Application of High Resolution Satellite Imagery to Characterize Individual-Based Environmental Heterogeneity in a Wild Blue Tit Population
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
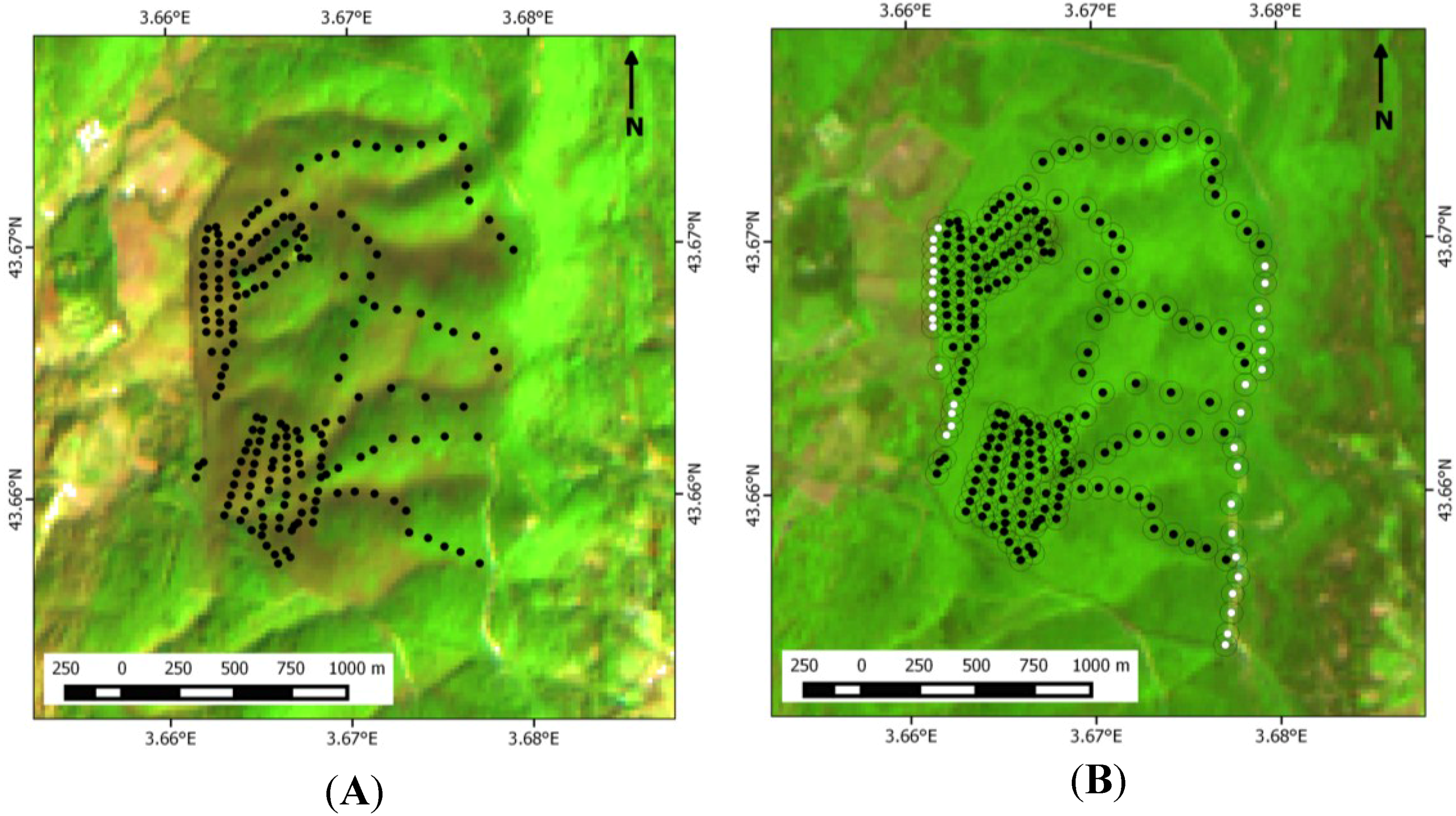
2.2. Ground Data Measurements
2.3. Image Data Acquisition and Processing
2.4. Statistical Analysis
3. Results
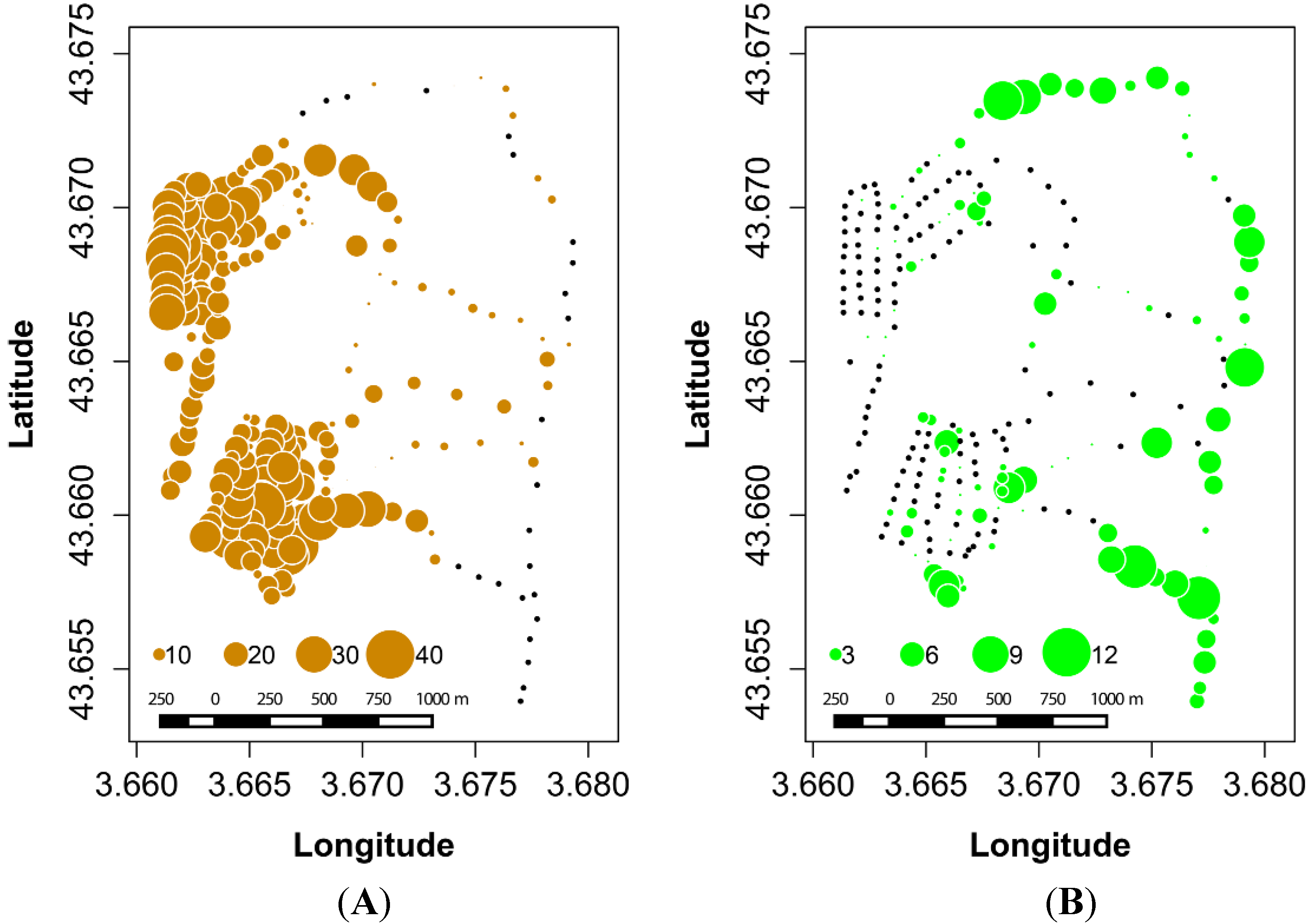
3.1. Ground Data on Oak Distribution across the Study Site
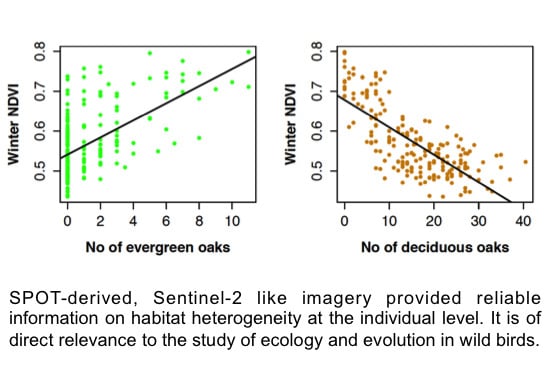
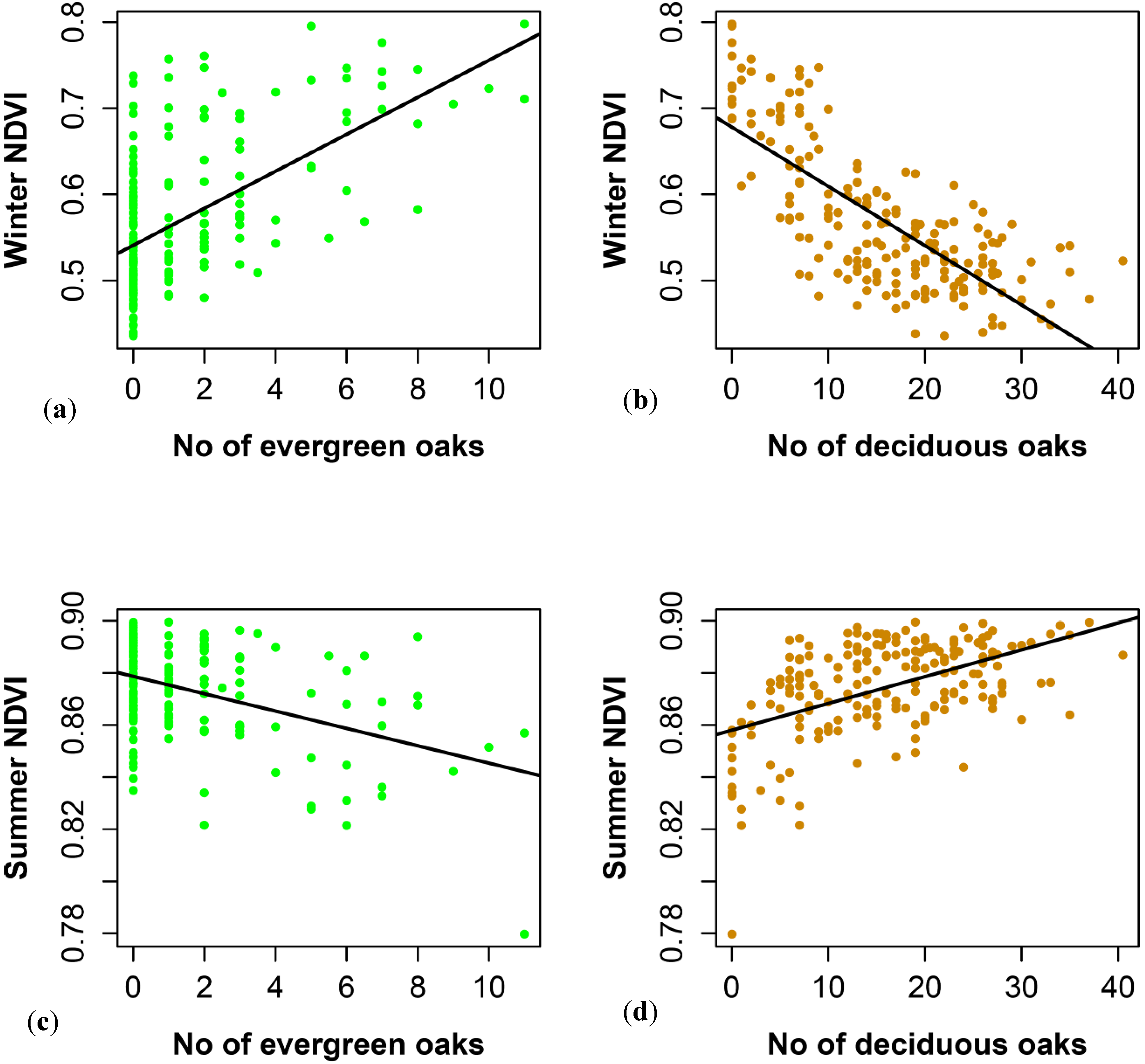
3.2. SPOT 4 (Take 5) Imagery as an Index of Vegetation Type is Accurately Validated with Ground Data from the Study Site
| NDVI Radius | Model | Deciduous Oaks (DeO) | Evergreen Oaks (EvO) | DeO + EvO |
|---|---|---|---|---|
| 10 m | Estimate | −0.00733 | 0.0243 | DeO: −0.00557, EvO: 0.01318 |
| SE | 0.00052 | 0.00228 | DeO: 0.00056, EvO: 0.00216 | |
| T value | −14.07 | 10.69 | DeO: −9.985, EvO: 6.105 | |
| p-value | <2 × 10−16 | <2 × 10−16 | DeO: <2 × 10−16; EvO: 5.63 × 10−9 | |
| % var. explained | 51% | 37% | 58% | |
| 50 m | Estimate | −0.00688 | 0.02148 | DeO: −0.00548, EvO: 0.01052 |
| SE | 0.00046 | 0.00210 | DeO: 0.00050, EvO: 0.00192 | |
| T value | −15.14 | 10.24 | DeO: −11.079, EvO: 5.489 | |
| p-value | 2 × 10−16 | 2 × 10−16 | DeO: 2 × 10−16, EvO: 1.27 × 10−7 | |
| % var. explained | 54% | 35% | 60% |
| NDVI Radius | Model | Deciduous Oaks (DeO) | Evergreen Oaks (EvO) | DeO + EvO |
|---|---|---|---|---|
| 10 m | Estimate | 0.00124 | −0.00361 | DeO: 0.00103, EvO: −0.00154 |
| SE | 0.00016 | 0.00065 | DeO: 0.00018, EvO: 0.00070 | |
| T value | 7.958 | −5.594 | DeO: 5.743, EvO: −2.207 | |
| p-value | 1.48 × 10−13 | 7.55 × 10−8 | DeO: 3.62 × 10−8, EvO: 0.0285 | |
| % var. explained | 24% | 14% | 26% | |
| 50 m | Estimate | 0.00103 | −0.00333 | DeO: 0.00080, EvO: −0.00174 |
| SE | 0.00013 | 0.00051 | DeO: 0.0001431, EvO: 0.0005540 | |
| T value | 8.215 | −6.531 | DeO: 5.575, EvO: −3.134 | |
| p-value | 3.07 × 10−14 | 5.71 × 10−10 | DeO: 8.34 × 10−8, EvO: 0.00199 | |
| % var. explained | 26% | 18% | 29% |
| NDVI Radius | Model | Deciduous Oaks (DeO) | Evergreen Oaks (EvO) | DeO + EvO |
|---|---|---|---|---|
| 50 m | Estimate | 0.00876 | −0.02709 | DeO: 0.00702, EvO: −0.01305 |
| SE | 0.00052 | 0.00248 | DeO: 0.00055, EvO: 0.00213 | |
| T value | 17.02 | −10.94 | DeO: 12.74, EvO: −6.12 | |
| p-value | <2 × 10−16 | <2 × 10−16 | DeO: < 2 × 10−16, EvO: 5.19 × 10−9 | |
| % var. explained | 60% | 38% | 66% |
3.3. Spatial Autocorrelation in Oak Composition is Visible in Ground-Based and NDVI-Based Correlograms
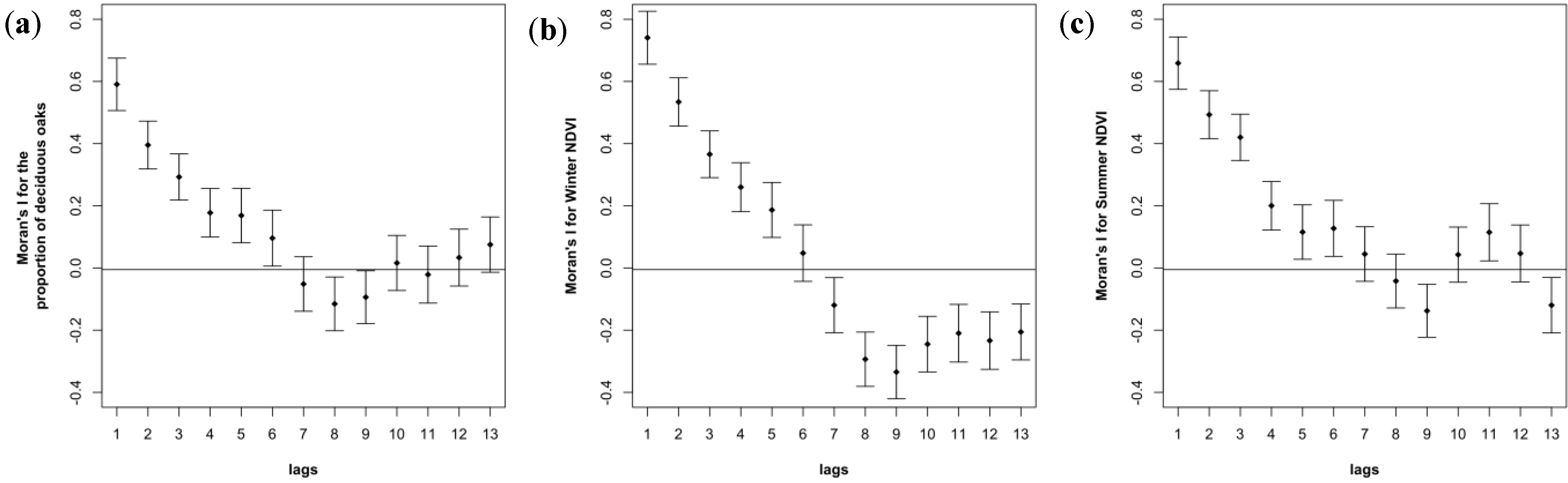
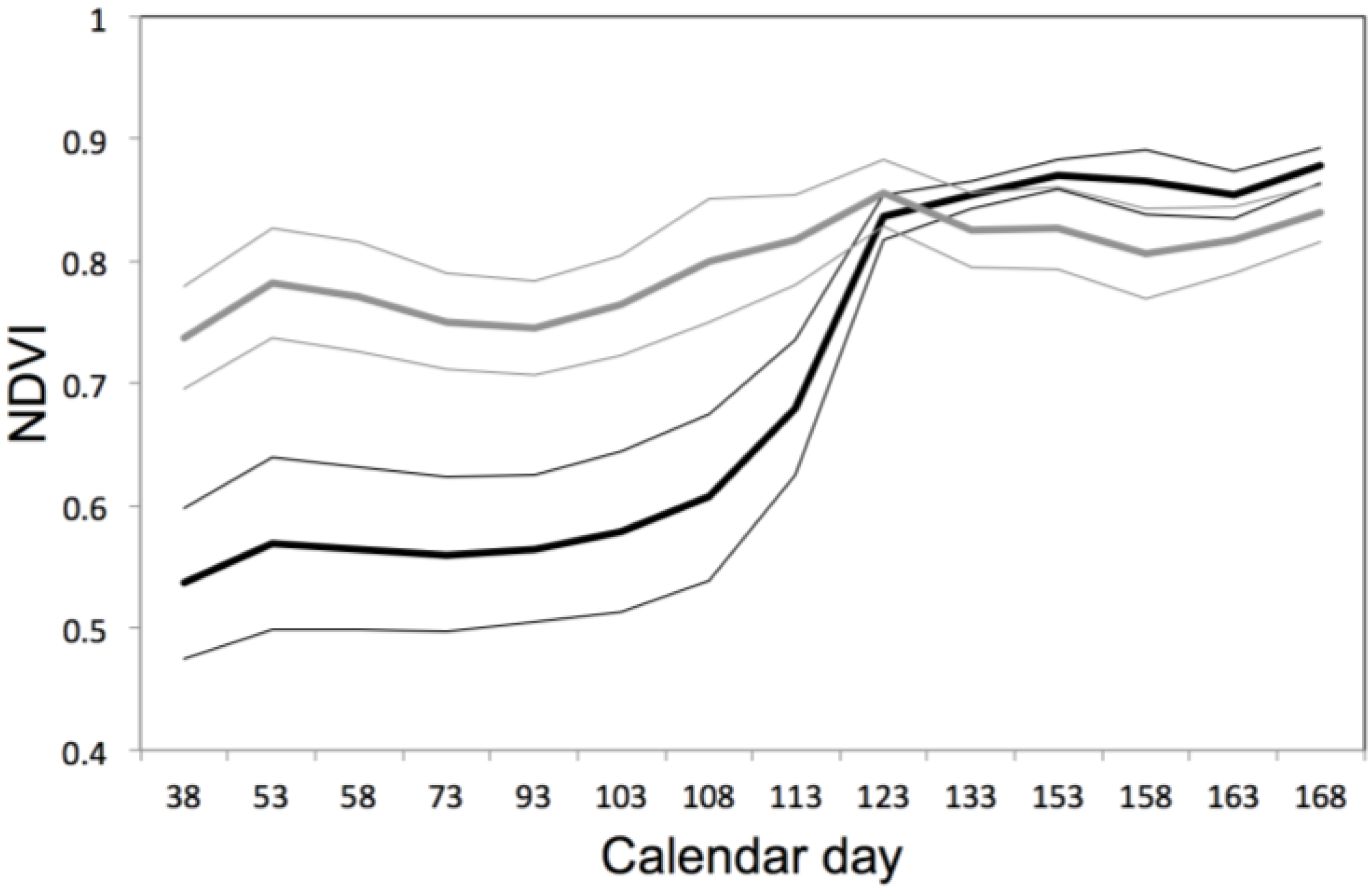
3.4. Contrasted Signatures of NDVI for Deciduous and Evergreen Oaks in a Temporal Time Series
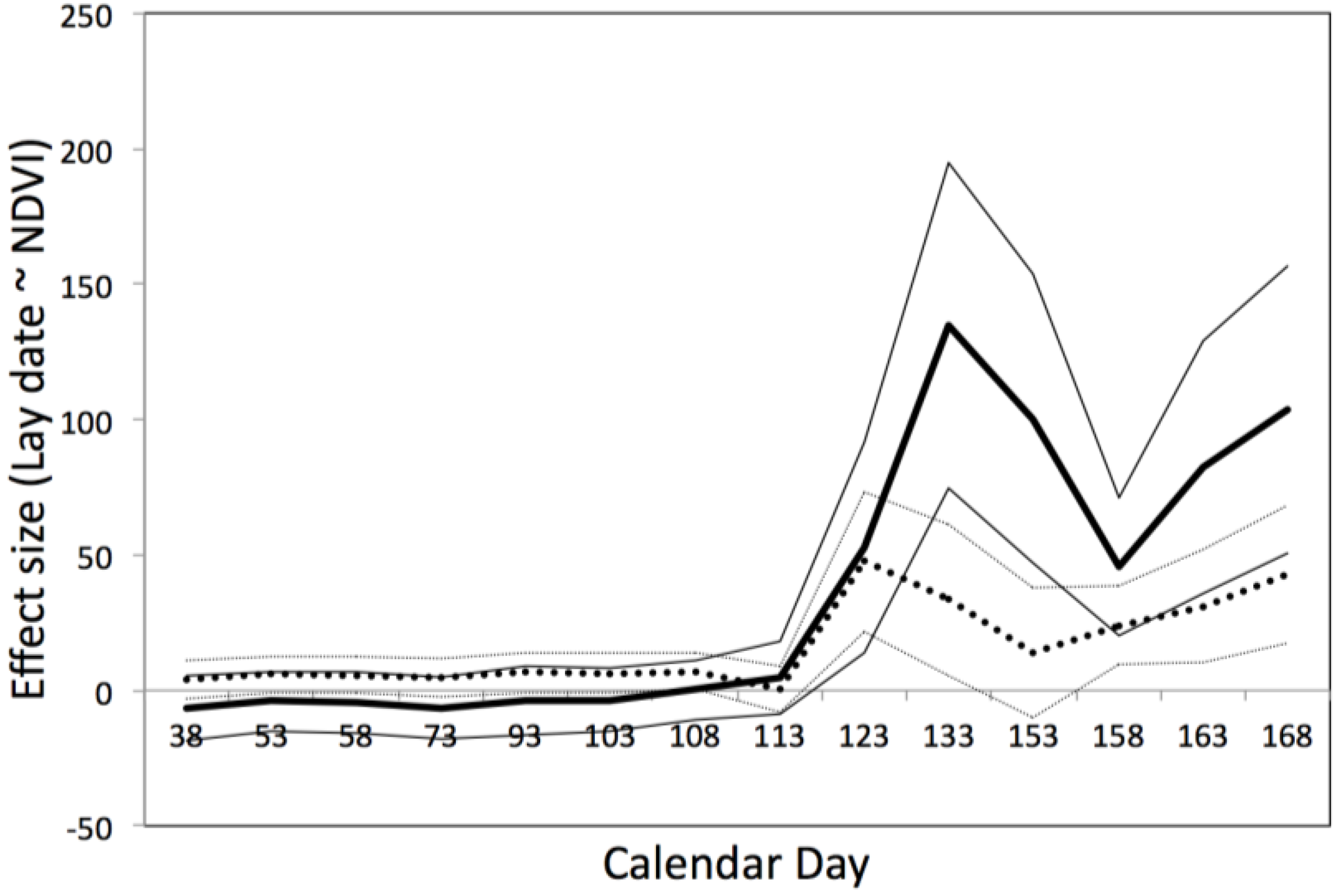
3.5. Predicting the Timing of Blue Tit Reproduction
4. Discussion
4.1. Winter NDVI Successfully Captured Oak Composition
4.2. Performance of Summer and Winter NDVI Values to Detect Environmental Heterogeneity
4.3. From Points in Space to Open-Ended Gradients: A Question of Scale
4.4. Matching Oak and Blue Tit Phenology
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Stearns, S.C. The Evolution of Life Histories; Oxford University Press: Oxford, UK, 1992. [Google Scholar]
- Parmesan, C. Influences of species, latitudes and methodologies on estimates of phenological response to global warming. Glob. Chang. Biol. 2007, 13, 1860–1872. [Google Scholar] [CrossRef]
- Both, C.; van Asch, M.; Bijlsma, R.G.; van den Burg, A.B.; Visser, M.E. Climate change and unequal phenological changes across four trophic levels: Constraints or adaptations? J. Anim. Ecol. 2009, 78, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Charmantier, A.; Doutrelant, C.; Dubuc-Messier, G.; Fargevieille, A.; Szulkin, M. Mediterranean blue tits as a case study of local adaptation. Evol. Appl. 2015, in press. [Google Scholar] [CrossRef]
- Blondel, J.; Thomas, D.W.; Charmantier, A.; Perret, P.; Bourgault, P.; Lambrechts, M.M. A thirty-year study of phenotypic and genetic variation of blue tits in mediterranean habitat mosaics. Bioscience 2006, 56, 661–673. [Google Scholar] [CrossRef]
- Blondel, J.; Dias, P.C.; Ferret, P.; Maistre, M.; Lambrechts, M.M. Selection-based biodiversity at a small spatial scale in a low-dispersing insular bird. Science 1999, 285, 1399–1402. [Google Scholar] [CrossRef] [PubMed]
- Charmantier, A.; Gienapp, P. Climate change and timing of avian breeding and migration: Evolutionary versus plastic changes. Evol. Appl. 2014, 7, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Both, C.; Bouwhuis, S.; Lessells, C.M.; Visser, M.E. Climate change and population declines in a long-distance migratory bird. Nature 2006, 441, 81–83. [Google Scholar] [CrossRef] [PubMed]
- Van Asch, M.; Visser, M.E. Phenology of forest caterpillars and their host trees: The importance of synchrony. Annu. Rev. Entomol. 2007, 52, 37–55. [Google Scholar] [CrossRef] [PubMed]
- Reed, T.E.; Grotan, V.; Jenouvrier, S.; Saether, B.-E.; Visser, M.E. Population growth in a wild bird is buffered against phenological mismatch. Science 2013, 340, 488–491. [Google Scholar] [CrossRef] [PubMed]
- Charmantier, A.; McCleery, R.H.; Cole, L.R.; Perrins, C.; Kruuk, L.E.B.; Sheldon, B.C. Adaptive phenotypic plasticity in response to climate change in a wild bird population. Science 2008, 320, 800–803. [Google Scholar] [CrossRef] [PubMed]
- Visser, M.E.; van Noordwijk, A.J.; Tinbergen, J.M.; Lessells, C.M. Warmer springs lead to mistimed reproduction in great tits (Parus major). Proc. R. Soc. B 1998, 265, 1867–1870. [Google Scholar] [CrossRef]
- Bourgault, P.; Thomas, D.; Perret, P.; Blondel, J. Spring vegetation phenology is a robust predictor of breeding date across broad landscapes: A multi-site approach using the corsican blue tit (Cyanistes caeruleus). Oecologia 2010, 162, 885–892. [Google Scholar] [CrossRef] [PubMed]
- Pettorelli, N.; Vik, J.O.; Mysterud, A.; Gaillard, J.M.; Tucker, C.J.; Stenseth, N.C. Using the satellite-derived ndvi to assess ecological responses to environmental change. Trends Ecol. Evol. 2005, 20, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Verhulst, S.; Tinbergen, J.M. Experimental evidence for a causal relationship between timing and success of reproduction in the great tit Parus major. J. Anim. Ecol. 1991, 60, 269–282. [Google Scholar] [CrossRef]
- Blondel, J.; Dias, P.C.; Maistre, M.; Perret, P. Habitat heterogeneity and life-history variation of mediterranean blue tits (Parus caeruleus). Auk 1993, 110, 511–520. [Google Scholar] [CrossRef]
- Banbura, J.; Blondel, J.; Dewildelambrechts, H.; Galan, M.J.; Maistre, M. Nestling diet variation in an insular mediterranean population of blue tits Parus caeruleus—Effects of years, territories and invididuals. Oecologia 1994, 100, 413–420. [Google Scholar] [CrossRef]
- Zandt, H.; Strijkstra, A.; Blondel, J.; van Balen, H. Food in two mediterranean blue tit populations: Do differences in caterpillar availability explain differences in timing of the breeding season? In Population Biology of Passerine Birds; Blondel, J., Gosler, A., Lebreton, J.D., Mccleery, R., Eds.; Springer-Verlag: Berlin/Heidelberg, Germany, 1990; pp. 145–155. [Google Scholar]
- Van Balen, J.H. A comparative study of the breeding ecology of the great tit Parus-major in different habitats. Ardea 1973, 61, 1–93. [Google Scholar]
- Visser, M.E.; Holleman, L.J.M.; Gienapp, P. Shifts in caterpillar biomass phenology due to climate change and its impact on the breeding biology of an insectivorous bird. Oecologia 2006, 147, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Visser, M.E.; Holleman, L.J.M. Warmer springs disrupt the synchrony of oak and winter moth phenology. Proc. R. Soc. B-Biol. Sci. 2001, 268, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Cole, E.F.; Long, P.; Zelazowski, P.; Szulkin, M.; Sheldon, B.C. Predicting bird phenology from space: Satellite-derived vegetation green-up signal uncovers spatial variation in phenological synchrony between birds and their environment. Ecol. Evol. 2015, in press. [Google Scholar] [CrossRef]
- Hinks, A.; Cole, E.F.; Fannon, K.; Wilkin, T.A.; Nakagawa, S.; Sheldon, B.C. Scale-dependent phenology synchrony between songbirds and their caterpillar food source. Am. Nat. 2015, 186, 84–97. [Google Scholar] [CrossRef] [PubMed]
- Hurley, M.A.; Hebblewhite, M.; Gaillard, J.M.; Dray, S.; Taylor, K.A.; Smith, W.K.; Zager, P.; Bonenfant, C. Functional analysis of normalized difference vegetation index curves reveals overwinter mule deer survival is driven by both spring and autumn phenology. Philos. Trans. R. Soc. B 2014, 369. [Google Scholar] [CrossRef] [PubMed]
- Tveraa, T.; Stien, A.; Bardsen, B.-J.; Fauchald, P. Population densities, vegetation green-up, and plant productivity: Impacts on reproductive success and juvenile body mass in reindeer. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Drusch, M.; Del Bello, U.; Carlier, S.; Colin, O.; Fernandez, V.; Gascon, F.; Hoersch, B.; Isola, C.; Laberinti, P.; Martimort, P.; et al. Sentinel-2: Esa’s optical high-resolution mission for gmes operational services. Remote Sens. Environ. 2012, 120, 25–36. [Google Scholar] [CrossRef]
- Blondel, J.; Isenmann, P.; Maistre, M.; Perret, P. What are the consequences of being a downy oak (Quercus pubescens) or a holm oak (Quercus-ilex) for breeding blue tits (Parus caeruleus). Vegetatio 1992, 100, 129–136. [Google Scholar] [CrossRef]
- Morin, X.; Roy, J.; Sonie, L.; Chuine, I. Changes in leaf phenology of three european oak species in response to experimental climate change. New Phytol. 2010, 186, 900–910. [Google Scholar] [CrossRef] [PubMed]
- Charmantier, A.; Blondel, J.; Perret, P.; Harmelin-Vivien, M. Tracing site-specific isotopic signatures along a blue tit Cyanistes caeruleus food chain. Ibis 2014, 156, 165–175. [Google Scholar] [CrossRef]
- Hagolle, O.; Huc, M.; Pascual, D.V.; Dedieu, G. A multi-temporal method for cloud detection, applied to Formosat-2, Venus, Landsat and Sentinel-2 images. Remote Sens. Environ. 2010, 114, 1747–1755. [Google Scholar] [CrossRef]
- Open foris geospatial toolkit: Free open-source solutions for environmental monitoring. Available online: http://www.openforis.org/ (accessed on 5 November 2014).
- QGIS Developmment Team. QGis Geographic Information System v. 2.8. Open Source Geospatial Foundation. Availbale online: http://qgis.osgeo.org (accessed on 1 April 2015).
- Team, R.D.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]
- Moran, P.A. The interpretation of statistical maps. J. R. Stat. Soc. Ser. B (Methodol.) 1948, 10, 243–251. [Google Scholar]
- Borcard, D.; Gillet, F.; Legendre, P. Numerical Ecology with R; Springer: New York, NY, USA, 2011; pp. 1–300. [Google Scholar]
- Marrot, P.; Garant, D.; Charmantier, A. Spatial autocorrelation in fitness affects the estimation of natural selection in the wild. Methods Ecol. Evol. 2015, in press. [Google Scholar] [CrossRef]
- Levin, S.A. The problem of pattern and scale in ecology. Ecology 1992, 73, 1943–1967. [Google Scholar] [CrossRef]
- Tremblay, I.; Thomas, D.; Blondel, J.; Perret, P.; Lambrechts, M.M. The effect of habitat quality on foraging patterns, provisioning rate and nestling growth in corsican blue tits Parus caeruleus. Ibis 2005, 147, 17–24. [Google Scholar] [CrossRef]
- Naef-Daenzer, B. Patch time allocation and patch sampling by foraging great and blue tits. Anim. Behav. 2000, 59, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Marshal, J.P.; Rajah, A.; Parrini, F.; Henley, M.; Henley, S.R.; Erasmus, B.F.N. Scale-dependent selection of greenness by african elephants in the kruger-private reserve transboundary region, South Africa. Eur. J. Wildl. Res. 2011, 57, 537–548. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szulkin, M.; Zelazowski, P.; Marrot, P.; Charmantier, A. Application of High Resolution Satellite Imagery to Characterize Individual-Based Environmental Heterogeneity in a Wild Blue Tit Population. Remote Sens. 2015, 7, 13319-13336. https://doi.org/10.3390/rs71013319
Szulkin M, Zelazowski P, Marrot P, Charmantier A. Application of High Resolution Satellite Imagery to Characterize Individual-Based Environmental Heterogeneity in a Wild Blue Tit Population. Remote Sensing. 2015; 7(10):13319-13336. https://doi.org/10.3390/rs71013319
Chicago/Turabian StyleSzulkin, Marta, Przemyslaw Zelazowski, Pascal Marrot, and Anne Charmantier. 2015. "Application of High Resolution Satellite Imagery to Characterize Individual-Based Environmental Heterogeneity in a Wild Blue Tit Population" Remote Sensing 7, no. 10: 13319-13336. https://doi.org/10.3390/rs71013319
APA StyleSzulkin, M., Zelazowski, P., Marrot, P., & Charmantier, A. (2015). Application of High Resolution Satellite Imagery to Characterize Individual-Based Environmental Heterogeneity in a Wild Blue Tit Population. Remote Sensing, 7(10), 13319-13336. https://doi.org/10.3390/rs71013319