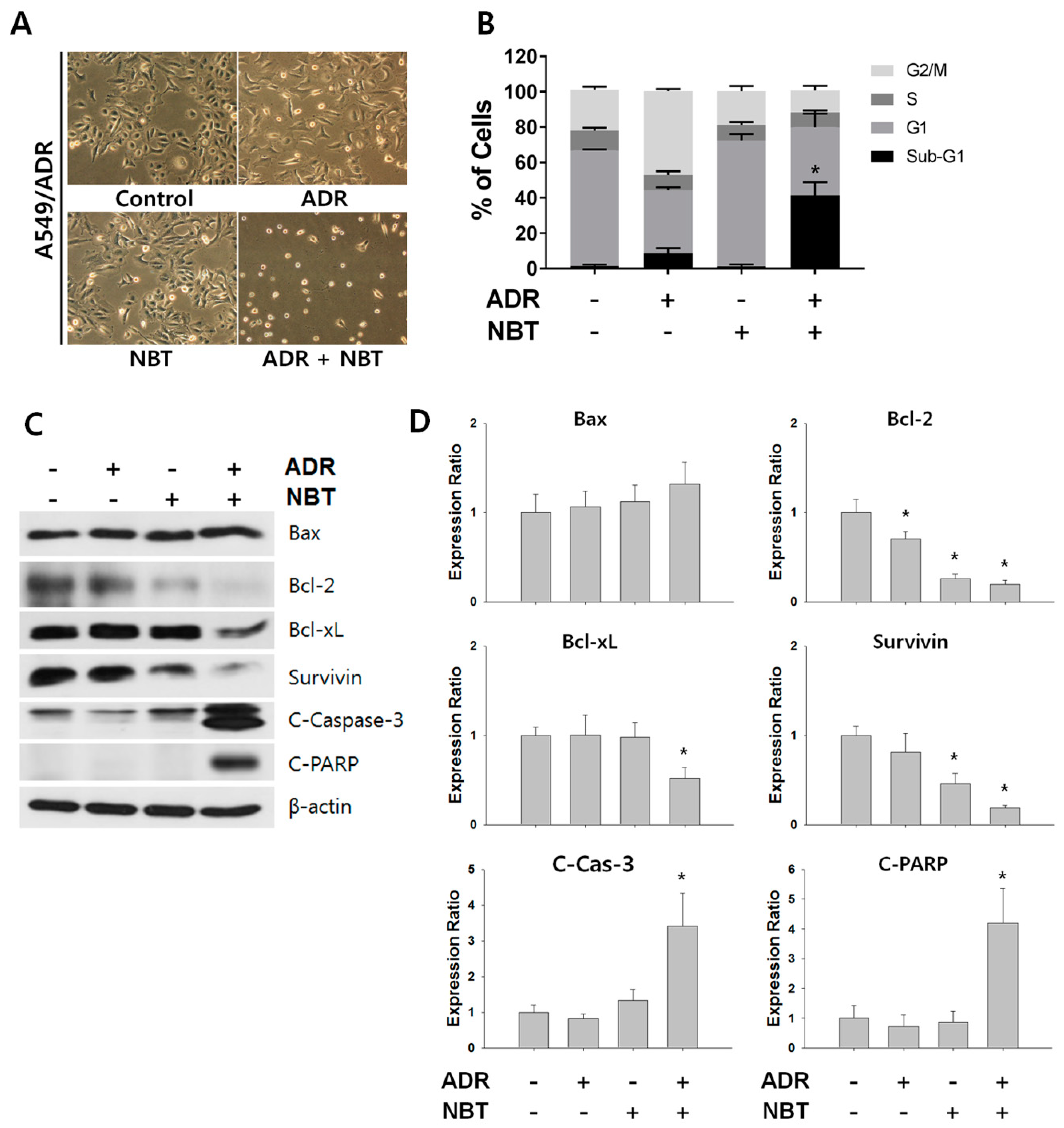
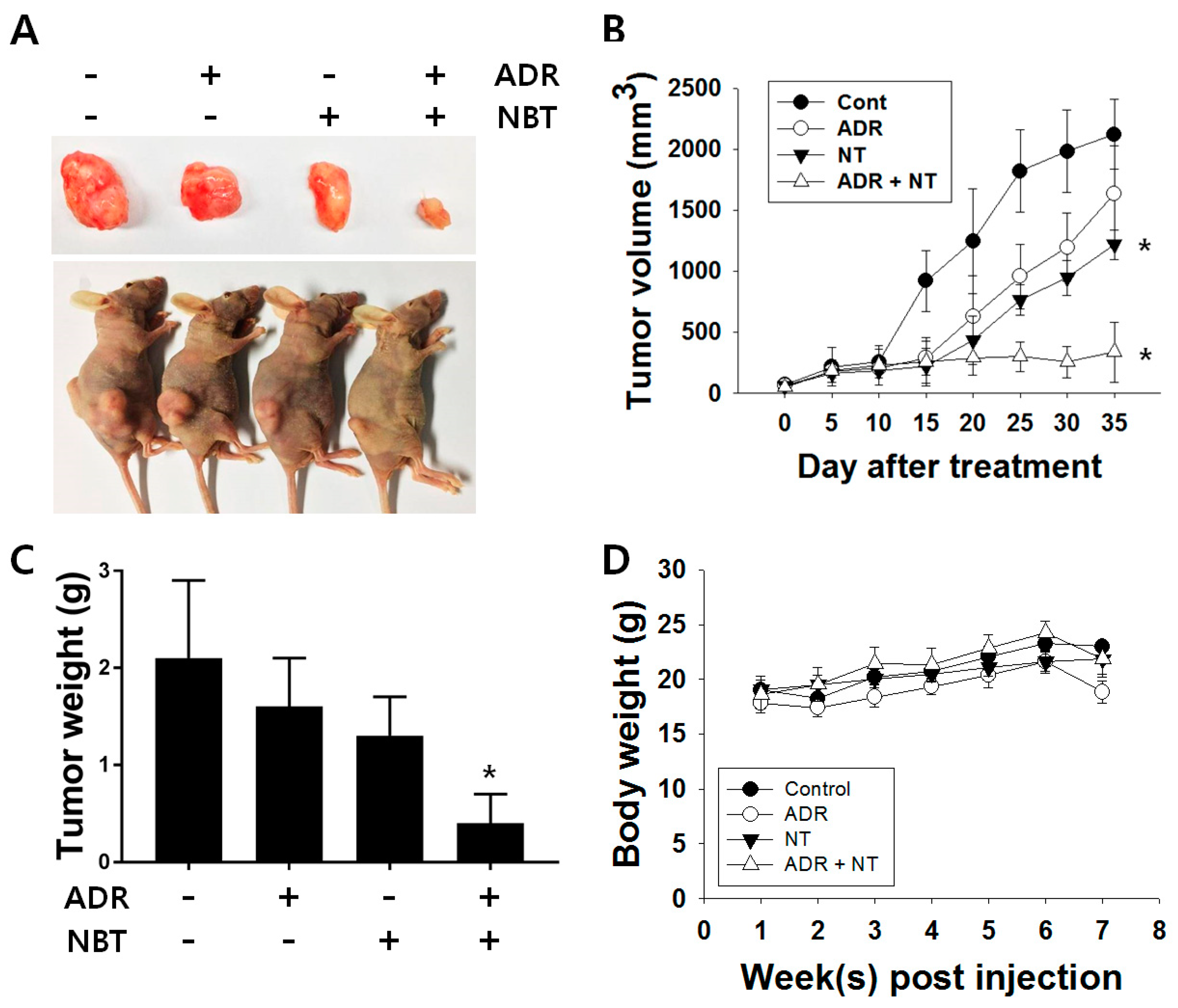
3.6. Combination Therapy with NBT and ADR Induced Apoptosis in A549/ADR Cells and Significantly Reduced Tumor Volume of A549/ADR Cells in Nude Mice
4. Discussion
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Soerjomataram, I.; Ervik, M.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.; Forman, D.; Bray, F. Cancer Incidence and Mortality Worldwide: Iarc Cancer Base no. 11.; International Agency for Research on Cancer: Paris, France, 2013. [Google Scholar]
- Cruz, C.S.D.; Tanoue, L.T.; Matthay, R.A. Lung cancer: Epidemiology, etiology, and prevention. Clin. Chest Med. 2011, 32, 605–644. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, S.; Belani, C. Systemic chemotherapy for advanced non-small cell lung cancer: Recent advances and future directions. Oncologist 2008, 13, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Le Chevalier, T. Adjuvant chemotherapy for resectable non-small-cell lung cancer: Where is it going? Ann. Oncol. 2010, 21, vii196–vii198. [Google Scholar] [CrossRef] [PubMed]
- Alfarouk, K.O.; Stock, C.-M.; Taylor, S.; Walsh, M.; Muddathir, A.K.; Verduzco, D.; Bashir, A.H.; Mohammed, O.Y.; Elhassan, G.O.; Harguindey, S. Resistance to cancer chemotherapy: Failure in drug response from adme to p-gp. Cancer Cell Int. 2015, 15, 71. [Google Scholar] [CrossRef] [PubMed]
- Abdolahpour, S.; Toliyat, T.; Omidfar, K.; Modjtahedi, H.; Wong, A.J.; Rasaee, M.J.; Kashanian, S.; Paknejad, M. Targeted delivery of doxorubicin into tumor cells by nanostructured lipid carriers conjugated to anti-egfrviii monoclonal antibody. Artif. Cells, Nanomed. Biotechnol. 2018, 46, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Vatsyayan, R.; Chaudhary, P.; Lelsani, P.C.R.; Singhal, P.; Awasthi, Y.C.; Awasthi, S.; Singhal, S.S. Role of rlip76 in doxorubicin resistance in lung cancer. Int. J. Oncol. 2009, 34, 1505–1511. [Google Scholar] [PubMed]
- Hu, J.; Zhang, X.; Wang, F.; Wang, X.; Yang, K.; Xu, M.; To, K.K.; Li, Q.; Fu, L. Effect of ceritinib (ldk378) on enhancement of chemotherapeutic agents in abcb1 and abcg2 overexpressing cells in vitro and in vivo. Oncotarget 2015, 6, 44643. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.; Bhardwaj, G.; Gerlach, J.; Mackie, J.; Grant, C.; Almquist, K.; Stewart, A.; Kurz, E.; Duncan, A.; Deeley, R. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science 1992, 258, 1650–1654. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.P. Targeting multidrug resistance protein 1 (mrp1, abcc1): Past, present, and future. Annu. Rev. Pharmacol. Toxicol. 2014, 54, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Manohar, C.F.; Bray, J.A.; Salwen, H.R.; Madafiglio, J.; Cheng, A.; Flemming, C.; Marshall, G.M.; Norris, M.D.; Haber, M.; Cohn, S.L. Mycn-mediated regulation of the mrp1 promoter in human neuroblastoma. Oncogene 2004, 23, 753. [Google Scholar] [CrossRef] [PubMed]
- Scotto, K.W. Transcriptional regulation of abc drug transporters. Oncogene 2003, 22, 7496. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Meng, Q.; Xie, Q.; Zhang, M. Effect and mechanism of resveratrol on drug resistance in human bladder cancer cells. Mol. Med. Rep. 2017, 15, 1179–1187. [Google Scholar] [CrossRef] [PubMed]
- Nogata, Y.; Sakamoto, K.; Shiratsuchi, H.; Ishii, T.; YANO, M.; Ohta, H. Flavonoid composition of fruit tissues of citrus species. Biosci. Biotechnol. Biochem. 2006, 70, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.Y.; Cho, M.; Ahn, K.S.; Cho, S.K. Nobiletin induces apoptosis and potentiates the effects of the anticancer drug 5-fluorouracil in p53-mutated snu-16 human gastric cancer cells. Nutr. Cancer 2013, 65, 286–295. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.H.; Kim, S.-M.; Nam, D.; Lee, J.-H.; Ahn, K.S.; Choi, S.-H.; Kim, S.-H.; Shim, B.S.; Chang, I.-M.; Ahn, K.S. Antimetastatic effect of nobiletin through the down-regulation of cxc chemokine receptor type 4 and matrix metallopeptidase-9. Pharm. Biol. 2012, 50, 1210–1218. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.-J.; Liu, J.-W.; Zhang, Q.-G.; Zhang, J.-J.; Xu, H.-T.; Liu, H.-J. Nobiletin inhibited hypoxia-induced epithelial-mesenchymal transition of lung cancer cells by inactivating of notch-1 signaling and switching on mir-200b. Die Pharm.-Int. J. Pharm. Sci. 2015, 70, 256–262. [Google Scholar]
- Ma, W.; Feng, S.; Yao, X.; Yuan, Z.; Liu, L.; Xie, Y. Nobiletin enhances the efficacy of chemotherapeutic agents in abcb1 overexpression cancer cells. Sci. Rep. 2015, 5, 18789. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Luo, X.; Qian, J.; Pang, X.; Song, J.; Qian, G.; Chen, J.; Chen, S. Fastuniq: A fast de novo duplicates removal tool for paired short reads. PLoS ONE 2012, 7, e52249. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. Star: Ultrafast universal rna-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from rna-seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. Edger: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. Rsem: Accurate transcript quantification from rna-seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat. Protoc. 2008, 4, 44. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2008, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hyun, H.B.; Moon, J.Y.; Cho, S.K. Quercetin suppresses cyr61-mediated multidrug resistance in human gastric adenocarcinoma ags cells. Molecules 2018, 23, 209. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T. Kegg for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Kovalev, A.; Tsvetaeva, D.; Grudinskaja, T. Role of abc-cassette transporters (mdr1, mrp1, bcrp) in the development of primary and acquired multiple drug resistance in patients with early and metastatic breast cancer. Exp. Oncol. 2013, 287–290. [Google Scholar]
- Luo, G.; Guan, X.; Zhou, L. Apoptotic effect of citrus fruit extract nobiletin on lung cancer cell line a549 in vitro and in vivo. Cancer Biol. Ther. 2008, 7, 966–973. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Lin, H.; Wang, Y.; Lv, W.; Zhang, S.; Qian, Y.; Deng, X.; Feng, N.; Yu, H.; Qian, B. D-limonene exhibits antitumor activity by inducing autophagy and apoptosis in lung cancer. OncoTargets Ther. 2018, 11, 1833. [Google Scholar] [CrossRef] [PubMed]
- Raphael, T.; Kuttan, G. Effect of naturally occurring monoterpenes carvone, limonene and perillic acid in the inhibition of experimental lung metastasis induced by b16f-10 melanoma cells. J. Exp. Clin. Cancer Res. CR 2003, 22, 419–424. [Google Scholar] [PubMed]
- Youn, H.; Jeong, J.-C.; Jeong, Y.S.; Kim, E.-J.; Um, S.-J. Quercetin potentiates apoptosis by inhibiting nuclear factor-kappab signaling in h460 lung cancer cells. Biol. Pharm. Bull. 2013, 36, 944–951. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Lee, E.J.; Min, K.H.; Hur, G.Y.; Lee, S.H.; Lee, S.Y.; Kim, J.H.; Shin, C.; Shim, J.J.; In, K.H. Quercetin enhances chemosensitivity to gemcitabine in lung cancer cells by inhibiting heat shock protein 70 expression. Clin. Lung Cancer 2015, 16, e235–e243. [Google Scholar] [CrossRef] [PubMed]
- Xia, R.; Sheng, X.; Xu, X.; Yu, C.; Lu, H. Hesperidin induces apoptosis and g0/g1 arrest in human non-small cell lung cancer a549 cells. Int. J. Mol. Med. 2018, 41, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Kohno, H.; Murakami, M.; Kagami, S.; El-Bayoumy, K. Suppressing effects of dietary supplementation of the organoselenium 1, 4-phenylenebis (methylene) selenocyanate and the citrus antioxidant auraptene on lung metastasis of melanoma cells in mice. Cancer Res. 2000, 60, 3713–3716. [Google Scholar] [PubMed]
- Luo, J. Glycogen synthase kinase 3β (gsk3β) in tumorigenesis and cancer chemotherapy. Cancer Lett. 2009, 273, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Hummel, I.; Klappe, K.; Ercan, C.; Kok, J.W. Mrp1 function and localization depend on cortical actin. Mol. Pharmacol. 2010. mol. 110.069013. [Google Scholar]
- Pec, M.K.; Aguirre, A.; Fernandez, J.J.; Souto, M.L.; Dorta, J.F.; Villar, J. Dehydrothyrsiferol does not modulate multidrug resistance-associated protein 1 resistance: A functional screening system for mrp1 substrates. Int. J. Mol. Med. 2002, 10, 605–608. [Google Scholar] [PubMed]
- Bristow, M.R.; Mason, J.W.; Billingham, M.E.; Daniels, J.R. Doxorubicin cardiomyopathy: Evaluation by phonocardiography, endomyocardial biopsy, and cardiac catheterization. Ann. Intern. Med. 1978, 88, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Zhang, C.; Wu, T.; Wang, Q.; Liu, J.; Dai, P. Transcriptome sequencing reveals key pathways and genes associated with cisplatin resistance in lung adenocarcinoma a549 cells. PLoS ONE 2017, 12, e0170609. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.G.; Ai, Y.W.; Yu, L.L.; Zhou, X.D.; Liu, J.; Li, J.H.; Xu, X.M.; Liu, S.; Chen, J.; Liu, F. Phosphoinositide 3-kinase/akt pathway plays an important role in chemoresistance of gastric cancer cells against etoposide and doxorubicin induced cell death. Int. J. Cancer 2008, 122, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.C.; Fahrmann, J.F.; Celiktas, M.; Aguilar, M.; Marini, K.D.; Jolly, M.K.; Katayama, H.; Wang, H.; Murage, E.N.; Dennison, J.B. Mcam mediates chemoresistance in small-cell lung cancer via the pi3k/akt/sox2 signaling pathway. Cancer Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Dan, H.C.; Park, S.; Yang, L.; Liu, Q.; Kaneko, S.; Ning, J.; He, L.; Yang, H.; Sun, M. Akt/pkb signaling mechanisms in cancer and chemoresistance. Front. Biosci. 2005, 10, 975. [Google Scholar] [CrossRef] [PubMed]
- Chesler, L.; Schlieve, C.; Goldenberg, D.D.; Kenney, A.; Kim, G.; McMillan, A.; Matthay, K.K.; Rowitch, D.; Weiss, W.A. Inhibition of phosphatidylinositol 3-kinase destabilizes mycn protein and blocks malignant progression in neuroblastoma. Cancer Res. 2006, 66, 8139–8146. [Google Scholar] [CrossRef] [PubMed]
- Bonvini, P.; Nguyen, P.; Trepel, J.; Neckers, L.M. In vivo degradation of n-myc in neuroblastoma cells is mediated by the 26s proteasome. Oncogene 1998, 16, 1131. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Wang, S.; Liu, Y.; Gu, J.; Gu, S.; Xu, Z.; Zhang, R.; Wang, Z.; Ma, H.; Chen, Y. Overexpression of mycn promotes proliferation of non-small cell lung cancer. Tumor Biol. 2016, 37, 12855–12866. [Google Scholar] [CrossRef] [PubMed]
- Knoepfler, P.S.; Kenney, A.M. Neural precursor cycling at sonic speed: N-myc pedals, gsk-3 brakes. Cell Cycle 2006, 5, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Sjostrom, S.K.; Finn, G.; Hahn, W.C.; Rowitch, D.H.; Kenney, A.M. The cdk1 complex plays a prime role in regulating n-myc phosphorylation and turnover in neural precursors. Dev. Cell 2005, 9, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Kenney, A.M.; Widlund, H.R.; Rowitch, D.H. Hedgehog and pi-3 kinase signaling converge on nmyc1 to promote cell cycle progression in cerebellar neuronal precursors. Development 2004, 131, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Vervoorts, J.; Lüscher-Firzlaff, J.; Lüscher, B. The ins and outs of myc regulation by posttranslational mechanisms. J. Biol. Chem. 2006, 281, 34725–34729. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, W.; Weiss, W. Myc proteins as therapeutic targets. Oncogene 2010, 29, 1249. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Pan, W. Gsk3: A multifaceted kinase in wnt signaling. Trends Biochem. Sci. 2010, 35, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Shu, W.; Guttentag, S.; Wang, Z.; Andl, T.; Ballard, P.; Lu, M.M.; Piccolo, S.; Birchmeier, W.; Whitsett, J.A.; Millar, S.E. Wnt/β-catenin signaling acts upstream of n-myc, bmp4, and fgf signaling to regulate proximal–distal patterning in the lung. Dev. Biol. 2005, 283, 226–239. [Google Scholar] [CrossRef] [PubMed]
- Szemes, M.; Greenhough, A.; Melegh, Z.; Malik, S.; Yuksel, A.; Catchpoole, D.; Gallacher, K.; Kollareddy, M.; Park, J.H.; Malik, K. Wnt signalling drives context-dependent differentiation or proliferation in neuroblastoma. Neoplasia 2018, 20, 335–350. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moon, J.Y.; Manh Hung, L.V.; Unno, T.; Cho, S.K. Nobiletin Enhances Chemosensitivity to Adriamycin through Modulation of the Akt/GSK3β/β–Catenin/MYCN/MRP1 Signaling Pathway in A549 Human Non-Small-Cell Lung Cancer Cells. Nutrients 2018, 10, 1829. https://doi.org/10.3390/nu10121829
Moon JY, Manh Hung LV, Unno T, Cho SK. Nobiletin Enhances Chemosensitivity to Adriamycin through Modulation of the Akt/GSK3β/β–Catenin/MYCN/MRP1 Signaling Pathway in A549 Human Non-Small-Cell Lung Cancer Cells. Nutrients. 2018; 10(12):1829. https://doi.org/10.3390/nu10121829
Chicago/Turabian StyleMoon, Jeong Yong, Le Van Manh Hung, Tatsuya Unno, and Somi Kim Cho. 2018. "Nobiletin Enhances Chemosensitivity to Adriamycin through Modulation of the Akt/GSK3β/β–Catenin/MYCN/MRP1 Signaling Pathway in A549 Human Non-Small-Cell Lung Cancer Cells" Nutrients 10, no. 12: 1829. https://doi.org/10.3390/nu10121829
APA StyleMoon, J. Y., Manh Hung, L. V., Unno, T., & Cho, S. K. (2018). Nobiletin Enhances Chemosensitivity to Adriamycin through Modulation of the Akt/GSK3β/β–Catenin/MYCN/MRP1 Signaling Pathway in A549 Human Non-Small-Cell Lung Cancer Cells. Nutrients, 10(12), 1829. https://doi.org/10.3390/nu10121829





