An Improved Method for Single Tree Trunk Extraction Based on LiDAR Data
Abstract
1. Introduction
2. Data and Methods
2.1. Data
2.1.1. Data Source
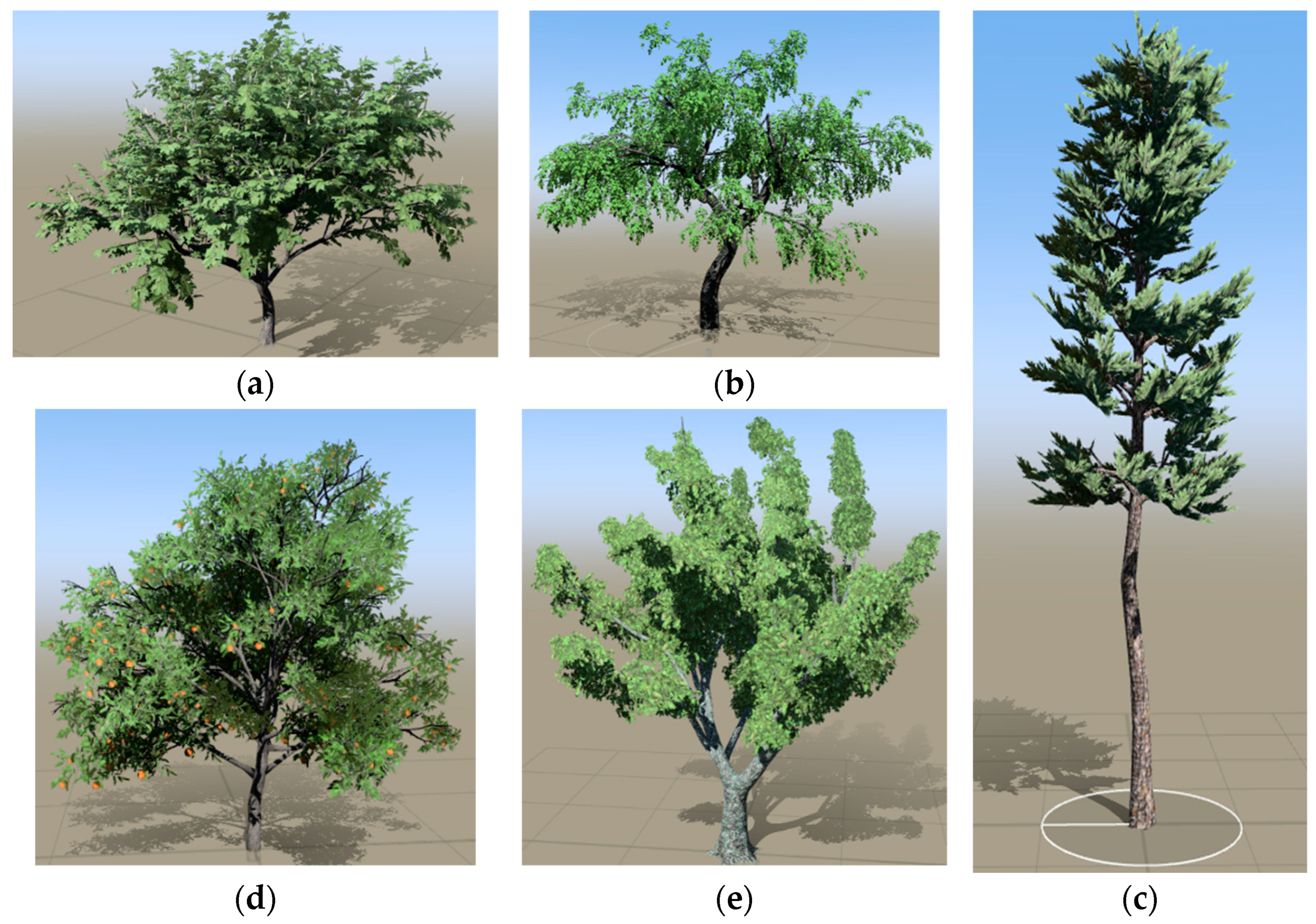
2.1.2. Point Cloud Data Simulation
2.2. Methods
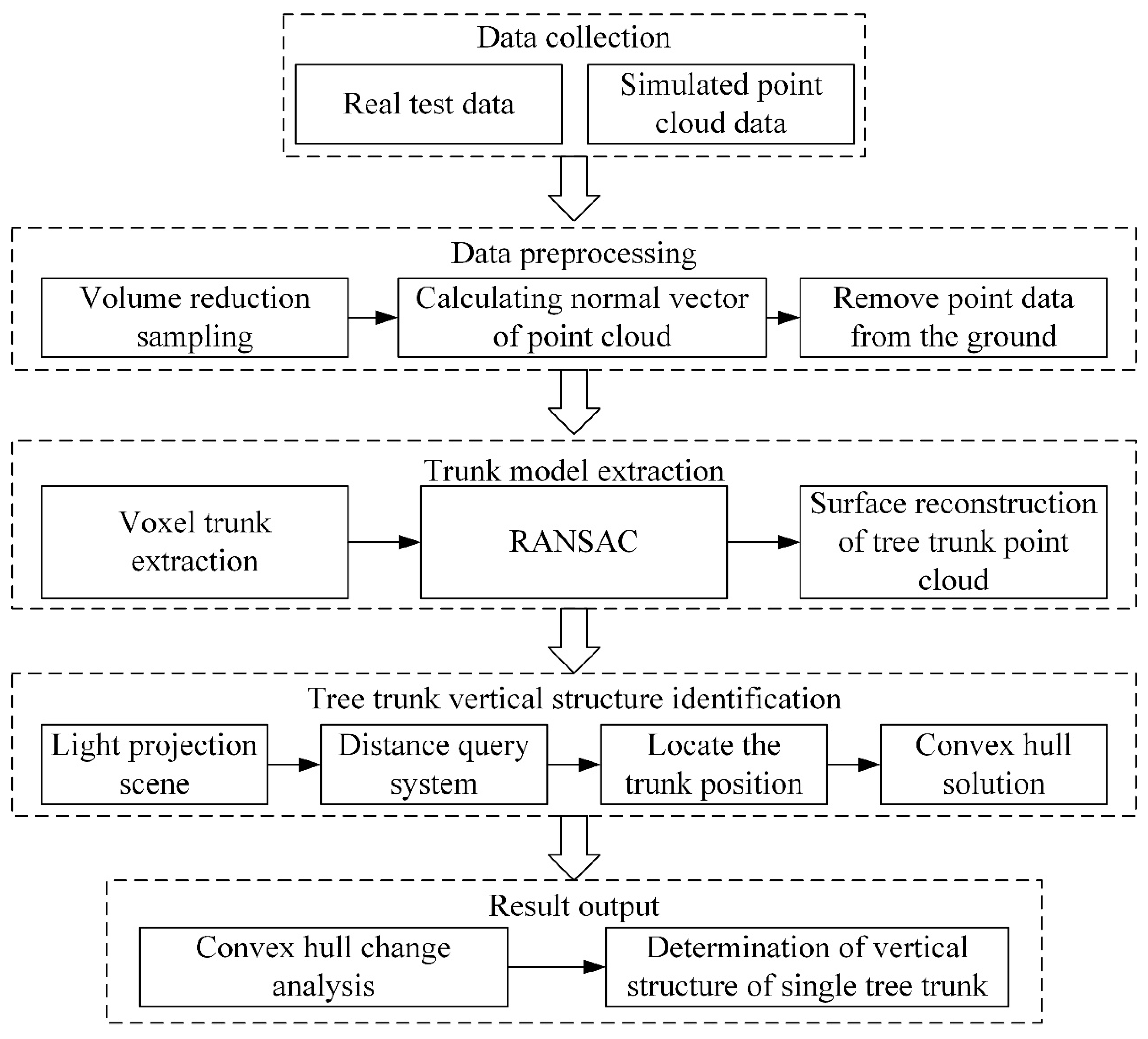
2.2.1. Method Flowchart
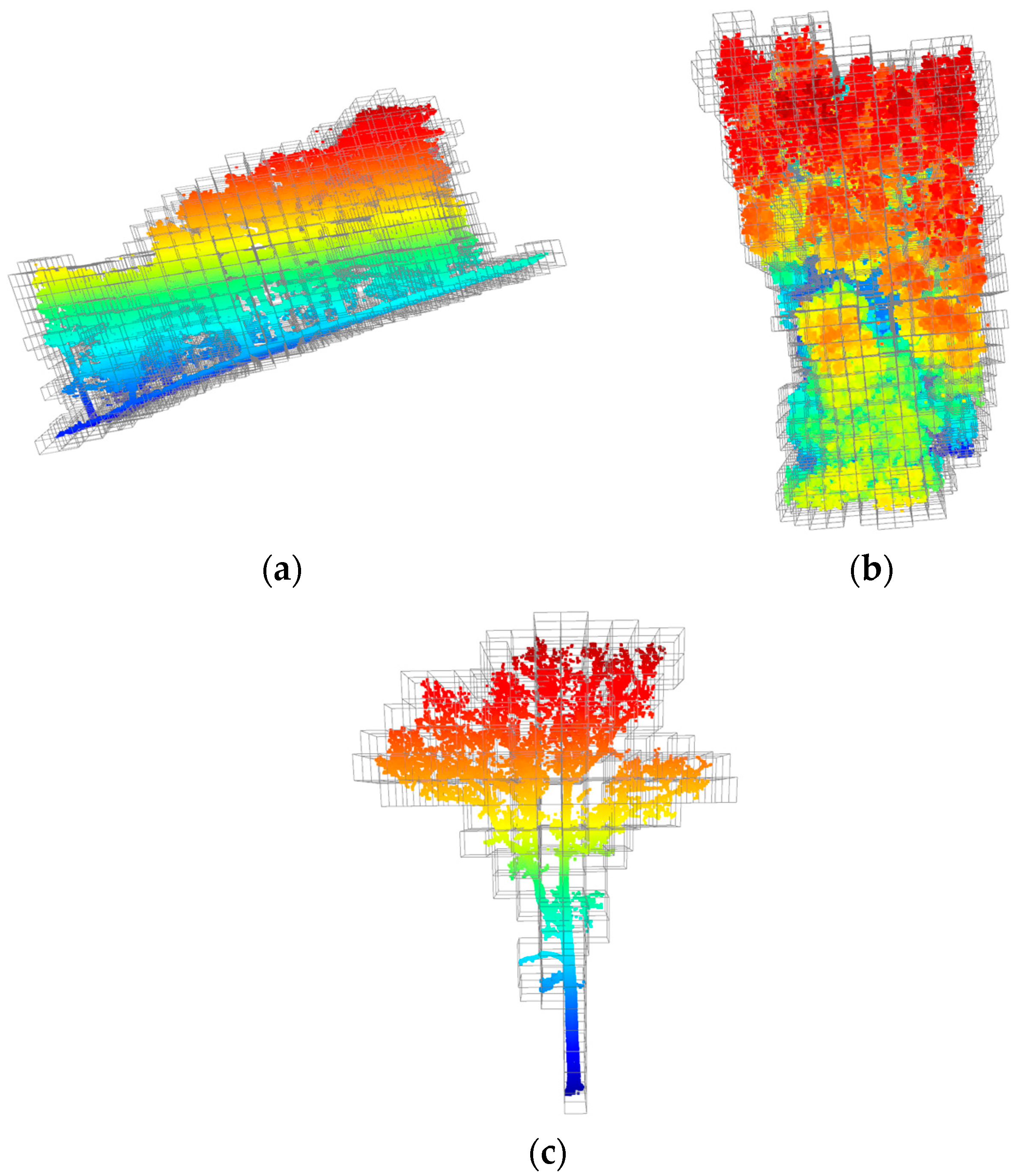
2.2.2. A Voxelized Trunk Extraction Algorithm
- (1)
- Voxelization: LiDAR point cloud data contains a large number of point elements used to describe the three-dimensional features in the scene. To reduce the amount of data processing, 3D voxels are constructed based on the XYZ coordinate system, and the point cloud data are divided into regular 3D grids, where each voxel is a cube with a side length of l. Any voxel can be indexed by the row (i), column (j), and layer (k), and equally divided in the XYZ coordinate system.
- (2)
- Voxel-based dimension analysis: After voxelizing the point cloud data, this paper uses principal component analysis (PCA) as the main method to analyze the voxel dimension. PCA is a widely accepted dimensional analysis method, and it is widely used to infer geometric types of point cloud data. The common inference types are the following three types of shapes: linear, planar, and spherical [41].
2.2.3. Extracting Discontinuous Vertical Structure and Robustness Test Based on RANSAC
| Algorithm 1 Extract the shapes in the point cloud |
do |
| if then |
end if until return |
2.2.4. Trunk Surface Reconstruction
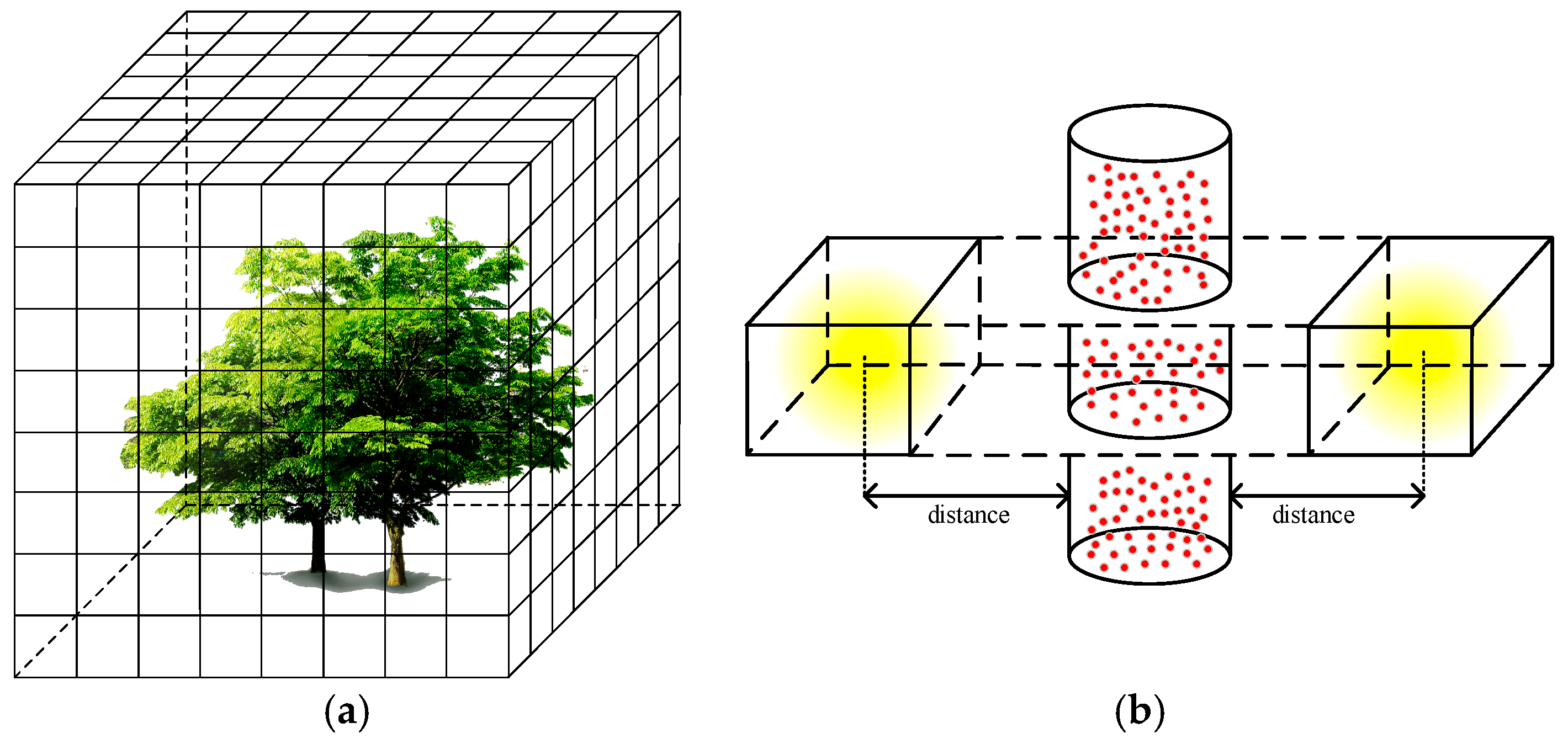
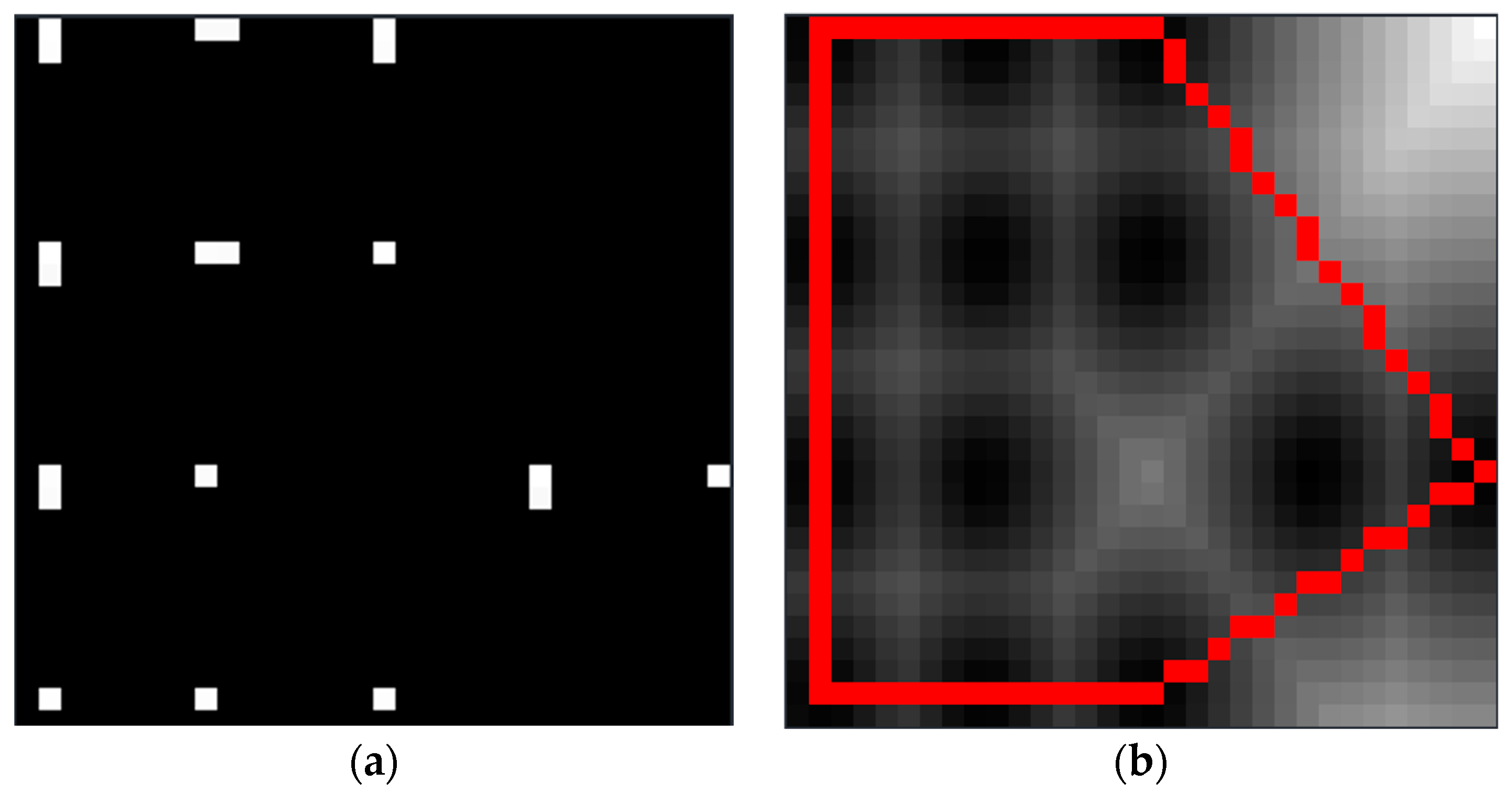
2.2.5. Locating the Trunk Position in the Light Projection Scene
2.2.6. Algorithm for Solving Trunk Positioning Point Convex Hull and Determination of Vertical Structure of Single Tree Trunk
3. Results
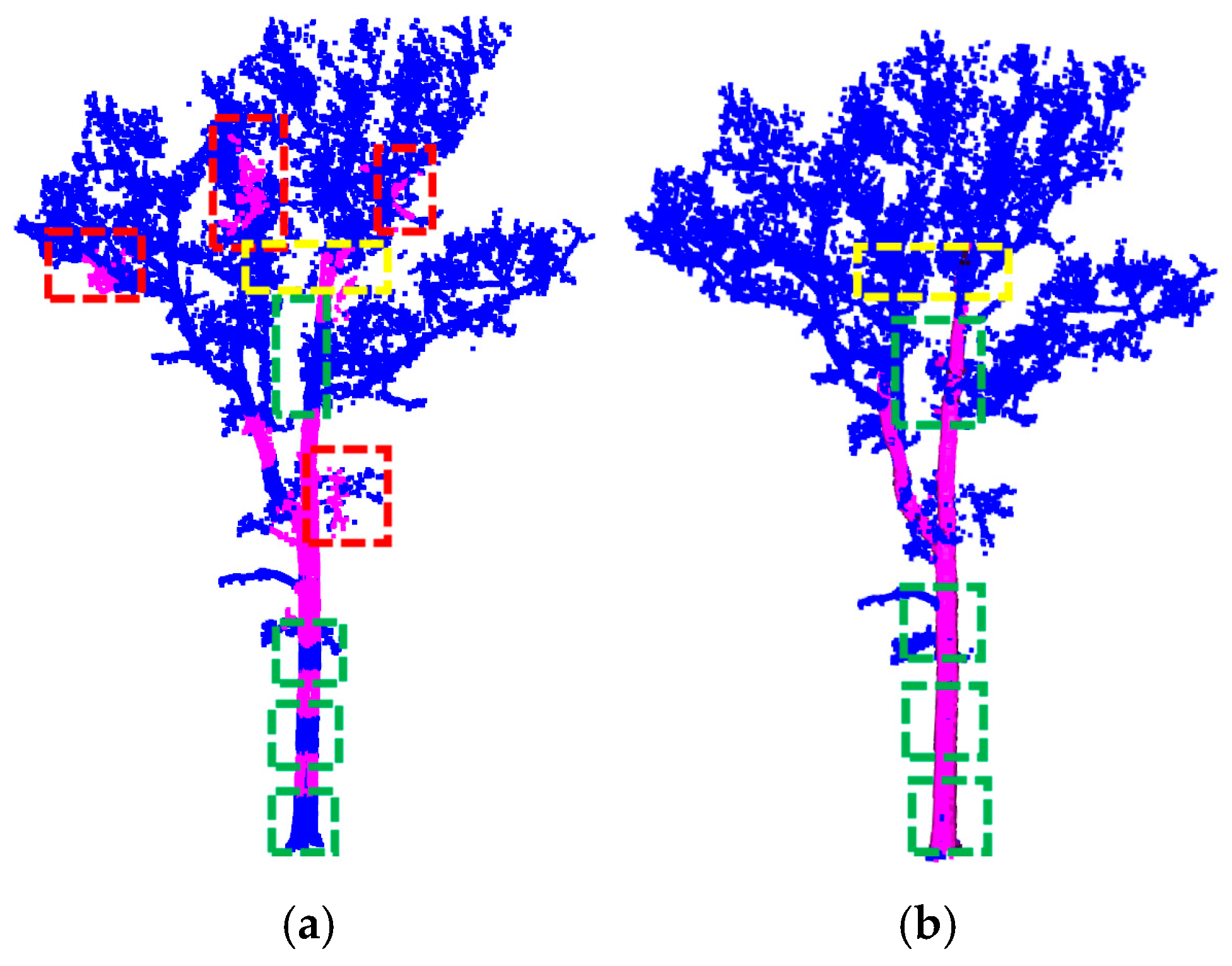
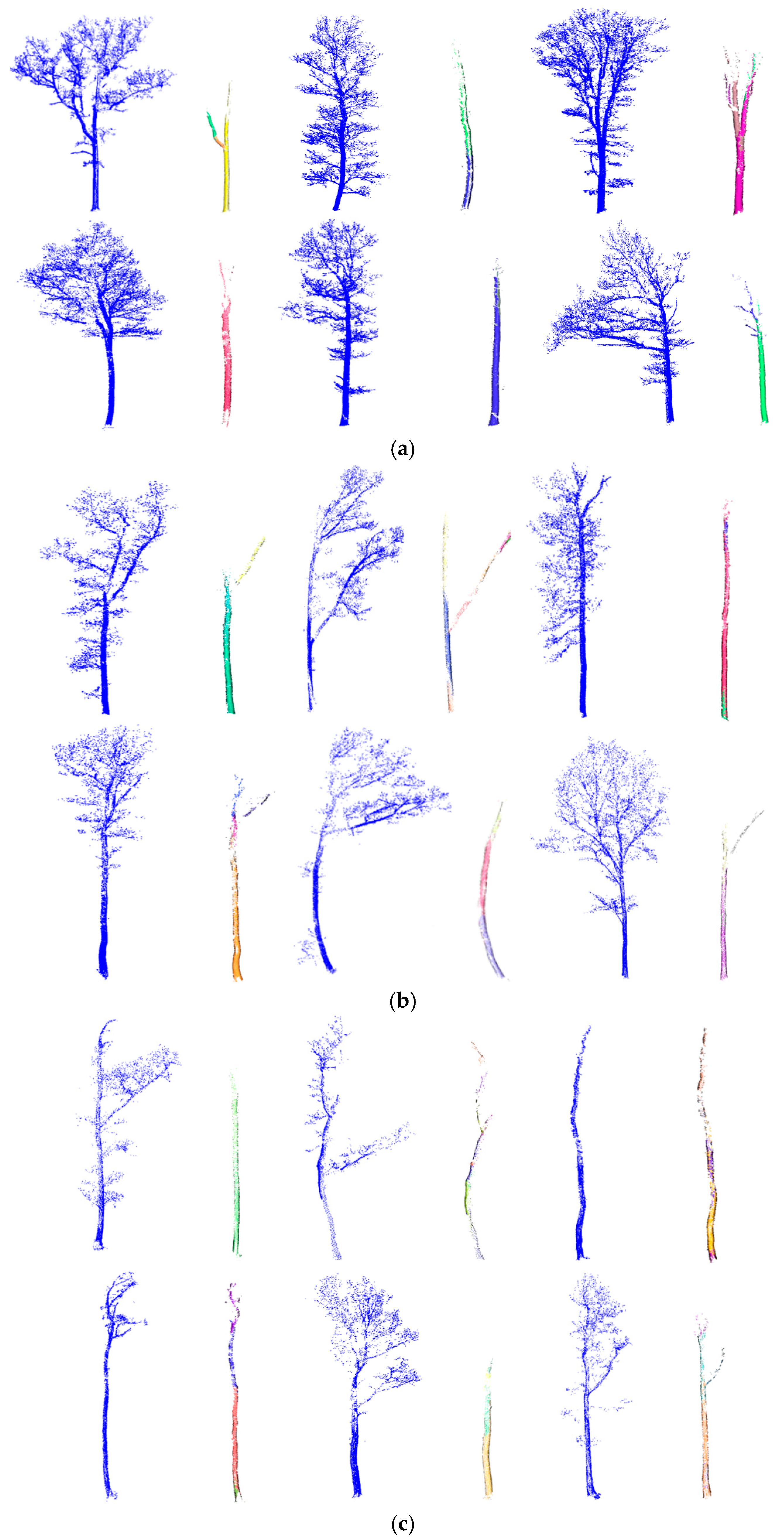
3.1. Single Trees
3.2. RANSAC Robustness Test
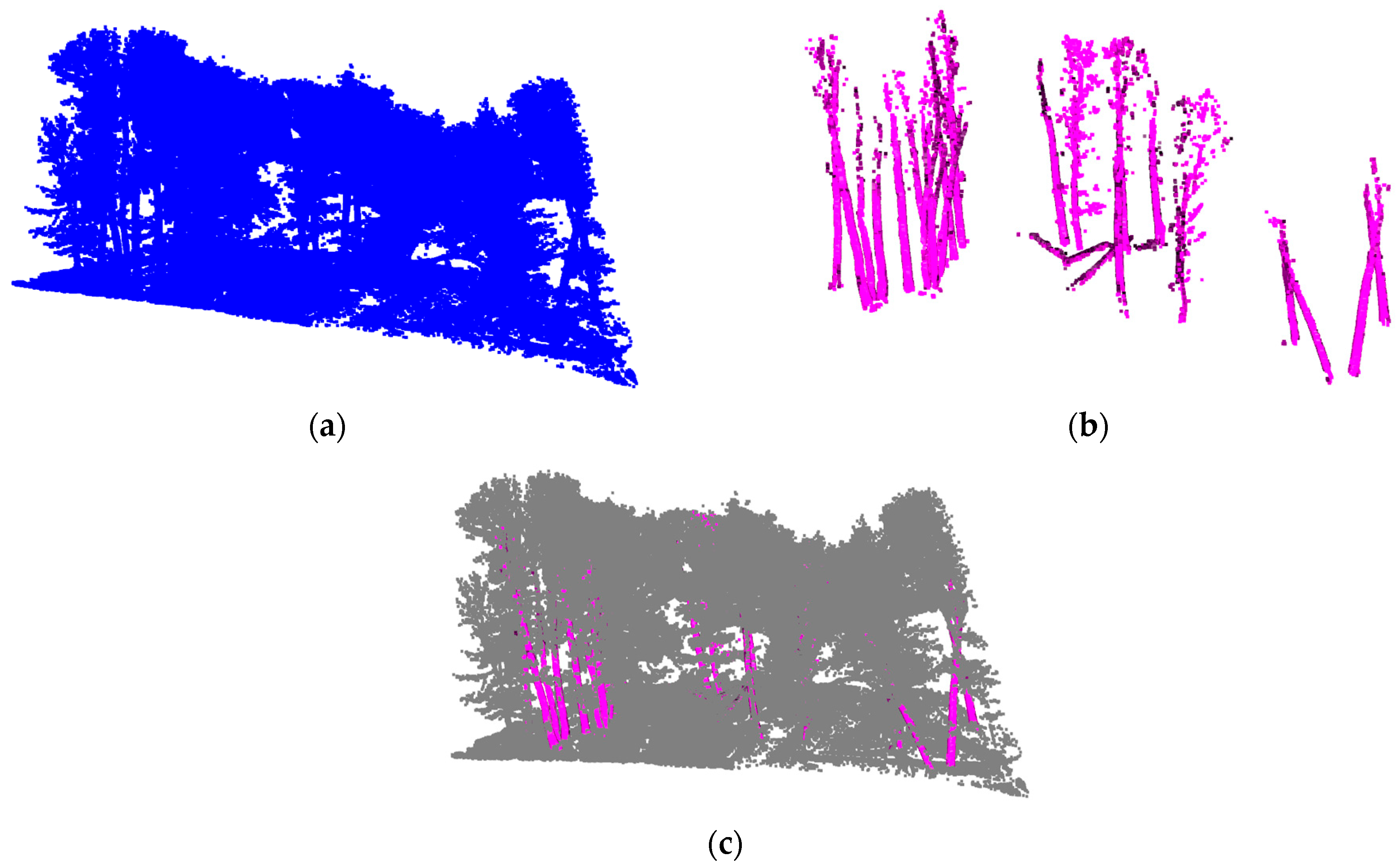
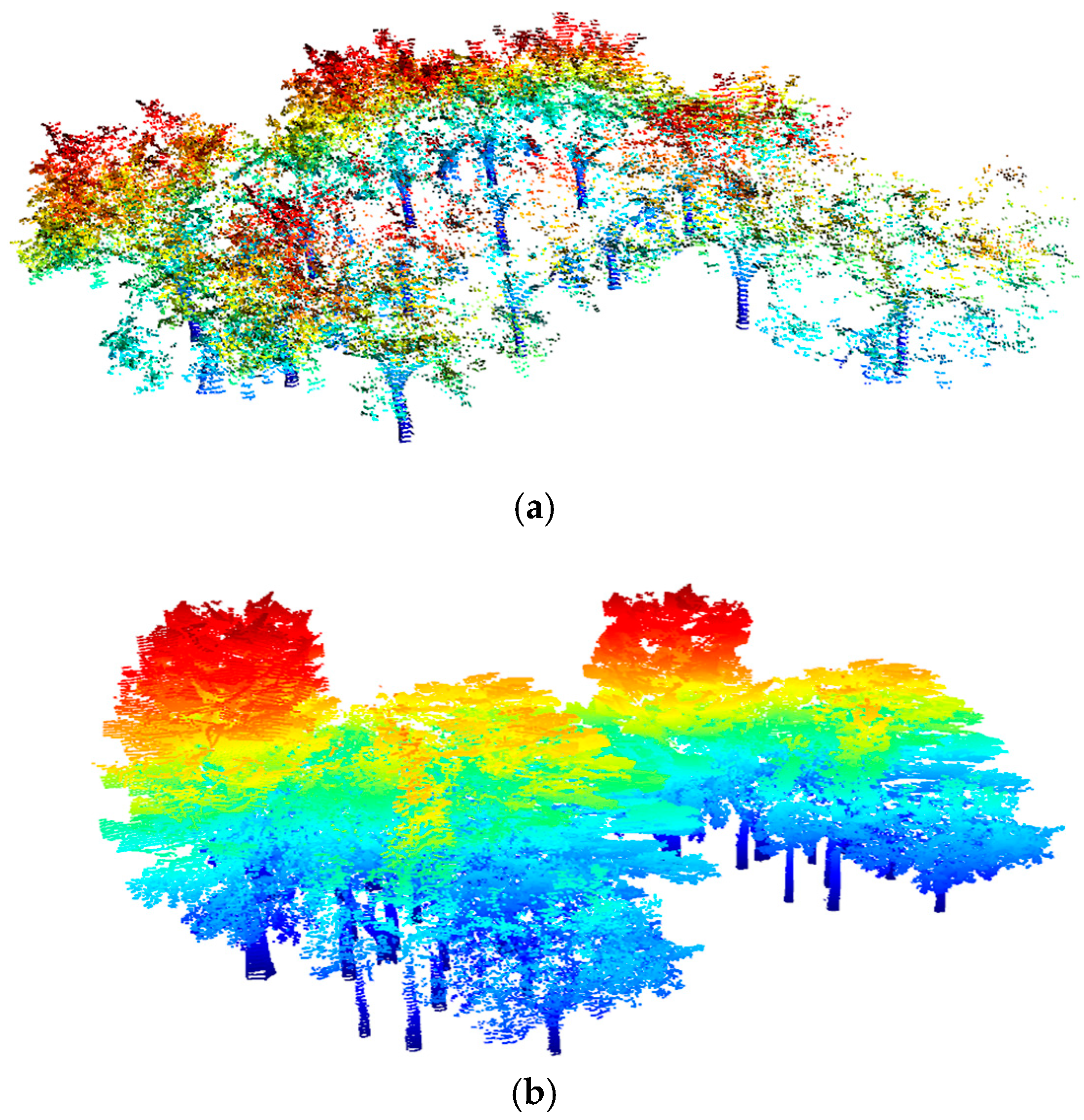
3.3. Trunk Locating
3.4. Vertical Structural Characteristics of Trunk
4. Discussion
5. Conclusions
- (1)
- Taking the linear shape solved by trunk recognition algorithm as the candidate shape of the RANSAC algorithm can effectively solve the connectivity problem of trunk point cloud in the vertical direction.
- (2)
- Introducing the light projection scene can extract and count the trunk position accurately.
- (3)
- In forest areas where the vertical structure of individual tree trunks differs in the light projection scene, the location points of the tree trunks show significant changes. Experimental data indicate that the rate of change exceeds 50%.
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ding, R. Importance of Forest Resources Investigation and Optimization Measures. World Trop. Agric. Inf. 2022, 4, 60–61. [Google Scholar]
- Xie, X.; Su, H.; Yang, Y.; Li, C.; Lu, F.; Luo, W.; Xu, Z. Estimation of stand parameters of eucalyptus plantation in Guangxi based on terrestrial laser scanner. For. Resour. Manag. 2022, 2, 100–108. [Google Scholar]
- Zhao, L. Application and prospect of airborne laser scanner in obtaining forest resource parameters. For. Sci. Technol. Inf. 2022, 54, 71–73. [Google Scholar]
- Dilixiati, B.; Halike, Y.; Yusufu, A.; Wei, J. Estimation of Populus euphratica leaf area index in the lower reaches of Tarim River by terrestrial laser scanner data. J. Northeast. For. Univ. 2020, 48, 46–50. [Google Scholar]
- Ross, N.; Jimenez, J.; Schnell, C.E.; Hartshorn, G.S.; Gregoire, T.G.; Oderwald, R. Canopy height models and airborne lasers to estimate forest biomass: Two problems. Int. J. Remote Sens. 2000, 21, 2153–2162. [Google Scholar]
- Luo, H.; Shu, Q.; Xi, L.; Huang, J.; Liu, Y.; Yang, Q. Research progress of estimating forest biomass by lidar. Green Technol. 2022, 24, 23–28. [Google Scholar]
- Chen, Q.; Qi, C. Lidar remote sensing of vegetation biomass. Remote Sens. Nat. Resour. 2013, 399, 399–420. [Google Scholar]
- Clark, M.L.; Roberts, D.A.; Ewel, J.J.; Clark, D.B. Estimation of tropical rain forest aboveground biomass with small-footprint lidar and hyperspectral sensors. Remote Sens. Environ. 2011, 115, 2931–2942. [Google Scholar]
- Wulder, M.A.; White, J.C.; Nelson, R.F.; Næsset, E.; Ørka, H.O.; Coops, N.C.; Gobakken, T. Lidar sampling for large-area forest characterization: A review. Remote Sens. Environ. 2012, 121, 196–209. [Google Scholar]
- Dong, P.; Chen, Q. LiDAR Remote Sensing and Applications; CRC Press: Boca Raton, FL, USA, 2017. [Google Scholar]
- Qi, Z.; Li, S.; Yue, W.; Liu, Q.; Li, Z. Natural forest gap identification based on drone laser scanner. J. Beijing For. Univ. 2022, 44, 44–53. [Google Scholar]
- Zhang, K. Identification of gaps in mangrove forests with airborne LIDAR. Remote Sens. Environ. 2008, 112, 2309–2325. [Google Scholar] [CrossRef]
- Zhu, B.; Jin, J.; Luo, H.; Long, F.; Li, C.; Yue, C. Inversion of average stand height byairborne laser scanner based on variable optimization. Jiangsu J. Agric. Sci. 2022, 38, 706–713. [Google Scholar]
- Wallace, A.; Nichol, C.; Woodhouse, I. Recovery of forest canopy parameters by inversion of multispectral LiDAR data. Remote Sens. 2012, 4, 509–531. [Google Scholar] [CrossRef]
- Moorthy, S.M.K.; Calders, K.; Vicari, M.B.; Verbeeck, H. Improved supervised learning-based approach for leaf and wood classification from LiDAR point clouds of forests. IEEE Trans. Geosci. Remote Sens. 2019, 58, 3057–3070. [Google Scholar] [CrossRef]
- Cao, L.; Coops, N.C.; Innes, J.L.; Dai, J.; Ruan, H.; She, G. Tree species classification in subtropical forests using small-footprint full-waveform LiDAR data. Int. J. Appl. Earth Obs. Geoinformation 2016, 49, 39–51. [Google Scholar] [CrossRef]
- Lou, Y.; Fan, Y.; Dai, Q.; Wang, Z.; Ku, W.; Zhao, M.; Yu, S. Relationship between the vertical structure of evergreen deciduous broad-leaved forest community and the overall species diversity of the community in Mount Tianmu. J. Ecol. 2021, 41, 8568–8577. [Google Scholar]
- Huang, W.; Pohjonen, V.; Johansson, S.; Nashanda, M.; Katigula, M.; Luukkanen, O. Species diversity, forest structure and species composition in Tanzanian tropical forests. For. Ecol. Manag. 2002, 173, 11–24. [Google Scholar] [CrossRef]
- Gui, X.; Lian, J.; Zhang, R.; Li, Y.; Shen, H.; Ni, Y.; Ye, W. Vertical structure and species diversity characteristics of subtropical evergreen broad-leaved forest community in Dinghushan. Biol. Divers. 2019, 27, 619–629. [Google Scholar]
- Jarron, L.R.; Coops, N.C.; MacKenzie, W.H.; Tompalski, P.; Dykstra, P. Detection of sub-canopy forest structure using airborne LiDAR. Remote Sens. Environ. 2020, 244, 111770. [Google Scholar] [CrossRef]
- de Almeida, D.R.A.; Zambrano, A.M.A.; Broadbent, E.N.; Wendt, A.L.; Foster, P.; Wilkinson, B.E.; Salk, C.; Papa, D.d.A.; Stark, S.C.; Valbuena, R.; et al. Detecting successional changes in tropical forest structure using GatorEye drone-borne lidar. Biotropica 2020, 52, 1155–1167. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, J.; Liu, G. Vertical Structure Classification of a Forest Sample Plot Based on Point Cloud Data. J. Indian Soc. Remote Sens. 2020, 48, 1215–1222. [Google Scholar]
- Putman, E.B.; Popescu, S.C.; Eriksson, M.; Zhou, T.; Klockow, P.; Vogel, J.; Moore, G.W. Detecting and quantifying standing dead tree structural loss with reconstructed tree models using voxelized terrestrial lidar data. Remote Sens. Environ. 2018, 209, 52–65. [Google Scholar]
- Taheriazad, L.; Moghadas, H.; Sanchez-Azofeifa, A. Calculation of leaf area index in a Canadian boreal forest using adaptive voxelization and terrestrial LiDAR. Int. J. Appl. Earth Obs. Geoinf. 2019, 83, 101923. [Google Scholar]
- Zhong, L.; Cheng, L.; Xu, H.; Wu, Y.; Chen, Y.; Li, M. Segmentation of individual trees from TLS and MLS data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 10, 774–787. [Google Scholar]
- Wang, D. Unsupervised semantic and instance segmentation of forest point clouds. ISPRS J. Photogramm. Remote Sens. 2020, 165, 86–97. [Google Scholar]
- Li, Z.; Wang, J.; Zhang, Z.; Jin, F.; Yang, J.; Sun, W.; Cao, Y. A Method Based on Improved iForest for Trunk Extraction and Denoising of Individual Street Trees. Remote Sens. 2022, 15, 115. [Google Scholar] [CrossRef]
- Neuville, R.; Bates, J.S.; Jonard, F. Estimating Forest Structure from UAV-Mounted LiDAR Point Cloud Using Machine Learning. Remote Sens. 2021, 13, 352. [Google Scholar] [CrossRef]
- Xu, S.; Sun, X.; Yun, J.; Wang, H. A New Clustering-Based Framework to the Stem Estimation and Growth Fitting of Street Trees From Mobile Laser Scanning Data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 3240–3250. [Google Scholar]
- Monnier, F.; Vallet, B.; Soheilian, B. Trees Detection From Laser Point Clouds Acquired in Dense Urban Areas by a Mobile Mapping System. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2012, 1, 245–250. [Google Scholar] [CrossRef]
- Lamprecht, S.; Stoffels, J.; Dotzler, S.; Haß, E.; Udelhoven, T. aTrunk—An ALS-Based Trunk Detection Algorithm. Remote Sens. 2015, 7, 9975–9997. [Google Scholar]
- Vandendaele, B.; Fournier, R.A.; Vepakomma, U.; Pelletier, G.; Lejeune, P.; Martin-Ducup, O. Estimation of Northern Hardwood Forest Inventory Attributes Using UAV Laser Scanning (ULS): Transferability of Laser Scanning Methods and Comparison of Automated Approaches at the Tree- and Stand-Level. Remote Sens. 2021, 13, 2796. [Google Scholar] [CrossRef]
- Wieser, M.; Mandlburger, G.; Hollaus, M.; Otepka, J.; Glira, P.; Pfeifer, N. A Case Study of UAS Borne Laser Scanning for Measurement of Tree Stem Diameter. Remote Sens. 2017, 9, 1154. [Google Scholar] [CrossRef]
- Brede, B.; Lau, A.; Bartholomeus, H.M.; Kooistra, L. Comparing RIEGL RiCOPTER UAV LiDAR Derived Canopy Height and DBH with Terrestrial LiDAR. Sensors 2017, 17, 2371. [Google Scholar] [CrossRef] [PubMed]
- Jaakkola, A.; Hyyppä, J.; Yu, X.; Kukko, A.; Kaartinen, H.; Liang, X.; Hyyppä, H.; Wang, Y. Autonomous Collection of Forest Field Reference—The Outlook and a First Step with UAV Laser Scanning. Remote Sens. 2017, 9, 785. [Google Scholar] [CrossRef]
- Trochta, J.; Krůček, M.; Vrška, T.; Král, K. 3D Forest: An application for descriptions of three-dimensional forest structures using terrestrial LiDAR. PLoS ONE 2017, 12, e0176871. [Google Scholar]
- Winiwarter, L.; Pena, A.M.E.; Zahs, V.; Weiser, H.; Searle, M.; Anders, K.; Höfle, B. Virtual Laser Scanning using HELIOS++-Applications in Machine Learning and Forestry. In Proceedings of the EGU General Assembly 2022, Vienna, Austria, 23–27 May 2022. [Google Scholar]
- Xiong, Q.; Huang, X.-Y. Speed Tree-Based Forest Simulation System. In Proceedings of the 2010 International Conference on Electrical and Control Engineering, Washington, DC, USA, 25–27 June 2010. [Google Scholar]
- Ayrey, E.; Fraver, S.; Kershaw, J.A., Jr.; Kenefic, L.S.; Hayes, D.; Weiskittel, A.R.; Roth, B.E. Layer Stacking: A Novel Algorithm for Individual Forest Tree Segmentation from LiDAR Point Clouds. Can. J. Remote Sens. 2017, 43, 16–27. [Google Scholar]
- Graham, R.L. An efficient algorith for determining the convex hull of a finite planar set. Inf. Process. Lett. 1972, 1, 132–133. [Google Scholar]
- Demantké, J.; Mallet, C.; David, N.; Vallet, B. Dimensionality based scale selection in 3D lidar point clouds. ISPRS–Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2011, 37, 97–102. [Google Scholar]
- Schnabel, R.; Wahl, R.; Klein, R. Efficient RANSAC for point-cloud shape detection. In Computer Graphics Forum; Blackwell Publishing Ltd.: Oxford, UK, 2007. [Google Scholar]
- Cao, D.; Wang, C.; Du, M.; Xi, X. A Multiscale Filtering Method for Airborne LiDAR Data Using Modified 3D Alpha Shape. Remote Sens. 2024, 16, 1443. [Google Scholar] [CrossRef]
- Hadas, E.; Borkowski, A.; Estornell, J.; Tymkow, P. Automatic estimation of olive tree dendrometric parameters based on airborne laser scanning data using alpha-shape and principal component analysis. GISci. Remote Sens. 2017, 54, 898–917. [Google Scholar]
- Zhou, Q.-Y.; Park, J.; Koltun, V. Open3D: A modern library for 3D data processing. arXiv 2018, arXiv:1801.09847. [Google Scholar]



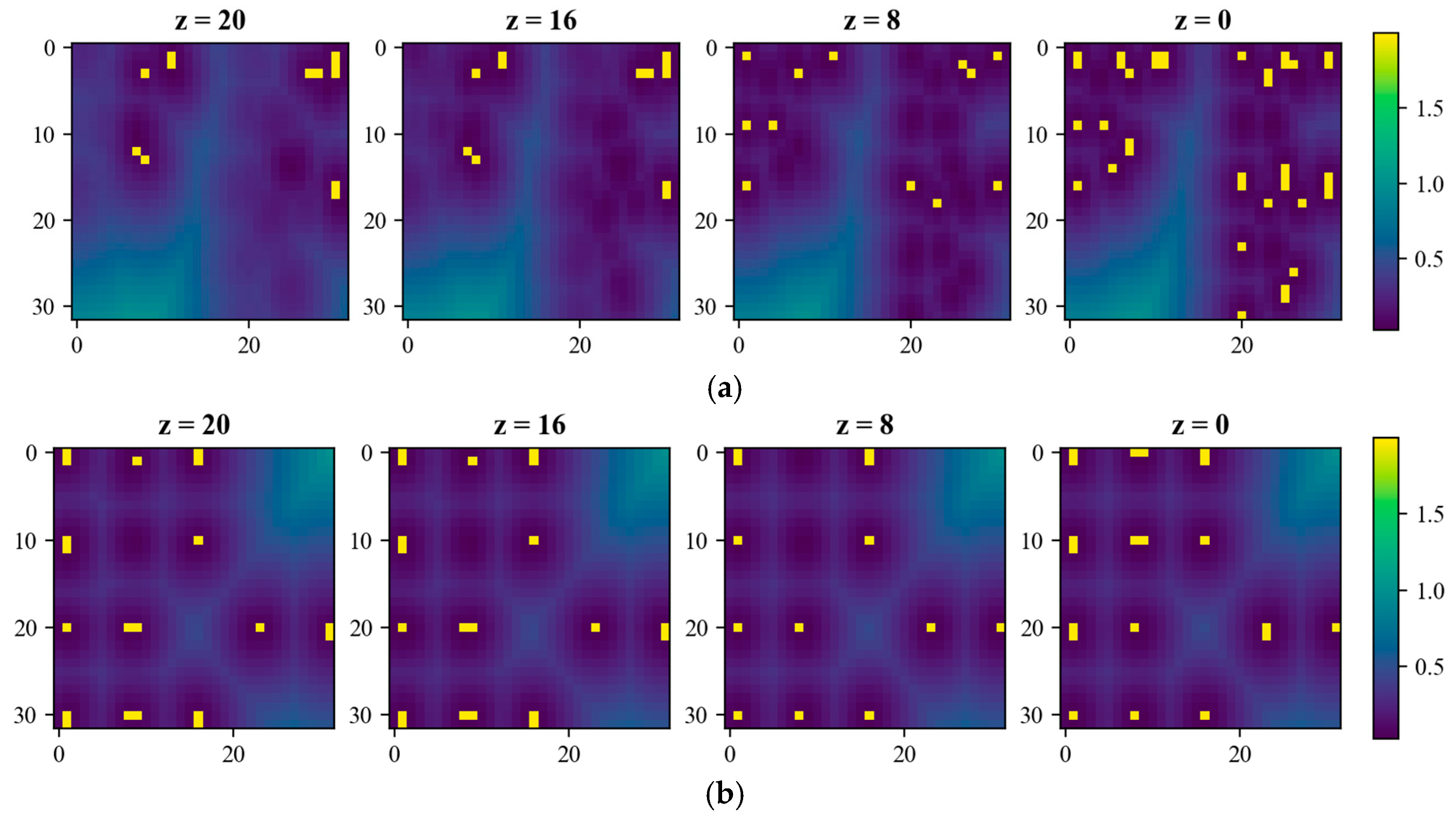
| Source | Data Volume | Density | Type | Angular Resolution |
|---|---|---|---|---|
| 3D Forest | 2 k~39 k | 20 kpt/m2 | Terrain, dead trees, coniferous broad leaves, fallen trees, etc. | / |
| Helios++ | 1920 k~53,912 k | 10 kpt/m2 | Needles, broad leaves, dead trees, etc. | 0.05° |
| Local Shape | Condition | Direction |
|---|---|---|
| Line | ||
| Surface | ||
| Sphere | No direction |
| Metrics | Alpha | ||||||||
| 0.1 | 0.15 | 0.2 | 0.25 | 0.3 | 0.35 | 0.4 | 0.45 | 0.5 | |
| TSC | 0.74 | 0.76 | 0.79 | 0.82 | 0.84 | 0.86 | 0.84 | 0.81 | 0.78 |
| CR | 0.75 | 0.78 | 0.81 | 0.85 | 0.88 | 0.91 | 0.88 | 0.84 | 0.80 |
| Group | | | |||||
|---|---|---|---|---|---|---|
| 0 | 10 k~39 k | 0.08~0.92 | 0.161~0.252 | 0.01 | 1~3 | |
| 1 | 5 k~10 k | 0.107 | 0.214 | 0.01 | 1~4 | |
| 2 | 0~5 k | 0.115 | 0.210 | 0.01 | 2~6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, J.; Ma, S.; Luan, G.; Dong, P.; Geng, R.; Zou, F.; Yin, J.; Zhao, Z. An Improved Method for Single Tree Trunk Extraction Based on LiDAR Data. Remote Sens. 2025, 17, 1271. https://doi.org/10.3390/rs17071271
Xia J, Ma S, Luan G, Dong P, Geng R, Zou F, Yin J, Zhao Z. An Improved Method for Single Tree Trunk Extraction Based on LiDAR Data. Remote Sensing. 2025; 17(7):1271. https://doi.org/10.3390/rs17071271
Chicago/Turabian StyleXia, Jisheng, Sunjie Ma, Guize Luan, Pinliang Dong, Rong Geng, Fuyan Zou, Junzhou Yin, and Zhifang Zhao. 2025. "An Improved Method for Single Tree Trunk Extraction Based on LiDAR Data" Remote Sensing 17, no. 7: 1271. https://doi.org/10.3390/rs17071271
APA StyleXia, J., Ma, S., Luan, G., Dong, P., Geng, R., Zou, F., Yin, J., & Zhao, Z. (2025). An Improved Method for Single Tree Trunk Extraction Based on LiDAR Data. Remote Sensing, 17(7), 1271. https://doi.org/10.3390/rs17071271







