Predicting Grassland Leaf Area Index in the Meadow Steppes of Northern China: A Comparative Study of Regression Approaches and Hybrid Geostatistical Methods
Abstract
:1. Introduction
2. Study Area and Data
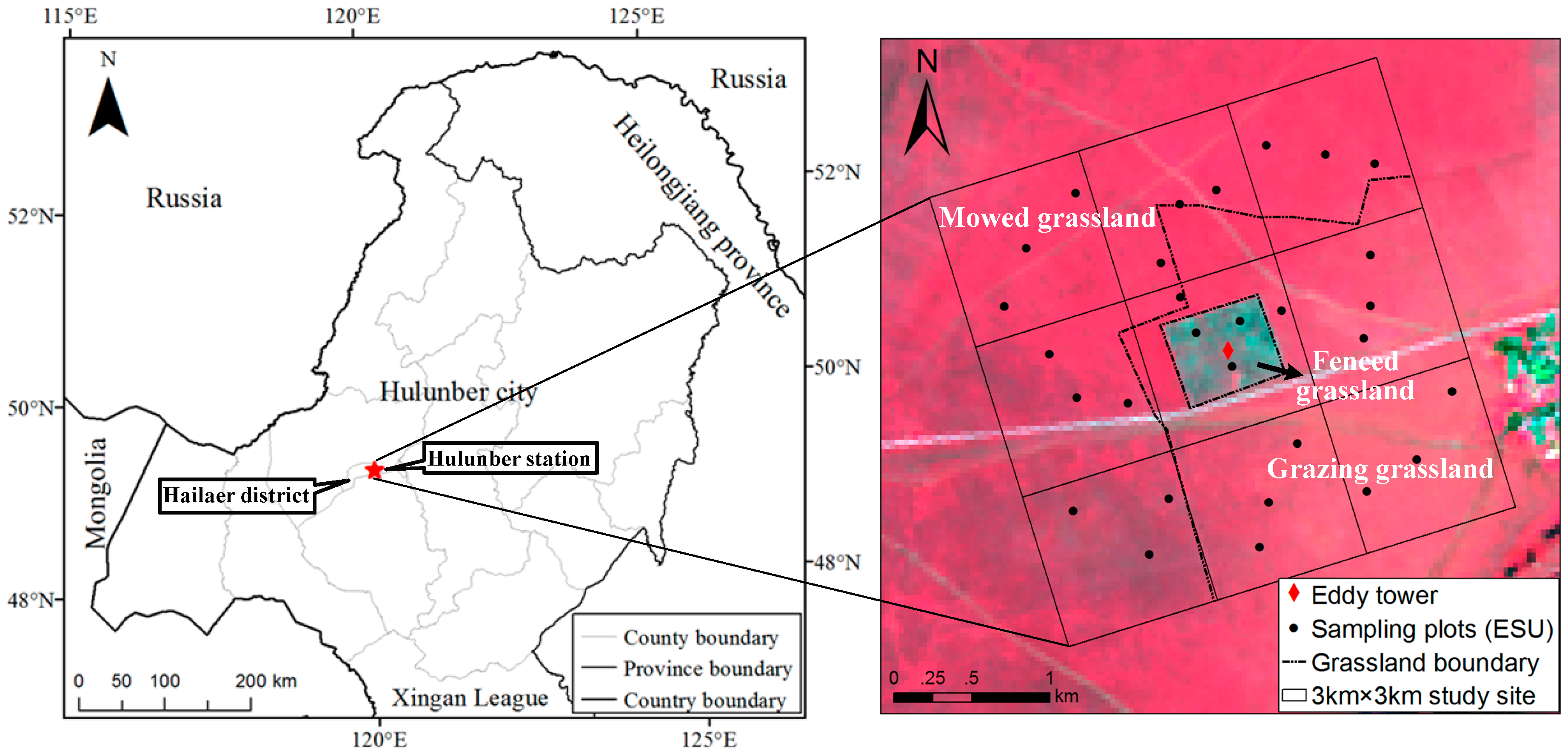
2.1. Study Site
2.2. Sampling Design and Field Measurements
2.3. Satellite Data
3. Methods
3.1. Partial Least Squares Regression (PLSR)
3.2. Artificial Neural Networks (ANNs)
3.3. Random Forests (RFs)
3.4. Regression Kriging (RK)
3.5. Random Forests Residuals Kriging (RFRK)
3.6. Model Implementation and Validation
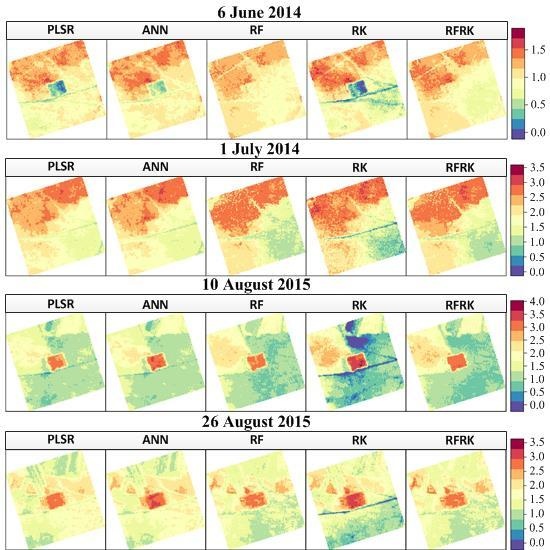
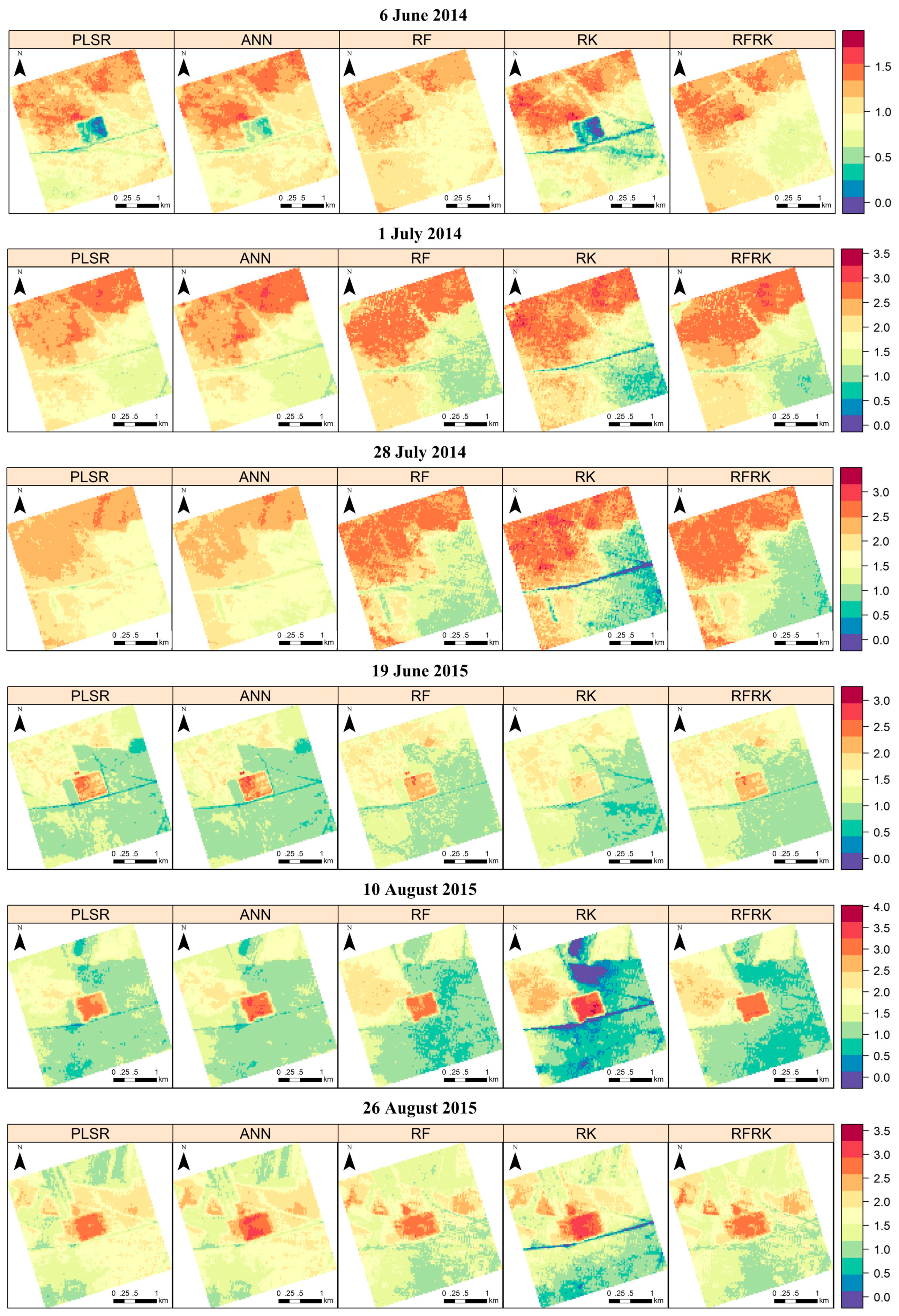
4. Results
4.1. Field LAI Measurements
4.2. LAI Spatial Prediction Based on the Four Reflectance Bands
4.3. Model Evaluation
5. Discussion
5.1. LAI Predictions Based on the Four Reflectance Bands and VIs
5.2. Model Performance in Different Grassland Types and at Different Growing Stages
5.3. Model Comparison and Study Limitations
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chen, J.M.; Black, T.A. Defining leaf area index for non-flat leaves. Plant Cell Environ. 1992, 15, 421–429. [Google Scholar] [CrossRef]
- Running, S.W.; Nemani, R.R.; Peterson, D.L.; Band, L.E.; Potts, D.F.; Pierce, L.L.; Spanner, M.A. Mapping regional forest evapotranspiration and photosynthesis by coupling satellite data with ecosystem simulation. Ecology 1989, 70, 1090–1101. [Google Scholar] [CrossRef]
- Sellers, P.J.; Dickinson, R.E.; Randall, D.A.; Betts, A.K.; Hall, F.G.; Berry, J.A.; Collatz, G.J.; Denning, A.S.; Mooney, H.A.; Nobre, C.A.; et al. Modeling the exchanges of energy, water, and carbon between continents and the atmosphere. Science 1997, 275, 502–509. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Woodcock, C.E.; Wang, Y.; Privette, J.L.; Shabanov, N.V.; Zhou, L.; Zhang, Y.; Buermann, W.; Dong, J.; Veikkanen, B.; et al. Multiscale analysis and validation of the MODIS LAI product: I. Uncertainty assessment. Remote Sens. Environ. 2002, 83, 414–430. [Google Scholar] [CrossRef]
- Wang, Y.; Woodcock, C.E.; Buermann, W.; Stenberg, P.; Voipio, P.; Smolander, H.; Häme, T.; Tian, Y.; Hu, J.; Knyazikhin, Y.; et al. Evaluation of the MODIS LAI algorithm at a coniferous forest site in Finland. Remote Sens. Environ. 2004, 91, 114–127. [Google Scholar] [CrossRef]
- Behera, S.K.; Behera, M.D.; Tuli, R. An indirect method of estimating leaf area index in a tropical deciduous forest of India. Ecol. Indic. 2015, 58, 356–364. [Google Scholar] [CrossRef]
- Olsoy, P.J.; Mitchell, J.J.; Levia, D.F.; Clark, P.E.; Glenn, N.F. Estimation of big sagebrush leaf area index with terrestrial laser scanning. Ecol. Indic. 2016, 61, 815–821. [Google Scholar] [CrossRef]
- Broge, N.H.; Leblanc, E. Comparing prediction power and stability of broadband and hyperspectral vegetation indices for estimation of green leaf area index and canopy chlorophyll density. Remote Sens. Environ. 2000, 76, 156–172. [Google Scholar] [CrossRef]
- Heiskanen, J. Estimating aboveground tree biomass and leaf area index in a mountain birch forest using ASTER satellite data. Int. J. Remote Sens. 2006, 27, 1135–1158. [Google Scholar] [CrossRef]
- Cho, M.A.; Skidmore, A.; Corsi, F.; van Wieren, S.E.; Sobhan, I. Estimation of green grass/herb biomass from airborne hyperspectral imagery using spectral indices and partial least squares regression. Int. J. Appl. Earth Obs. 2007, 9, 414–424. [Google Scholar] [CrossRef]
- Weiss, M.; Baret, F. Evaluation of canopy biophysical variable retrieval performances from the accumulation of large swath satellite data. Remote Sens. Environ. 1999, 70, 293–306. [Google Scholar] [CrossRef]
- Verrelst, J.; Schaepman, M.E.; Koetz, B.; Kneubühler, M. Angular sensitivity analysis of vegetation indices derived from CHRIS/PROBA data. Remote Sens. Environ. 2008, 112, 2341–2353. [Google Scholar] [CrossRef]
- Verrelst, J.; Schaepman, M.E.; Malenovský, Z.; Clevers, J.G.P.W. Effects of woody elements on simulated canopy reflectance: Implications for forest chlorophyll content retrieval. Remote Sens. Environ. 2010, 114, 647–656. [Google Scholar] [CrossRef]
- Kimes, D.S.; Nelson, R.F.; Manry, M.T.; Fung, A.K. Review article: Attributes of neural networks for extracting continuous vegetation variables from optical and radar measurements. Int. J. Remote Sens. 1998, 19, 2639–2663. [Google Scholar] [CrossRef]
- Durbha, S.S.; King, R.L.; Younan, N.H. Support vector machines regression for retrieval of leaf area index from multiangle imaging spectroradiometer. Remote Sens. Environ. 2007, 107, 348–361. [Google Scholar] [CrossRef]
- Vuolo, F.; Neugebauer, N.; Bolognesi, S.; Atzberger, C.; Urso, G. Estimation of leaf area index using DEIMOS-1 data: Application and transferability of a semi-empirical relationship between two agricultural areas. Remote Sens. 2013, 5, 1274–1291. [Google Scholar] [CrossRef]
- Liang, L.; Di, L.; Zhang, L.; Deng, M.; Qin, Z.; Zhao, S.; Lin, H. Estimation of crop LAI using hyperspectral vegetation indices and a hybrid inversion method. Remote Sens. Environ. 2015, 165, 123–134. [Google Scholar] [CrossRef]
- Verrelst, J.; Camps-Valls, G.; Muñoz-Marí, J.; Rivera, J.P.; Veroustraete, F.; Clevers, J.G.P.W.; Moreno, J. Optical remote sensing and the retrieval of terrestrial vegetation bio-geophysical properties—A review. ISPRS J. Photogramm. 2015, 108, 273–290. [Google Scholar] [CrossRef]
- Verrelst, J.; Rivera, J.P.; Veroustraete, F.; Muñoz-Marí, J.; Clevers, J.G.P.W.; Camps-Valls, G.; Moreno, J. Experimental Sentinel-2 LAI estimation using parametric, non-parametric and physical retrieval methods—A comparison. ISPRS J. Photogramm. 2015, 108, 260–272. [Google Scholar] [CrossRef]
- Williams, M.; Bell, R.; Spadavecchia, L.; Street, L.E.; Van Wijk, M.T. Upscaling leaf area index in an arctic landscape through multiscale observations. Glob. Chang. Biol. 2008, 14, 1517–1530. [Google Scholar] [CrossRef]
- Berterretche, M.; Hudak, A.T.; Cohen, W.B.; Maiersperger, T.K.; Gower, S.T.; Dungan, J. Comparison of regression and geostatistical methods for mapping leaf area index (LAI) with Landsat ETM+ data over a boreal forest. Remote Sens. Environ. 2005, 96, 49–61. [Google Scholar] [CrossRef]
- Martinez, B.; Cassiraga, E.; Camacho, F.; Garcia-Haro, J. Geostatistics for mapping leaf area index over a cropland landscape: Efficiency sampling assessment. Remote Sens. 2010, 2, 2584–2606. [Google Scholar] [CrossRef]
- Castillo-Santiago, M.Á.; Ghilardi, A.; Oyama, K.; Hernández-Stefanoni, J.L.; Torres, I.; Flamenco-Sandoval, A.; Fernández, A.; Mas, J.-F. Estimating the spatial distribution of woody biomass suitable for charcoal making from remote sensing and geostatistics in central Mexico. Energy Sustain. Dev. 2013, 17, 177–188. [Google Scholar] [CrossRef]
- Galeana-Pizaña, J.M.; López-Caloca, A.; López-Quiroz, P.; Silván-Cárdenas, J.L.; Couturier, S. Modeling the spatial distribution of above-ground carbon in mexican coniferous forests using remote sensing and a geostatistical approach. Int. J. Appl. Earth Obs. 2014, 30, 179–189. [Google Scholar] [CrossRef]
- Heuvelink, G.B.M.; Griffith, D.A. Space-Time geostatistics for geography: A case study of radiation monitoring across parts of germany. Geogr. Anal. 2010, 42, 161–179. [Google Scholar] [CrossRef]
- Ge, Y.; Liang, Y.; Wang, J.; Zhao, Q.; Liu, S. Upscaling sensible heat fluxes with area-to-area regression kriging. IEEE Geosci. Remote Sens. Lett. 2015, 12, 656–660. [Google Scholar]
- Hernández-Stefanoni, J.L.; Alberto Gallardo-Cruz, J.; Meave, J.A.; Dupuy, J.M. Combining geostatistical models and remotely sensed data to improve tropical tree richness mapping. Ecol. Indic. 2011, 11, 1046–1056. [Google Scholar] [CrossRef]
- Viana, H.; Aranha, J.; Lopes, D.; Cohen, W.B. Estimation of crown biomass of pinus pinaster stands and shrubland above-ground biomass using forest inventory data, remotely sensed imagery and spatial prediction models. Ecol. Model. 2012, 226, 22–35. [Google Scholar] [CrossRef]
- Guo, P.; Li, M.; Luo, W.; Tang, Q.; Liu, Z.; Lin, Z. Digital mapping of soil organic matter for rubber plantation at regional scale: An application of random forest plus residuals kriging approach. Geoderma 2015, 237–238, 49–59. [Google Scholar] [CrossRef]
- Were, K.; Bui, D.T.; Dick, Ø.B.; Singh, B.R. A comparative assessment of support vector regression, artificial neural networks, and random forests for predicting and mapping soil organic carbon stocks across an afromontane landscape. Ecol. Indic. 2015, 52, 394–403. [Google Scholar] [CrossRef]
- Yang, F.; Sun, J.; Fang, H.; Yao, Z.; Zhang, J.; Zhu, Y.; Song, K.; Wang, Z.; Hu, M. Comparison of different methods for corn LAI estimation over northeastern China. Int. J. Appl. Earth Obs. 2012, 18, 462–471. [Google Scholar]
- Hengl, T.; Heuvelink, G.B.M.; Rossiter, D.G. About regression-kriging: From equations to case studies. Comput. Geosci. 2007, 33, 1301–1315. [Google Scholar] [CrossRef]
- Tang, H.; Li, Z.; Zhu, Z.; Chen, B.; Zhang, B.; Xin, X. Variability and climate change trend in vegetation phenology of recent decades in the greater Khingan mountain area, northeastern China. Remote Sens. 2015, 7, 11914–11932. [Google Scholar] [CrossRef]
- Wu, Q.; Jin, Y.; Bao, Y.; Hai, Q.; Yan, R.; Chen, B.; Zhang, H.; Zhang, B.; Li, Z.; Li, X.; et al. Comparison of two inversion methods for leaf area index using HJ-1 satellite data in a temperate meadow steppe. Int. J. Remote Sens. 2015, 36, 1–16. [Google Scholar] [CrossRef]
- De Kauwe, M.G.; Disney, M.I.; Quaife, T.; Lewis, P.; Williams, M. An assessment of the MODIS collection 5 leaf area index product for a region of mixed coniferous forest. Remote Sens. Environ. 2011, 115, 767–780. [Google Scholar] [CrossRef]
- VAlidation of Land European Remote sensing Instruments. Available online: http://w3.avignon.inra.fr/valeri/ (accessed on 18 January 2016).
- Li, Z.; Tang, H.; Xin, X.; Zhang, B.; Wang, D. Assessment of the MODIS LAI product using ground measurement data and HJ-1A/1A imagery in the meadow steppe of Hulunber, China. Remote Sens. 2014, 6, 6242–6265. [Google Scholar] [CrossRef]
- Chen, J.M.; Pavlic, G.; Brown, L.; Cihlar, J.; Leblanc, S.G.; White, H.P.; Hall, R.J.; Peddle, D.R.; King, D.J.; Trofymow, J.A.; et al. Derivation and validation of Canada-wide coarse-resolution leaf area index maps using high-resolution satellite imagery and ground measurements. Remote Sens. Environ. 2002, 80, 165–184. [Google Scholar] [CrossRef]
- Welles, J.M.; Norman, J.M. Instrument for indirect measurement of canopy architecture. Agron. J. 1991, 83, 818–825. [Google Scholar] [CrossRef]
- Fang, H.; Li, W.; Wei, S.; Jiang, C. Seasonal variation of leaf area index (LAI) over paddy rice fields in NE China: Intercomparison of destructive sampling, LAI-2200, digital hemispherical photography (DHP), and ACCUPAR methods. Agric. For. Meteorol. 2014, 198–199, 126–141. [Google Scholar] [CrossRef]
- China Centre for Resources Satellite Data and Application. Available online: http://www.cresda.com (accessed on 18 January 2016).
- Bo, J.; Shunlin, L.; Townshend, J.R.; Dodson, Z.M. Assessment of the radiometric performance of Chinese HJ-1 satellite CCD instruments. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2013, 6, 840–850. [Google Scholar]
- Agrawal, G.; Sarup, J.; Bhopal, M. Comparision of QUAC and FLAASH atmospheric correction modules on EO-1 hyperion data of Sanchi. Int. J. Adv. Eng. Sci. Technol. 2011, 4, 178–186. [Google Scholar]
- Baret, F.; Guyot, G. Potentials and limits of vegetation indices for LAI and APAR assessment. Remote Sens. Environ. 1991, 35, 161–173. [Google Scholar] [CrossRef]
- Rouse, J.W., Jr.; Haas, R.; Schell, J.; Deering, D. Monitoring vegetation systems in the great plains with Erts. NASA Spec. Publ. 1974, 351, 309. [Google Scholar]
- Jordan, C.F. Derivation of leaf-area index from quality of light on the forest floor. Ecology 1969, 50, 663–666. [Google Scholar] [CrossRef]
- Kaufman, Y.J.; Tanre, D. Atmospherically resistant vegetation index (ARVI) for EOS-MODIS. IEEE Trans. Geosci. Remote Sens. 1992, 30, 261–270. [Google Scholar] [CrossRef]
- Gitelson, A.A. Wide dynamic range vegetation index for remote quantification of biophysical characteristics of vegetation. J. Plant Physiol. 2004, 161, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Darvishzadeh, R.; Atzberger, C.; Skidmore, A.; Schlerf, M. Mapping grassland leaf area index with airborne hyperspectral imagery: A comparison study of statistical approaches and inversion of radiative transfer models. ISPRS J. Photogramm. 2011, 66, 894–906. [Google Scholar] [CrossRef]
- Geladi, P.; Kowalski, B.R. Partial least-squares regression: A tutorial. Anal. Chim. Acta 1986, 185, 1–17. [Google Scholar] [CrossRef]
- Pandit, C.M.; Filippelli, G.M.; Li, L. Estimation of heavy-metal contamination in soil using reflectance spectroscopy and partial least-squares regression. Int. J. Remote Sens. 2010, 31, 4111–4123. [Google Scholar] [CrossRef]
- Mevik, B.-H.; Wehrens, R. The PLS package: Principal component and partial least squares regression in R. J. Stat. Softw. 2007, 18, 1–24. [Google Scholar] [CrossRef]
- Lek, S.; Guégan, J.F. Artificial neural networks as a tool in ecological modelling, an introduction. Ecol. Model. 1999, 120, 65–73. [Google Scholar] [CrossRef]
- Lek, S.; Giraudel, J.L.; Guégan, J.F. Neuronal networks: Algorithms and architectures for ecologists and evolutionary ecologists. In Artificial Neuronal Networks; Lek, S., Guégan, J.-F., Eds.; Springer Berlin Heidelberg: Berlin, Germany; Heidelberg, Germany, 2000; pp. 3–27. [Google Scholar]
- Team, R.C. R: A Language and Environment for Statistical Computing. 2013. Available online: http://cran.fiocruz.br/web/packages/dplR/vignettes/timeseries-dplR.pdf (accessed on 4 April 2016).
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by randomforest. R News 2002, 2, 18–22. [Google Scholar]
- Prasad, A.M.; Iverson, L.R.; Liaw, A. Newer classification and regression tree techniques: Bagging and random forests for ecological prediction. Ecosystems 2006, 9, 181–199. [Google Scholar] [CrossRef]
- Heung, B.; Bulmer, C.E.; Schmidt, M.G. Predictive soil parent material mapping at a regional-scale: A random forest approach. Geoderma 2014, 214–215, 141–154. [Google Scholar] [CrossRef]
- Svetnik, V.; Liaw, A.; Tong, C.; Culberson, J.C.; Sheridan, R.P.; Feuston, B.P. Random forest: A classification and regression tool for compound classification and QSAR modeling. J. Chem. Inf. Comput. Sci. 2003, 43, 1947–1958. [Google Scholar] [CrossRef] [PubMed]
- Hengl, T.; Heuvelink, G.B.M.; Stein, A. A generic framework for spatial prediction of soil variables based on regression-kriging. Geoderma 2004, 120, 75–93. [Google Scholar] [CrossRef]
- Odeh, I.O.A.; McBratney, A.B.; Chittleborough, D.J. Further results on prediction of soil properties from terrain attributes: Heterotopic cokriging and regression-kriging. Geoderma 1995, 67, 215–226. [Google Scholar] [CrossRef]
- Pebesma, E.J. Multivariable geostatistics in S: The gstat package. Comput. Geosci. 2004, 30, 683–691. [Google Scholar] [CrossRef]
- Hultquist, C.; Chen, G.; Zhao, K. A comparison of gaussian process regression, random forests and support vector regression for burn severity assessment in diseased forests. Remote Sens. Lett. 2014, 5, 723–732. [Google Scholar] [CrossRef]
- Genuer, R.; Poggi, J.-M.; Tuleau-Malot, C. Variable selection using random forests. Pattern Recogn. Lett. 2010, 31, 2225–2236. [Google Scholar] [CrossRef]
- Genuer, R.; Poggi, J.-M.; Tuleau, C. Random Forests: Some Methodological Insights. 2008. Available online: http://arxiv.org/pdf/0811.3619v1.pdf (accessed on 4 April 2016).
- Huang, C.; Townshend, J.R.G. A stepwise regression tree for nonlinear approximation: Applications to estimating subpixel land cover. Int. J. Remote Sens. 2003, 24, 75–90. [Google Scholar] [CrossRef]
- Deng, Y.; Liu, X.N.; Yan, R.R.; Wang, X.; Yang, G.X.; Ren, Z.C.; Xin, X.P. Soil respiration of Hulunber meadow steppe and response of its controlling factors to different grazing intensities. Acta Pratacult. Sin. 2013, 22, 22–29. (In Chinese) [Google Scholar]
- Yan, R.; Tang, H.; Xin, X.; Chen, B.; Murray, P.J.; Yan, Y.; Wang, X.; Yang, G. Grazing intensity and driving factors affect soil nitrous oxide fluxes during the growing seasons in the Hulunber meadow steppe of China. Environ. Res. Lett. 2016, 11, 054004. [Google Scholar] [CrossRef]
- Yan, R.; Xin, X.; Zhang, B.; Yan, Y.; Yang, G. Influence of cattle grazing gradient on plant community characteristics in Hulunber meadow steppe. Chin. J. Grassl. 2010, 32, 62–67. (In Chinese) [Google Scholar]
- Propastin, P. Modifying geographically weighted regression for estimating aboveground biomass in tropical rainforests by multispectral remote sensing data. Int. J. Appl. Earth Obs. 2012, 18, 82–90. [Google Scholar] [CrossRef]
- Ramoelo, A.; Cho, M.A.; Mathieu, R.; Madonsela, S.; van de Kerchove, R.; Kaszta, Z.; Wolff, E. Monitoring grass nutrients and biomass as indicators of rangeland quality and quantity using random forest modelling and Worldview-2 data. Int. J. Appl. Earth Obs. 2015, 43, 43–54. [Google Scholar] [CrossRef]
- Papale, D.; Black, T.A.; Carvalhais, N.; Cescatti, A.; Chen, J.; Jung, M.; Kiely, G.; Lasslop, G.; Mahecha, M.D.; Margolis, H.; et al. Effect of spatial sampling from european flux towers for estimating carbon and water fluxes with artificial neural networks. J. Geophys. Res. Biogeosci. 2015, 120, 1941–1957. [Google Scholar] [CrossRef]
- Isik, F.; Ozden, G. Estimating compaction parameters of fine- and coarse-grained soils by means of artificial neural networks. Environ. Earth Sci. 2013, 69, 2287–2297. [Google Scholar] [CrossRef]
- Hajjem, A.; Bellavance, F.; Larocque, D. Mixed-effects random forest for clustered data. J. Stat. Comput. Sim. 2014, 84, 1313–1328. [Google Scholar] [CrossRef]
- Pope, G.; Treitz, P. Leaf area index (LAI) estimation in boreal mixedwood forest of Ontario, Canada using light detection and ranging (LIDAR) and WorldView-2 imagery. Remote Sens. 2013, 5, 5040–5063. [Google Scholar] [CrossRef]
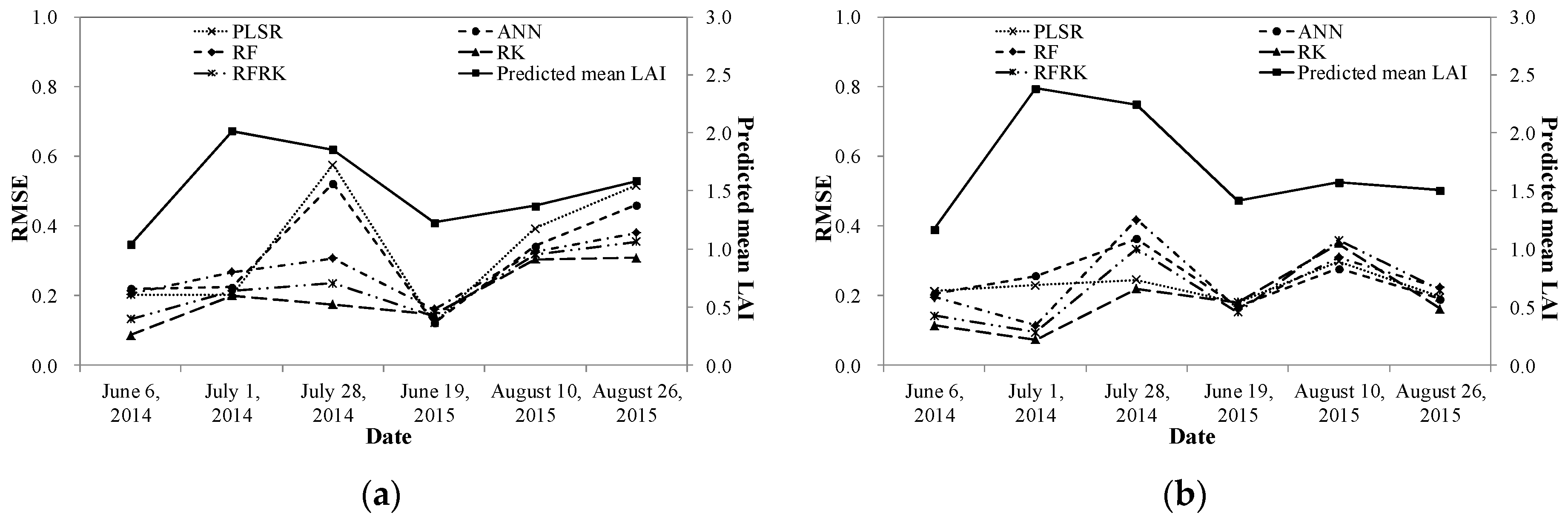
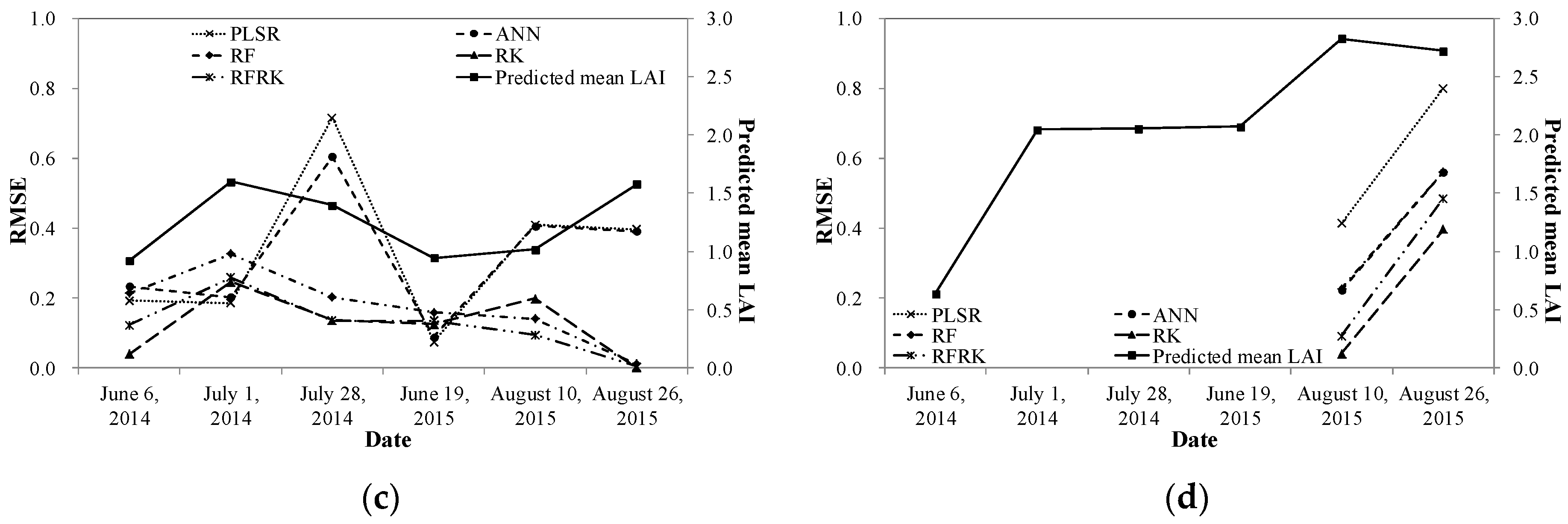
| Experiment Date | 6 June 2014 | 1 July 2014 | 28 July 2014 | 19 June 2015 | 10 August 2015 | 26 August 2015 |
|---|---|---|---|---|---|---|
| Sensor for HJ Images | HJ1B CCD1 | HJ1B CCD2 | HJ1A CCD1 | HJ1B CCD1 | HJ1B CCD1 | HJ1B CCD2 |
| HJ Image Acquisition Date | 6 June 2014 | 29 June 2014 | 29 July 2014 | 15 June 2015 | 10 August 2015 | 26 August 2015 |
| Date | 6 June 2014 | 1 July 2014 | 28 July 2014 | 19 June 2015 | 10 August 2015 | 26 August 2015 | |
|---|---|---|---|---|---|---|---|
| No. of training data | 16 | 18 | 17 | 18 | 19 | 16 | |
| No. of validation data | 4 | 8 | 5 | 6 | 6 | 5 | |
| Mowed grassland | Mean | 1.32 | 2.56 | 2.35 | 1.57 | 1.80 | 1.43 |
| Min | 1.00 | 2.01 | 1.45 | 1.19 | 0.88 | 1.05 | |
| Max | 1.50 | 3.22 | 2.75 | 2.03 | 2.56 | 2.12 | |
| Stdev | 0.19 | 0.30 | 0.42 | 0.24 | 0.50 | 0.30 | |
| Grazing grassland | Mean | 0.94 | 1.68 | 1.21 | 0.93 | 0.83 | 0.83 |
| Min | 0.61 | 0.81 | 0.69 | 0.77 | 0.69 | 0.77 | |
| Max | 1.45 | 2.73 | 2.64 | 1.17 | 1.02 | 0.89 | |
| Stdev | 0.30 | 0.59 | 0.64 | 0.13 | 0.10 | 0.05 | |
| Fenced grassland | Mean | - 2 | - | - | - | 3.40 | 3.50 |
| Min | - | - | - | - | 3.40 | 3.44 | |
| Max | - | - | - | - | 3.41 | 3.56 | |
| Stdev | - | - | - | - | 0.01 | 0.08 | |
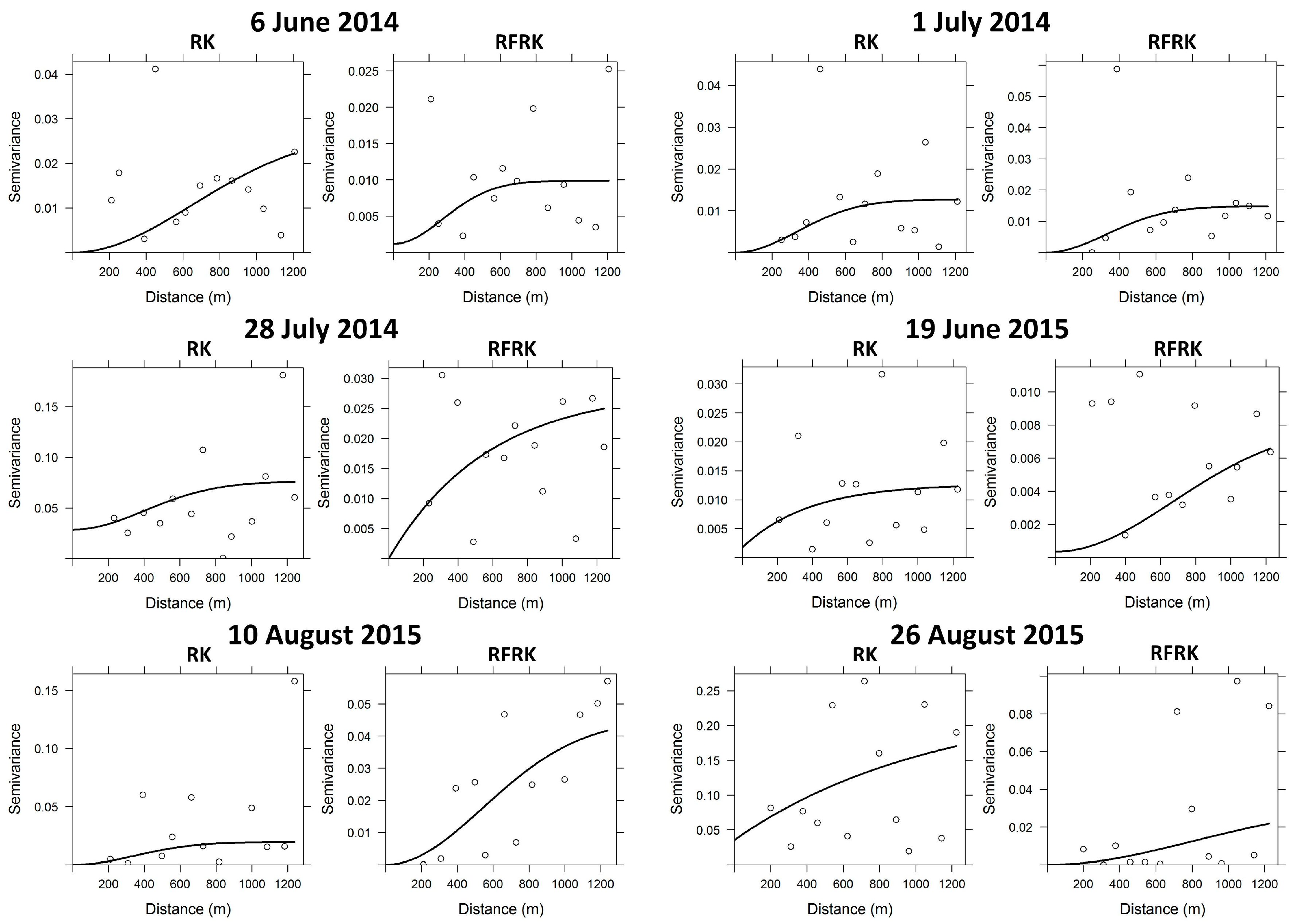
| Date | RK | RFRK | ||||||
|---|---|---|---|---|---|---|---|---|
| Model | Nugget | Sill | Range | Model | Nugget | Sill | Range | |
| 6 June 2014 | Gaussian | 0.00 | 0.03 | 918.86 | Gaussian | 0.00 | 0.01 | 461.98 |
| 1 July 2014 | Gaussian | 0.00 | 0.02 | 560.13 | Gaussian | 0.00 | 0.02 | 609.90 |
| 28 July 2014 | Gaussian | 0.03 | 0.02 | 480.70 | Exponential | 0.00 | 0.03 | 595.52 |
| 19 June 2015 | Exponential | 0.00 | 0.03 | 2230.32 | Gaussian | 0.00 | 0.01 | 839.06 |
| 10 August 2015 | Gaussian | 0.00 | 0.02 | 441.76 | Gaussian | 0.00 | 0.05 | 769.84 |
| 26 August 2015 | Exponential | 0.04 | 0.20 | 1095.47 | Gaussian | 0.00 | 0.03 | 1105.05 |
| 6 June 2014 | 1 July 2014 | ||||||||
| Min | Max | Mean | Stdev | Min | Max | Mean | Stdev | ||
| Measured LAI | 0.61 | 1.50 | 1.10 | 0.31 | Measured LAI | 0.81 | 3.22 | 2.15 | 0.63 |
| PLSR | 0.00 | 1.64 | 1.02 | 0.25 | PLSR | 1.00 | 3.01 | 2.04 | 0.45 |
| ANN | 0.30 | 1.66 | 1.08 | 0.22 | ANN | 0.95 | 3.20 | 2.02 | 0.49 |
| RF | 0.78 | 1.48 | 1.07 | 0.16 | RF | 0.99 | 2.98 | 1.98 | 0.59 |
| RK | 0.00 | 1.77 | 0.96 | 0.33 | RK | 0.09 | 3.36 | 2.03 | 0.65 |
| RFRK | 0.60 | 1.58 | 1.05 | 0.21 | RFRK | 0.79 | 3.21 | 2.00 | 0.60 |
| 28 July 2014 | 19 June 2015 | ||||||||
| Min | Max | Mean | Stdev | Min | Max | Mean | Stdev | ||
| Measured LAI | 0.69 | 2.75 | 1.80 | 0.82 | Measured LAI | 0.77 | 2.03 | 1.27 | 0.38 |
| PLSR | 0.97 | 2.64 | 1.96 | 0.30 | PLSR | 0.00 | 2.86 | 1.19 | 0.37 |
| ANN | 0.82 | 2.64 | 1.86 | 0.33 | ANN | 0.20 | 3.05 | 1.16 | 0.38 |
| RF | 0.87 | 2.63 | 1.83 | 0.61 | RF | 0.65 | 2.80 | 1.29 | 0.35 |
| RK | 0.00 | 3.26 | 1.80 | 0.78 | RK | 0.10 | 2.11 | 1.23 | 0.35 |
| RFRK | 0.68 | 2.74 | 1.83 | 0.68 | RFRK | 0.60 | 2.79 | 1.28 | 0.36 |
| 10 August 2015 | 26 August 2015 | ||||||||
| Min | Max | Mean | Stdev | Min | Max | Mean | Stdev | ||
| Measured LAI | 0.69 | 3.41 | 1.51 | 0.82 | Measured LAI | 0.77 | 3.56 | 1.49 | 0.75 |
| PLSR | 0.10 | 3.11 | 1.38 | 0.43 | PLSR | 0.00 | 3.04 | 1.64 | 0.38 |
| ANN | 0.40 | 3.35 | 1.43 | 0.46 | ANN | 0.00 | 3.24 | 1.68 | 0.40 |
| RF | 0.80 | 3.01 | 1.41 | 0.57 | RF | 0.00 | 3.00 | 1.57 | 0.43 |
| RK | 0.00 | 3.76 | 1.25 | 0.79 | RK | 0.00 | 3.40 | 1.47 | 0.57 |
| RFRK | 0.65 | 3.26 | 1.39 | 0.62 | RFRK | 0.00 | 2.98 | 1.58 | 0.44 |
| MAE | RMSE | R2 | |
|---|---|---|---|
| PLSR | 0.27 | 0.35 | 0.77 |
| ANN | 0.26 | 0.33 | 0.81 |
| RF | 0.21 | 0.27 | 0.89 |
| RK | 0.16 | 0.21 | 0.92 |
| RFRK | 0.17 | 0.23 | 0.91 |
| PLSR | ANN | RF | RK | RFRK | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MAE | RMSE | MAE | RMSE | MAE | RMSE | MAE | RMSE | MAE | RMSE | |
| Four bands | 0.274 | 0.353 | 0.264 | 0.328 | 0.209 | 0.269 | 0.164 | 0.212 | 0.173 | 0.231 |
| Four bands + SR | 0.216 | 0.279 | 0.221 | 0.277 | 0.197 | 0.252 | 0.180 | 0.261 | 0.150 | 0.217 |
| Four bands + one VI | 0.233 | 0.308 | 0.225 | 0.287 | 0.197 | 0.257 | 0.182 | 0.251 | 0.174 | 0.255 |
| Four bands + two VIs | 0.220 | 0.283 | 0.222 | 0.281 | 0.194 | 0.260 | 0.188 | 0.285 | 0.170 | 0.227 |
| Four bands + three VIs | 0.217 | 0.280 | 0.221 | 0.278 | 0.199 | 0.267 | 0.195 | 0.300 | 0.181 | 0.241 |
| Four bands + four VIs | 0.217 | 0.280 | 0.221 | 0.277 | 0.201 | 0.274 | 0.205 | 0.315 | 0.180 | 0.252 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Wang, J.; Tang, H.; Huang, C.; Yang, F.; Chen, B.; Wang, X.; Xin, X.; Ge, Y. Predicting Grassland Leaf Area Index in the Meadow Steppes of Northern China: A Comparative Study of Regression Approaches and Hybrid Geostatistical Methods. Remote Sens. 2016, 8, 632. https://doi.org/10.3390/rs8080632
Li Z, Wang J, Tang H, Huang C, Yang F, Chen B, Wang X, Xin X, Ge Y. Predicting Grassland Leaf Area Index in the Meadow Steppes of Northern China: A Comparative Study of Regression Approaches and Hybrid Geostatistical Methods. Remote Sensing. 2016; 8(8):632. https://doi.org/10.3390/rs8080632
Chicago/Turabian StyleLi, Zhenwang, Jianghao Wang, Huan Tang, Chengquan Huang, Fan Yang, Baorui Chen, Xu Wang, Xiaoping Xin, and Yong Ge. 2016. "Predicting Grassland Leaf Area Index in the Meadow Steppes of Northern China: A Comparative Study of Regression Approaches and Hybrid Geostatistical Methods" Remote Sensing 8, no. 8: 632. https://doi.org/10.3390/rs8080632
APA StyleLi, Z., Wang, J., Tang, H., Huang, C., Yang, F., Chen, B., Wang, X., Xin, X., & Ge, Y. (2016). Predicting Grassland Leaf Area Index in the Meadow Steppes of Northern China: A Comparative Study of Regression Approaches and Hybrid Geostatistical Methods. Remote Sensing, 8(8), 632. https://doi.org/10.3390/rs8080632