Abstract
The accurate identification of rice growth stages is critical for precision agriculture, crop management, and yield estimation. Remote sensing technologies, particularly multimodal approaches that integrate high spatial and hyperspectral resolution imagery, have demonstrated great potential in large-scale crop monitoring. Multimodal data fusion offers complementary and enriched spectral–spatial information, providing novel pathways for crop growth stage recognition in complex agricultural scenarios. However, the lack of publicly available multimodal datasets specifically designed for rice growth stage identification remains a significant bottleneck that limits the development and evaluation of relevant methods. To address this gap, we present RiceStageSeg, a multimodal benchmark dataset captured by unmanned aerial vehicles (UAVs), designed to support the development and assessment of segmentation models for rice growth monitoring. RiceStageSeg contains paired centimeter-level RGB and 10-band multispectral (MS) images acquired during several critical rice growth stages, including jointing and heading. Each image is accompanied by fine-grained, pixel-level annotations that distinguish between the different growth stages. We establish baseline experiments using several state-of-the-art semantic segmentation models under both unimodal (RGB-only, MS-only) and multimodal (RGB + MS fusion) settings. The experimental results demonstrate that multimodal feature-level fusion outperforms unimodal approaches in segmentation accuracy. RiceStageSeg offers a standardized benchmark to advance future research in multimodal semantic segmentation for agricultural remote sensing. The dataset will be made publicly available on GitHub v0.11.0 (accessed on 1 August 2025).
1. Introduction
The accurate monitoring of crop growth stages is a critical component of precision agricultural management and yield forecasting, playing a pivotal role in guiding field operations, optimizing resource allocation, and improving agricultural productivity [1]. As one of the world’s three staple cereals, rice growth development directly influences agronomic practices, including irrigation, fertilization, and pest and disease management strategies [2]. Consequently, the accurate identification of rice at critical growth stages, such as jointing, heading, and maturity, is essential for safeguarding food security and mitigating agricultural risks under climate variability [3,4].
Traditional methods for determining rice growth stages largely rely on manual field observations, which—despite offering reasonable accuracy at small scales—are plagued by prolonged data collection cycles, high labor and financial costs, significant subjective bias, and limited spatial coverage. The rapid advancement of remote sensing technologies has led to the maturation of multisource image–based crop phenotyping techniques. Particularly, the fusion of multimodal remote sensing imagery, such as RGB and multispectral data, which integrates rich spectral information and spatial–structural characteristics, has emerged as a promising, large-scale, time-efficient, and non-invasive solution for growth monitoring [5].
Extensive research has been conducted on identifying rice cultivation distribution and growth information using satellite remote sensing imagery. For instance, Xu et al. [6] utilized Sentinel-2 data and employed semantic segmentation networks, such as U-Net, DeepLabv3, and Swin Transformer, to identify rice-growing areas, demonstrating the potential of the Transformer architecture in crop remote sensing. Fatchurrachman et al. [7] used Sentinel-1 and Sentinel-2 time series with k-means clustering to achieve the high-precision mapping of rice paddy distribution and phenology in Malaysia. Additionally, Wang et al. [8] processed an SAR VH polarization time series with the dynamic time warping (DTW) algorithm to successfully map rice phenology across six counties in Northeast China. In another study, Zhao et al. [9] proposed an improved phenology-based rice mapping algorithm that integrates both optical and SAR (radar) time-series data, enhancing the robustness of rice maps.
With the rapid advancement of UAV remote sensing technology, centimeter-level high-resolution imagery has shown remarkable potential for monitoring and identifying crop phenological stages. For instance, Wang et al. [10] employed fixed-wing UAVs equipped with multispectral sensors to acquire high-resolution images of rice fields, integrating vegetation indices (e.g., CIRE, NDRE) and texture features into an artificial neural network model to estimate the leaf area index (LAI), the above-ground biomass (AGB), and the plant nitrogen concentration (PNC). The model achieved R2 values of 0.86, 0.92, and 0.86 for these indicators, respectively, with significantly reduced RMSE. Yang et al. [4] proposed a method for identifying the main growth stages (phenology) of rice crops using UAV RGB images and a CNN model. This method can perform large-scale identification using a single image, and the model predictions are highly consistent with ground measurements, with an accuracy rate of 83.9% and a mean absolute error (MAE) of 0.18. In another study [11], spatiotemporal UAV imagery was used to derive vegetation indices, plant height (PH), canopy cover (CC), and days after transplanting (DAT) for AGB estimation. Models based on partial least squares regression (PLSR) outperformed others in experimental plots, while Elastic Net (ENet) models yielded superior accuracy under field conditions. In addition, in the study by Mia et al. [12], a multimodal deep learning model combining UAV multispectral imagery with weekly weather data was used for rice yield prediction. The results showed that the model employing an AlexNet image feature extractor and integrating weekly meteorological information achieved an RMSE of approximately 0.859 t/ha and an RMSPE of 14%.
Despite the progress achieved by the aforementioned methods, current mainstream approaches predominantly rely on single modality data for modeling. This practice has significant limitations in capturing the complex spectral dimensions and spatial structures inherent in agricultural scenes [13,14]. Meanwhile, publicly available data resources for the research community remain scarce. In particular, there is a lack of standardized datasets featuring high spatial resolution, multimodal inputs, and fine-grained semantic annotations. This scarcity severely constrains the verifiability of methodological innovations and the fairness of cross-model comparisons. Furthermore, the existing models often lack a systematic framework for leveraging the informational complementarity between different modalities during the modeling process, and an effective fusion mechanism tailored for agricultural remote sensing scenarios has yet to be established. The combination of these issues makes it difficult to further improve the accuracy and robustness of rice phenology monitoring, highlighting an urgent need for more targeted model designs and data support.
To address the aforementioned challenges, this paper introduces RiceStageSeg, a multimodal benchmark dataset. We systematically collected the RGB and multispectral imagery of rice at key growth stages and produced fine-grained, pixel-level semantic segmentation annotations. This dataset is designed to support a comprehensive evaluation of semantic segmentation models under different modalities and fusion strategies. Furthermore, this paper establishes baseline experiments using several mainstream semantic segmentation models (e.g., U-Net, DeepLabv3+, Swin-Unet). These experiments systematically compare the performance of single modality versus multimodality inputs to validate the advantages of multimodal fusion in enhancing the accuracy of phenology identification.
The main contributions of this paper are summarized as follows:
- We construct and publicly release a high-quality multimodal semantic segmentation dataset, RiceStageSeg, which covers the multiple key growth stages of rice, and includes both RGB and multispectral imagery.
- We propose and evaluate multiple multimodal fusion strategies based on state-of-the-art semantic segmentation models, and provide reproducible benchmark experiments.
- We explore the complementary nature of information across different remote sensing modalities and analyze fusion mechanisms, offering insights for multimodal modeling in agricultural remote sensing.
2. Related Work
2.1. Crop Growth Stage Monitoring Methods
Ground-based manual observation is the most traditional method for crop phenology monitoring, which typically relies on fixed-point, periodic recording of crop changes at key growth stages, such as sowing, emergence, jointing, heading, and maturity. Although this method offers high accuracy on a small scale and is particularly suitable for specific crops and small-scale studies, it is labor-intensive, has limited spatial coverage, and systematic observation is difficult to sustain in complex terrain, such as alpine tundra. The GCPE dataset constructed by Mori et al. (2023) [15] significantly improved data consistency and usability by integrating ground-based growth data for crops, such as maize and wheat, from multiple countries. However, the construction and maintenance of this dataset remain highly dependent on manual operations, making it difficult to meet the demands of large-scale automated agricultural monitoring [15]. Furthermore, manual records are susceptible to subjective judgment, which further limits their application in large-scale agricultural monitoring.
Traditional remote sensing approaches for vegetation phenology monitoring rely heavily on time-series data, typically derived from high-revisit-cycle MODIS satellite imagery. These methods involve calculating vegetation indices, such as the normalized difference vegetation index (NDVI), the enhanced vegetation index (EVI), and solar-induced chlorophyll fluorescence (SIF), followed by constructing vegetation growth curves over time. Subsequent data processing steps, including data transformation and functional fitting, are applied for noise reduction, after which phenological information is extracted using threshold-based methods, curve feature analysis, or shape-model approaches [16,17,18]. Such techniques are highly valuable for large-scale (e.g., national or global) vegetation phenology assessments and for studying historical phenological changes. However, due to the relatively coarse spatial resolution of satellite imagery and their reliance solely on time-series spectral information, these methods are limited in providing timely and precise phenology retrieval when newly acquired imagery is available or when time-series data are incomplete.
In recent years, methods integrating deep learning with remote sensing data have garnered widespread attention. For example, Qu et al. [19] proposed the CTANet model, which incorporates convolutional attention architecture (CAA) and temporal attention architecture (TAA), integrating spatial, channel, and temporal attention modules to address the challenges of crop spatial heterogeneity as well as spectral and temporal variations. The model achieved an overall accuracy of 93.9% in classifying maize, rice, and soybean, with F1 scores of 95.6%, 95.7%, and 94.7%, respectively. Zhou et al. [20] developed rice yield prediction models across multiple varieties by combining UAV-based multispectral imagery with machine learning and deep learning algorithms. The multitemporal 2D CNN (CNN-M2D) model achieved the best performance, with an RRMSE of 8.13% and an R2 of 0.73, demonstrating strong robustness in multivariety scenarios, and effectively capturing spatiotemporal yield variations. Furthermore, with the application of UAV remote sensing technology, submeter high resolution offers a significant advantage for monitoring early growth stages, especially for monitoring the green-up and tillering stages of rice, where it has demonstrated clear superiority over satellite-based remote sensing methods [21,22,23].
2.2. Crop Growth Stage Dataset
Remote sensing datasets are fundamental to crop phenology monitoring and the evaluation of semantic segmentation algorithms, providing crucial support for model training, validation, and cross-regional applications [24]. In recent years, several remote sensing datasets related to crop phenology have been released, advancing the research and technological development in the field of crop monitoring. RiceAtlas [25] is a global database of rice crop calendars that contains information on planting and harvesting dates for rice across multiple countries. This database provides vital data support for the spatiotemporal analysis of rice phenology, and is particularly valuable for the research on rice production patterns and the impacts of climate change. Additionally, the Crop Calendar dataset [26] covers the planting and harvesting dates for major global crops (e.g., maize, wheat, and soybean), and is widely used in the studies on agricultural production models and the effects of climate change on crops. This dataset enables researchers to conduct in-depth analyses of the growth cycles of different crops and their relationships with environmental factors.
For rice phenology research, the SICKLE dataset [27] was developed as a multimodal, multitemporal resource combining Landsat-8, Sentinel-1, and Sentinel-2 imagery from 2018 to 2021 over the Cauvery Delta region in India. Annotated with key phenological parameters, such as sowing, transplanting, and harvesting dates, as well as variety and yield data, it provides 351 labeled paddy samples from 145 plots. This dataset supports tasks, including rice phenology identification, crop type mapping, and yield prediction, offering a valuable benchmark for developing and evaluating remote sensing–based models. Meanwhile, Sen4AgriNet [28] is a multimodal remote sensing dataset that combines Sentinel-1 (radar) and Sentinel-2 (optical) data, primarily designed for multicrop classification and time-series analysis. This dataset facilitates the dynamic monitoring of crop phenology under varying environmental conditions, offering a more comprehensive data source for crop growth monitoring. The PASTIS-R dataset [29] incorporates spatially aligned Sentinel-2 optical and Sentinel-1 C-band radar time series, enabling multimodal analysis for agricultural applications. It provides semantic and instance annotations for tasks such as parcel classification, pixel-based segmentation, and panoptic parcel segmentation. By integrating data from multiple sensors, this dataset provides higher resolution and precision for the accurate monitoring and analysis of crop growth stages. The Agricultural-Vision dataset [30] focuses on agricultural anomaly detection, providing RGB and multispectral images annotated with nine types of agricultural anomalies, such as water stress, weeds, and disease spots. This dataset not only contributes to agricultural health monitoring but offers valuable support for crop growth pattern recognition.
Although the existing datasets for crop phenology monitoring provide valuable resources for agricultural remote sensing, the majority are still predominantly single modality, with the use of optical and radar imagery being the most common. Multimodal datasets, particularly comprehensive ones that integrate data from diverse sensors (e.g., optical remote sensing, radar, and meteorological data), remain relatively scarce. Future research must focus more on fusing multisource data to enhance the accuracy and adaptability of crop phenology monitoring. The development of such multimodal datasets will help overcome the limitations of traditional datasets and further advance the application and development of crop phenology monitoring technologies.
2.3. Remote Sensing Semantic Segmentation
In recent years, the application of semantic segmentation in agricultural remote sensing has expanded significantly, finding widespread use in tasks such as crop type identification, farmland parcel boundary extraction, crop stress monitoring, and precision agriculture management. By performing pixel-level classification on remote sensing imagery, semantic segmentation technology can effectively extract the distribution of different crops within agricultural areas, thereby providing a scientific basis for agricultural production management. Sheng et al. [31] developed the HBRNet, a network that fuses a Swin Transformer backbone with a boundary enhancement module, achieving an mIoU of 75.15% on the DeepGlobe dataset—a 9.2% improvement over the traditional U-Net. These studies have demonstrated that the advancement of deep learning methods has led to substantial improvements in the accuracy of crop classification and boundary extraction from remote sensing images.
To further enhance model performance in complex agricultural environments, multimodal semantic segmentation methods have garnered increasing attention. These methods can fuse data from different sensors to enrich feature representations and to improve model robustness. Such approaches hold promising application prospects in agricultural remote sensing, especially in challenging natural conditions, such as cloudy weather and varying illumination. The U3M model proposed by Li et al. [32] employed an unbiased multiscale fusion strategy. By integrating multiple data sources in multimodal remote sensing scenarios, it successfully enhanced segmentation accuracy and demonstrated superior robustness in crop growth monitoring, with an accuracy gain of over 8%. The LoGoCAF framework, developed by Zhang et al. [33], fuses hyperspectral and thermal infrared data and incorporates a cross-modal attention mechanism, significantly mitigating noise in nighttime remote sensing imagery and increasing the Dice coefficient by 6.8%. Furthermore, the Mirror U-Net model designed by Marinov et al. [34] for medical image segmentation strengthens the discriminative capability for edge regions through its multimodal branches and multitask loss structure. This design holds significant relevance for remote sensing applications, having shown a 4.3% improvement in the Dice coefficient. The application of these methods in agricultural remote sensing—particularly for tasks such as the precise monitoring of crop growth stages, crop classification, and boundary extraction—has demonstrated immense potential and practical value.
3. RiceStageSeg Dataset
This section introduces the construction process of the RiceStageSeg dataset, including the study area, the acquisition and processing of multisource remote sensing data, and the generation of pixel-level growth stage labels.
3.1. Description of the Study Area
This study was conducted in the rice-producing regions of Chongqing Municipality, located in southwestern China. The selected study areas include Nanchuan District, Yongchuan District, Tongnan District, and Kaizhou District (Figure 1), which together represent the major rice cultivation zones in Chongqing. Geographically, Chongqing lies in the upper reaches of the Yangtze River, spanning from 105°11′E to 110°11′E and 28°10′N to 32°13′N. The region features a subtropical monsoon climate with an average annual temperature of 16–18 °C and annual precipitation ranging from 1000 to 1400 mm, making it warm, humid, and highly suitable for rice cultivation. The topography of the study area is complex, predominantly consisting of hilly and mountainous terrain, with rice paddies primarily distributed in river valleys and on gentle slopes. The main cropping system is single-season rice, although double-season rice or rice–rapeseed rotation is also practiced in some areas. The selected districts are characterized by extensive planting areas, high yields, and typical features of the southwestern rice-growing zone. Moreover, due to the influence of local climate and terrain, rice phenology in these areas exhibits distinct spatiotemporal heterogeneity, providing rich data and representative conditions for this study. The selection of these regions not only reflects the typical cultivation characteristics of the southwestern rice zone, but offers a solid scientific basis for the accurate identification of key rice growth stages.
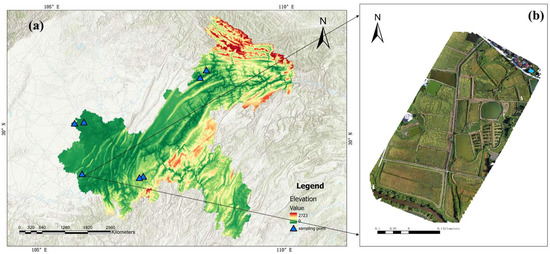
Figure 1.
(a) Map of Chongqing Municipality with marked study areas; (b) Laishu study site in Yongchuan District. All maps are projected using WGS84 UTM Zone 49N.
3.2. Data Acquisition and Preprocessing
High-resolution RGB imagery was acquired using the DJI Matrice 300 RTK multirotor UAV. This UAV, developed by DJI Innovations Co., Ltd. (Shenzhen, China), offers high operational efficiency, stable flight performance, strong maneuverability, and the ability to capture diverse image types. Its capacity to take off and land on varied terrains makes it particularly well-suited for rapid aerial surveys in rural areas. The M300 RTK weighs approximately 6.3 kg, has a diagonal wheelbase of 895 mm, and a maximum flight speed of 23 m/s. Data acquisition was conducted on three occasions: 13–14 June, 18–20 July, and 12–14 Augus 2023, yielding a total of 16 UAV image sets. The UAV was simultaneously equipped with a Zenmuse H20 RGB camera and a MicaSense RedEdge-MX Dual multispectral sensor, enabling the synchronized capture of RGB and multispectral imagery. Following the Standards for Agrometeorological Observations in China, the collected rice field imagery was classified into six key growth stages in chronological order as follows: green-up, tillering, jointing, heading, milky, and maturity stages (as illustrated in Figure 2). Image stitching and processing were performed using Agisoft Metashape Professional v1.6.4, following a standard workflow, including importing image folders, radiometric calibration using a reflectance panel, photo alignment, camera optimization, dense point cloud generation, digital elevation model (DEM) construction, orthomosaic generation, reflectance normalization, and image export. Detailed specifications for the two sensor modalities (RGB and multispectral) are provided in Table 1 and Table 2, respectively.
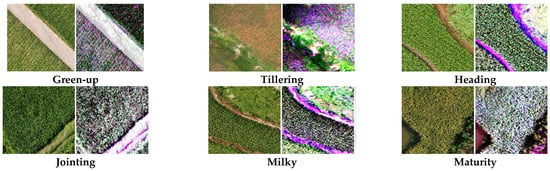
Figure 2.
Rice Growth Stage Visualization: The left column shows the RGB images, and the right column displays the corresponding multispectral images.

Table 1.
RGB Imagery Information Display Table.

Table 2.
Multispectral Imagery Information Display Table. The 10 spectral bands have the following center wavelengths and bandwidths (in nm): 444 (28), 475 (32), 531 (14), 560 (27), 650 (16), 668 (14), 705 (10), 717 (12), 740 (18), and 842 (57).
3.3. Data Annotation
The data annotation was performed based on high-resolution RGB orthomosaics acquired by the UAVs. Manual visual interpretation was conducted using ArcGIS Pro version 2.5.2 by two image analysts with expertise in remote sensing and agronomy. The annotators delineated rice growth stages according to image characteristics, such as texture, color, canopy structure, and field shape, supplemented by the acquisition dates and corresponding meteorological records of each study area. The images were classified into seven categories as follows: green-up, tillering, jointing, heading, milky, maturity, and background.
To ensure annotation quality, all labeled results underwent cross-validation by both interpreters, and field revisits were conducted in selected plots to verify the temporal accuracy and spatial consistency of the labels. Regions with disagreement or ambiguity in stage identification were excluded from the final annotation set to maintain the reliability of the data used for downstream model training. The example of the annotation results is shown in Figure 3.
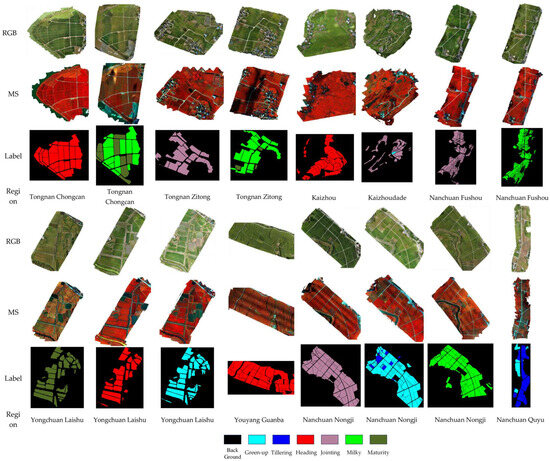
Figure 3.
Visualization of RGB, Multispectral, and Annotation Maps. The false-color image was generated from 10-band NIR data by assigning Band 10 (717 nm) to red, Band 3 (531 nm) to green, and Band 1 (444 nm) to blue. This particular combination was selected to enhance specific features of interest in the study area and to provide better visual contrast for interpretation.
The distribution of each category across all images is summarized in Table 3. It can be observed that our dataset is rich in volume and well-balanced across a wide range of rice growth stages.

Table 3.
Category-wise Pixel Counts.
The dataset will be made publicly available at https://github.com/SHANOAL/RiceStageSeg.git (accessed on 1 August 2025).
4. Baseline Experiments
4.1. Experimental Settings
To establish standard baselines on the RiceStageSeg dataset and to systematically evaluate the impact of different input modalities on rice growth stage segmentation performance, we selected several mainstream semantic segmentation networks as baseline models, including Swin Transformer [35], HRNet [36], UPerNet [37], PSPNet [38], U-Net [39], and ViT [40].
Each model was trained and tested under three input configurations as follows: using only RGB images (RGB-only), using only multispectral images (MS-only), and using a multimodal combination of RGB and MS images (RGB + MS).
In the multimodal fusion experiments, we adopted the traditional early fusion strategy, directly concatenating RGB and multispectral (MS) images along the channel dimension to integrate information from both modalities for segmentation tasks.
All experiments in this study were conducted using the MMSegmentation framework to ensure fair and consistent comparisons across different models. The training was configured with the SGD optimizer (learning rate = 0.001, momentum = 0.9, weight decay = 0.0009) wrapped by the OptimWrapper. A polynomial learning rate (PolyLR) scheduler was employed, with a minimum learning rate of 1 × 10−4, power of 0.9, and a total of 8000 iterations, where the schedule was applied per iteration (by_epoch = False). The loss function used for all experiments was cross-entropy loss, and the batch size was set to 16.
For data preparation, image patches of size 128 × 128 pixels were generated for both training and evaluation. The dataset was randomly divided into training and test sets with an 8:2 ratio, ensuring no overlap between patches during dataset generation. No data augmentation strategies were applied in this study. Model performance was comprehensively evaluated using Intersection over Union (IoU) and F1 score as the evaluation metrics, as follows:
where TP, FP, and FN denote true positives, false positives, and false negatives, respectively; Precision is defined as TP/(TP + FP), and Recall is defined as TP/(TP + FN).
Table 4 outlines the software and hardware configurations used in all experiments.

Table 4.
Experimental Software and Hardware Environment.
4.2. Unimodal Segmentation
In the single modality setting, we trained and evaluated the Swin Transformer, PSPNet, UPerNet, HRNet, U-Net, and ViT models using either RGB images or multispectral (MS) images as the sole input. The RGB-only experiments primarily assessed the performance of visible-light imagery in segmenting rice growth stages, while the MS-only experiments explored the potential of multispectral imagery in distinguishing different growth phases. These single modality experiments enabled us to analyze the differences in feature representation capabilities between modalities, and also provided essential baseline references for evaluating the effectiveness of subsequent multimodal fusion strategies.
4.2.1. RGB-Only Segmentation
In this section, we evaluate the semantic segmentation performance under the RGB-only input setting. Specifically, we use RGB imagery as the sole input modality to train and test four mainstream semantic segmentation models—Swin Transformer, PSPNet, UPerNet, HRNet, U-Net, and ViT—on the RiceStageSeg dataset, aiming to establish a baseline performance for the RGB modality.
The experimental results and key configuration parameters for each model are summarized in Table 5, while the visualized segmentation outcomes are presented in Figure 4.

Table 5.
Configuration and Segmentation Performance Comparison of Models under RGB-only Input.
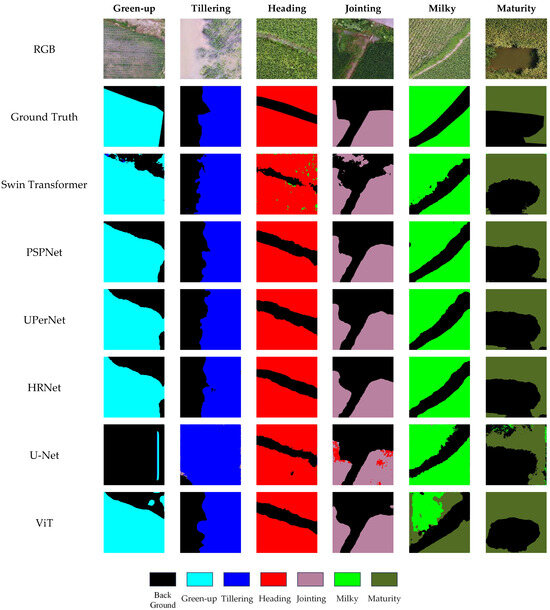
Figure 4.
Segmentation visualization results under RGB-only input.
Based on the results in Table 5, all tested models under the RGB-only input setting achieved strong segmentation performance on the RiceStageSeg dataset, with stable recognition across most rice growth stages. This demonstrates both the adaptability of mainstream semantic segmentation architectures to RGB imagery and the high quality of the RiceStageSeg dataset in terms of annotation accuracy, sample diversity, and class separability.
Among the models, PSPNet achieved the highest overall performance (mIoU 85.31%, F1 92.63%), showing excellent robustness in capturing global context and multiscale information, particularly in mid-to-late growth stages, such as milky and maturity. UPerNet also performed competitively (mIoU 83.23%, F1 90.15%), confirming the advantage of its ResNet backbone combined with multiscale feature integration. HRNet demonstrated strong boundary delineation and accurate recognition of small or transitional areas (mIoU 82.28%), benefiting from its architecture that maintains high-resolution representations throughout the network.
By contrast, the Swin Transformer and ViT, while still achieving satisfactory performance (mIoU 78.56% and 81.25%, respectively), showed slightly lower overall accuracy than the CNN-based models. This may be attributed to the limited dataset size and training resources, which can hinder the training stability and full potential of Transformer-based architectures. Nonetheless, their strong global modeling capability allowed them to perform well in certain stages, such as heading. U-Net, despite its simpler architecture, achieved competitive accuracy in some phases (e.g., heading with IoU 90.16%), confirming its effectiveness in semantic segmentation tasks with fewer parameters.
Overall, these results highlight that the RGB data alone can provide sufficient spatial and texture information to support accurate rice phenology segmentation. However, variations in performance across models and growth stages suggest room for further improvement, particularly through the integration of additional modalities, such as multispectral data, to enhance stage differentiation.
4.2.2. Multispectral-Only Segmentation
In this section, we evaluate the semantic segmentation performance under the MS-only input setting. Specifically, we used multispectral (MS) imagery as the sole input modality to train and test four mainstream semantic segmentation models on the RiceStageSeg dataset: Swin Transformer, PSPNet, UPerNet, HRNet, U-Net, and ViT. This was conducted to establish a performance baseline for the MS modality. The experimental results and the key configuration parameters for each model are presented in Table 6, and the visualized segmentation results are illustrated in Figure 5.

Table 6.
Comparison of Configurations and Segmentation Performance for Various Models with MS-only Input.
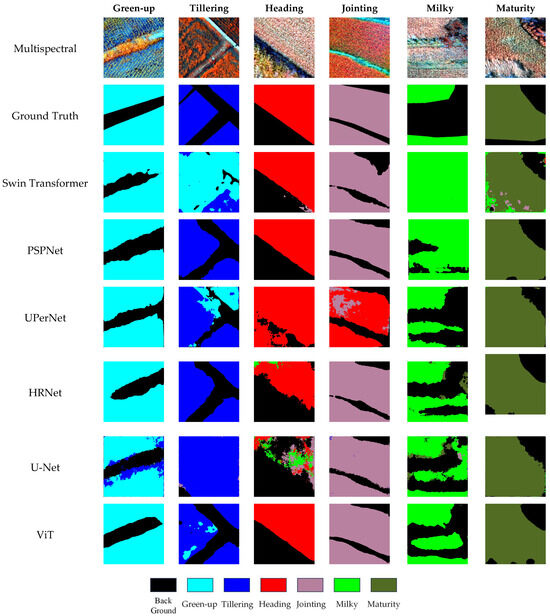
Figure 5.
Segmentation visualization results under MS-only input.
Based on the results in Table 6, using only multispectral (MS) imagery as input generally leads to lower segmentation performance compared to RGB-only configurations. This suggests that the single MS modality has limitations in representing spatial details, likely due to its lower spatial resolution. However, the MS data still demonstrate distinctive advantages, particularly in identifying certain mid-to-late growth stages, such as jointing, milky, and maturity. This is because the MS imagery captures non-visible spectral features, such as canopy reflectance patterns and subtle structural variations, which are not easily detected by the RGB sensors.
Among the models, PSPNet and HRNet maintained relatively stable performance with MS-only input, achieving mIoU scores of 85.77% and 83.72%, respectively. This indicates that their architectures can partially compensate for the lower spatial resolution of the MS data and extract meaningful spectral features. By contrast, UPerNet and Swin Transformer experienced more pronounced performance drops (mIoU 83.67% and 79.18%, respectively), suggesting that these models rely more heavily on high-quality spatial details and rich semantic information, which are less prominent in the MS imagery. U-Net and ViT showed the lowest performance, with mIoU scores of 67.07% and 71.16%, respectively, highlighting their limited ability to effectively exploit spectral information in the absence of strong spatial cues.
Overall, while the MS data alone underperform relative to the RGB, they provide complementary spectral information that is valuable for phenology segmentation. These findings reinforce the potential of integrating the MS with the RGB data to leverage the strengths of both modalities, laying a solid foundation for the development of more effective multimodal fusion strategies.
4.3. RGB + MS Fusion Segmentation
In an early fusion strategy, we utilized all 13 available feature channels as inputs, combining 3 RGB channels with 10 multispectral bands. This approach aims to fully utilize the spatial detail of the RGB data and the spectral richness of the multispectral data. This fused input was then used to train and test each baseline model. The early fusion approach requires no major modifications to the model architecture, making it easy to implement and broadly compatible across different network designs.
Table 7 presents the segmentation performance and key configuration parameters of the four baseline models under the early fusion strategy, while the corresponding visualized segmentation results are shown in Figure 6.

Table 7.
Comparison of Segmentation Performance for Various Models Using an Early Fusion Strategy with the RGB and MS Inputs.
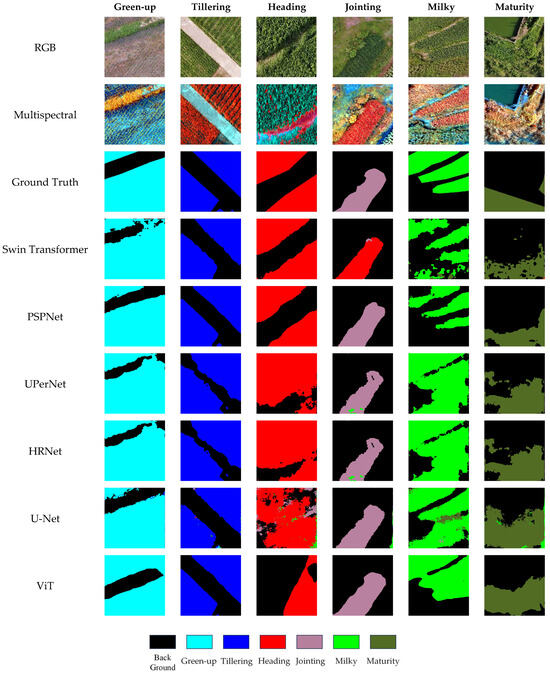
Figure 6.
Segmentation visualization results under RGB + Multispectral early fusion input.
The experimental results in Table 7 show that early fusion of the RGB and multispectral data has heterogeneous effects across different model architectures. UPerNet achieved the highest overall performance under early fusion, with an mIoU of 91.03% and an average F1 score of 95.29%, clearly outperforming several other models. This demonstrates that UPerNet effectively leverages complementary information from both modalities while maintaining robustness to potential noise introduced by fusion. Similarly, PSPNet also performed well, reaching the mIoU (89.63%) and F1 score (94.51%), indicating its strong capacity to exploit cross-modal features when both the RGB spatial details and the multispectral richness are available.
HRNet maintained moderate performance, with an mIoU of 88.97% and F1 of 94.14%. While its high-resolution feature streams help preserve detailed information, the architecture may still experience slight interference from the heterogeneity of fused features. Swin Transformer and ViT, however, showed lower performance (mIoU of 86.30% and 84.01%, respectively), suggesting that transformer-based backbones without explicit cross-modal alignment may suffer from redundant or conflicting information when features are simply concatenated. U-Net yielded the lowest scores (mIoU of 79.09%, F1 of 88.27%), likely because its relatively simple architecture struggles to integrate multimodal information effectively.
Overall, these results confirm that fusing RGB and multispectral data is beneficial, as several models (notably UPerNet and PSPNet) achieved better segmentation accuracy than when using single modalities. At the same time, the differences across models highlight that the success of early fusion heavily depends on the network’s ability to manage cross-modal complementarity and to suppress redundancy. These findings further motivate the exploration of advanced fusion mechanisms to fully unlock the potential of multimodal data.
4.4. Results Summary
By synthesizing the experimental results across the three input configurations, we observe that the early fusion of RGB and multispectral (MS) data can effectively enhance segmentation performance compared to single modality inputs. This improvement demonstrates that combining the spatial detail of the RGB with the spectral richness of the MS data provides a more comprehensive representation of rice growth stages. However, not all existing fusion models fully leverage this potential. Some approaches fail to adequately align or integrate cross-modal features, leading to redundancy or even performance degradation. These findings highlight both the promise of multimodal fusion and the necessity of developing architectures specifically tailored to RGB–MS integration, which will be the focus of our future work.
5. Discussion
5.1. Consistency of Phenological Stage Classification Across Temporal Variations
To further examine whether using single-temporal data is sufficient for accurate rice phenological stage identification, we analyzed the consistency of model performance across observations taken at different times within the same phenological window. Specifically, we selected two UAV images, Image 5 and Image 11, which were acquired from different regions and at different times but represent the same rice growth stage.
The experimental results, as presented in Table 8, Table 9 and Table 10, show no significant differences in accuracy, F1 score, or IoU between these two images. This consistency indicates that our model successfully captures and generalizes the stable texture and spectral features associated with rice phenology, regardless of moderate temporal variations within the same growth stage. These findings suggest that single-temporal data are sufficient for reliable phenological stage classification under the conditions examined in this study.

Table 8.
Segmentation Performance Comparison of Different Models on Image 5 and Image 11 Under RGB Input Configurations.

Table 9.
Segmentation Performance Comparison of Different Models on Image 5 and Image 11 Under MS Input Configurations.

Table 10.
Segmentation Performance Comparison of Different Models on Image 5 and Image 11 Under Early Fusion Input Configurations.
5.2. Analysis of Multispectral Modality Performance and Its Potential
The use of multispectral images alone in the experimental section yielded poor results, as the labels were annotated based on the RGB images, and thus had the same resolution as the RGB data. By contrast, the spatial resolution of the multispectral images is typically only half that of the RGB images, placing them at a disadvantage in this regard. Our research indicates that spatial resolution plays a critical role in identifying rice growth stages. Although the results from the multispectral images are generally inferior to those from the RGB images, this does not mean that the spectral information is unimportant. A reasonable explanation is that the baseline models and pre-trained weights used in our experiments were primarily developed for the RGB data and lack the mechanisms to fully utilize spectral correlations. In future work, we will continue to explore the interaction between spatial resolution and spectral resolution on model performance. This aligns with our original intention in releasing the multimodal benchmark dataset: to empower researchers to explore deeper and more compelling research questions through our dataset and benchmark results.
To further explore the potential of multispectral (MS) data, we incorporated several recently proposed remote sensing foundation models, including HyperSIGMA [41], SpectralGPT [42], and SatMAE [43], using their base configurations as the backbone and the UperHead as the decoder. These models are pretrained on large-scale remote sensing datasets, enabling them to capture rich spectral–spatial relationships that the standard RGB-oriented models may fail to exploit.
The integration of these foundation models significantly improved the segmentation performance when using the MS data, as shown in Table 11. This demonstrates that the lower performance of the MS data in baseline experiments is not due to the lack of valuable spectral information, but rather to the limited capability of conventional models to utilize it effectively. Consequently, with models specifically designed or pretrained to handle spectral data, the advantages of the multispectral imagery can be fully realized (especially the HyperSIGMA and SpectralGPT models), further reinforcing the value of spectral information in rice phenology identification.

Table 11.
Experimental Results of Remote Sensing Foundation Models Applied to Multispectral Imagery.
6. Conclusions
In this study, we proposed RiceStageSeg, a dedicated multimodal remote sensing benchmark dataset for semantic segmentation of rice growth stages. The dataset comprises precisely co-registered RGB and multispectral (MS) imagery, accompanied by pixel-level annotations for various growth stages. Covering multiple critical growth phases, RiceStageSeg features high spatial resolution and spatiotemporal diversity, providing a standardized platform for training and evaluating agricultural remote sensing segmentation models.
Based on this dataset, we systematically evaluated several mainstream semantic segmentation models under different input modalities. The experimental results demonstrate that the RGB modality alone achieves strong segmentation performance; although the MS modality is less effective when used alone, it exhibits complementary advantages in certain stages. Early fusion strategies leveraged the complementary strengths of RGB imagery’s high spatial resolution and multispectral imagery’s rich spectral information. Most models significantly improved segmentation accuracy, demonstrating the advantages of multimodal semantic segmentation.
RiceStageSeg not only fills the gap in publicly available multimodal datasets for rice phenology segmentation but provides a solid data foundation for future research in multimodal model design, modality selection strategies, and cross-season transferability. In future work, we plan to expand the temporal dimension of the dataset and incorporate heterogeneous modalities, such as radar data, aiming to further advance the intelligent applications of agricultural remote sensing in precision farming and disaster response.
Author Contributions
Conceptualization, J.Z. and T.C.; methodology, J.D. and T.C.; software, T.C. and Q.M.; validation, T.C.; formal analysis, J.Z. and T.C.; investigation, J.D.; resources, J.Z. and Y.C.; data curation, Y.L.; writing—original draft preparation, J.Z. and T.C.; writing—review and editing, J.Z. and T.C.; visualization, J.Z. and T.C.; supervision, E.S.; project administration, E.S.; funding acquisition, J.Z. and E.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, grant number 42175193, open found project of CMA Key Open Laboratory of Transforming Climate Resources to Economy, grant number 2023006.
Data Availability Statement
The dataset will be made publicly available at: https://github.com/SHANOAL/RiceStageSeg.git (accessed on accessed on 1 August 2025).
Acknowledgments
Thanks are also due to the National Natural Science Foundation of China (Grant No. 42175193) and the CMA Key Open Laboratory of Transforming Climate Resources to Economy (Grant No. 2023006) for the financial support.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Serrano, L.; Filella, I.; Penuelas, J. Remote sensing of biomass and yield of winter wheat under different nitrogen supplies. Crop Sci. 2000, 40, 723–731. [Google Scholar] [CrossRef]
- Singh, S.; Tyagi, V.; Naresh, R.K. Effect of Irrigation Scheduling and Fertility Management on Growth and Yield Parameters of Basmati Rice. Int. J. Plant Soil Sci. 2023, 35, 503–518. [Google Scholar] [CrossRef]
- Sakamoto, T.; Yokozawa, M.; Toritani, H.; Shibayama, M.; Ishitsuka, N.; Ohno, H. A crop phenology detection method using time-series MODIS data. Remote Sens. Environ. 2005, 96, 366–374. [Google Scholar] [CrossRef]
- Yang, Q.; Shi, L.; Han, J.; Yu, J.; Huang, K. A Near Real-Time Deep Learning Approach for Detecting Rice Phenology Based on UAV Images. Agric. For. Meteorol. 2020, 287, 107938. [Google Scholar] [CrossRef]
- Zeng, L.; Wardlow, B.D.; Xiang, D.; Hu, S.; Li, D. A review of vegetation phenological metrics extraction using time-series, multispectral satellite data. Remote Sens. Environ. 2020, 237, 111511. [Google Scholar] [CrossRef]
- Xu, H.; Song, J.; Zhu, Y. Evaluation and Comparison of Semantic Segmentation Networks for Rice Identification Based on Sentinel-2 Imagery. Remote Sens. 2023, 15, 1499. [Google Scholar] [CrossRef]
- Fatchurrachman; Rudiyanto; Soh, N.C.; Shah, R.M.; Giap, S.G.E.; Setiawan, B.I.; Minasny, B. High-resolution mapping of paddy rice extent and growth stages across peninsular malaysia using a fusion of sentinel-1 and 2 time series data in google earth engine. Remote Sens. 2022, 14, 1875. [Google Scholar] [CrossRef]
- Wang, M.; Wang, J.; Chen, L.; Du, Z. Mapping paddy rice and rice phenology with Sentinel-1 SAR time series using a unified dynamic programming framework. Open Geosci. 2022, 14, 414–428. [Google Scholar] [CrossRef]
- Zhao, Z.; Dong, J.; Zhang, G.; Yang, J.; Liu, R.; Wu, B.; Xiao, X. Improved Phenology-Based Rice Mapping Algorithm by Integrating Optical and Radar Data. Remote Sens. Environ. 2024, 315, 114460. [Google Scholar] [CrossRef]
- Wang, W.K.; Zhang, J.Y.; Hui, W.; Cao, Q.; Tian, Y.C.; Zhu, Y.; Cao, W.X.; Liu, X.J. Non-Destructive Monitoring of Rice Growth Key Indicators Based on Fixed-Wing UAV Multispectral Images. Sci. Agric. Sin. 2023, 56, 4175–4191. [Google Scholar]
- Dai, Y.; Yu, S.; Ma, T.; Ding, J.; Chen, K.; Zeng, G.; Xie, A.; He, P.; Peng, S.; Zhang, M.; et al. Improving the estimation of rice above-ground biomass based on spatio-temporal UAV imagery and phenological stages. Front. Plant Sci. 2024, 15, 1328834. [Google Scholar] [CrossRef]
- Mia, M.S.; Tanabe, R.; Habibi, L.N.; Hashimoto, N.; Homma, K.; Maki, M.; Matsui, T.; Tanaka, T.S.T. Multimodal Deep Learning for Rice Yield Prediction Using UAV-Based Multispectral Imagery and Weather Data. Remote Sens. 2023, 15, 2511. [Google Scholar] [CrossRef]
- Hong, D.; Gao, L.; Yokoya, N.; Yao, J.; Chanussot, J.; Du, Q. More diverse means better: Multimodal deep learning meets remote-sensing imagery classification. IEEE Trans. Geosci. Remote Sens. 2020, 59, 4340–4354. [Google Scholar] [CrossRef]
- Deng, J.; Hong, D.; Li, C.; Yao, J.; Yang, Z.; Zhang, Z.; Chanussot, J. RustQNet: Multimodal deep learning for quantitative inversion of wheat stripe rust disease index. Comput. Electron. Agric. 2024, 225, 109245. [Google Scholar] [CrossRef]
- Mori, A.; Doi, Y.; Iizumi, T. GCPE: The global dataset of crop growth events for agricultural and earth system modeling. J. Agric. Meteorol. 2023, 79, 120–129. [Google Scholar] [CrossRef]
- Sakamoto, T.; Gitelson, A.A.; Arkebauer, T.J. MODIS-based corn grain yield estimation model incorporating crop phenology information. Remote Sens. Environ. 2013, 131, 215–231. [Google Scholar] [CrossRef]
- d’Andrimont, R.; Taymans, M.; Lemoine, G.; Ceglar, A.; Yordanov, M.; van der Velde, M. Detecting flowering phenology in oil seed rape parcels with Sentinel-1 and-2 time series. Remote Sens. Environ. 2020, 239, 111660. [Google Scholar] [CrossRef]
- Jeong, S.J.; Schimel, D.; Frankenberg, C.; Drewry, D.T.; Fisher, J.B.; Verma, M.; Berry, J.A.; Lee, J.-E.; Joiner, J. Application of satellite solar-induced chlorophyll fluorescence to understanding large-scale variations in vegetation phenology and function over northern high latitude forests. Remote Sens. Environ. 2017, 190, 178–187. [Google Scholar] [CrossRef]
- Qu, T.; Wang, H.; Li, X.; Luo, D.; Yang, Y.; Liu, J.; Zhang, Y. A fine crop classification model based on multitemporal Sentinel-2 images. Int. J. Appl. Earth Obs. Geoinf. 2024, 134, 104172. [Google Scholar] [CrossRef]
- Zhou, H.; Huang, F.; Lou, W.; Gu, Q.; Ye, Z.; Hu, H.; Zhang, X. Yield prediction through UAV-based multispectral imaging and deep learning in rice breeding trials. Agric. Syst. 2025, 223, 104214. [Google Scholar] [CrossRef]
- Shi, Y.; Han, L.; Zhang, X.; Sobeih, T.; Gaiser, T.; Thuy, N.H.; Behrend, D.; Srivastava, A.K.; Halder, K.; Ewert, F. Deep Learning Meets Process-Based Models: A Hybrid Approach to Agricultural Challenges. arXiv 2025, arXiv:2504.16141. [Google Scholar] [CrossRef]
- Zhang, C.; Kovacs, J.M. The application of small unmanned aerial systems for precision agriculture: A review. Precis. Agric. 2012, 13, 693–712. [Google Scholar] [CrossRef]
- Song, Y.; Wang, J.; Shang, J.; Liao, C. Using UAV-based SOPC derived LAI and SAFY model for biomass and yield estimation of winter wheat. Remote Sens. 2020, 12, 2378. [Google Scholar] [CrossRef]
- Munteanu, A.; Neagul, M. Semantic Segmentation of Vegetation in Remote Sensing Imagery Using Deep Learning. arXiv 2022, arXiv:2209.14364. [Google Scholar] [CrossRef]
- Laborte, A.G.; Gutierrez, M.A.; Balanza, J.G.; Saito, K.; Zwart, S.J.; Boschetti, M.; Murty, M.; Villano, L.; Aunario, J.K.; Reinke, R. RiceAtlas, a spatial database of global rice calendars and production. Sci. Data 2017, 4, 170074. [Google Scholar] [CrossRef]
- Kotsuki, S.; Tanaka, K. SACRA—A method for the estimation of global high-resolution crop calendars from a satellite-sensed NDVI. Hydrol. Earth Syst. Sci. 2015, 19, 4441–4461. [Google Scholar] [CrossRef]
- Sani, D.; Mahato, S.; Saini, S.; Agarwal, H.K.; Devshali, C.C.; Anand, S.; Arora, G.; Jayaraman, T. SICKLE: A multi-sensor satellite imagery dataset annotated with multiple key cropping parameters. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2024; pp. 5995–6004. [Google Scholar]
- Sykas, D.; Papoutsis, I.; Zografakis, D. Sen4AgriNet: A Harmonized Multi-Country, Multi-Temporal Benchmark Dataset for Agricultural Earth Observation Machine Learning Applications. In Proceedings of the 2021 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Brussels, Belgium, 11–16 July 2021; pp. 5830–5833. [Google Scholar]
- Garnot, V.S.F.; Landrieu, L.; Chehata, N. Multi-modal temporal attention models for crop mapping from satellite time series. ISPRS J. Photogramm. Remote Sens. 2022, 187, 294–305. [Google Scholar] [CrossRef]
- Chiu, M.T.; Xu, X.; Wei, Y.; Huang, Z.; Schwing, A.G.; Brunner, R.; Khachatrian, H.; Karapetyan, H.; Dozier, I.; Rose, G. Agriculture-vision: A large aerial image database for agricultural pattern analysis. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 19–24 June 2020; pp. 2828–2838. [Google Scholar]
- Sheng, J.; Sun, Y.; Huang, H.; Xu, W.; Pei, H.; Zhang, W.; Wu, X. HBRNet: Boundary Enhancement Segmentation Network for Cropland Extraction in High-Resolution Remote Sensing Images. Agriculture 2022, 12, 1284. [Google Scholar] [CrossRef]
- Li, B.; Zhang, D.; Zhao, Z.; Gao, J.; Li, X. U3M: Unbiased Multiscale Modal Fusion Model for Multimodal Semantic Segmentation. Pattern Recognit. 2025, 168, 111801. [Google Scholar] [CrossRef]
- Zhang, X.; Yokoya, N.; Gu, X.; Tian, Q.; Bruzzone, L. Local-to-Global Cross-Modal Attention-Aware Fusion for HSI-X Semantic Segmentation. IEEE Trans. Geosci. Remote Sens. 2024, 62, 5531817. [Google Scholar] [CrossRef]
- Marinov, Z.; Reiß, S.; Kersting, D.; Kleesiek, J.; Stiefelhagen, R. Mirror u-net: Marrying multimodal fission with multi-task learning for semantic segmentation in medical imaging. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Paris, France, 2–3 October 2023; pp. 2283–2293. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Sun, K.; Xiao, B.; Liu, D.; Wang, J. Deep high-resolution representation learning for human pose estimation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 5693–5703. [Google Scholar]
- Xiao, T.; Liu, Y.; Zhou, B.; Jiang, Y.; Sun, J. Unified perceptual parsing for scene understanding. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 418–434. [Google Scholar]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2881–2890. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Wang, D.; Hu, M.; Jin, Y.; Miao, Y.; Yang, J.; Xu, Y.; Qin, X.; Ma, J.; Sun, L.; Li, C.; et al. Hypersigma: Hyperspectral intelligence comprehension foundation model. IEEE Trans. Pattern Anal. Mach. Intell. arXiv 2025, arXiv:2406.11519. [Google Scholar] [CrossRef]
- Hong, D.; Zhang, B.; Li, X.; Li, Y.; Li, C.; Yao, J.; Yokoya, N.; Li, H.; Ghamisi, P.; Jia, X.; et al. SpectralGPT: Spectral remote sensing foundation model. arXiv 2023, arXiv:2311.07113. [Google Scholar] [CrossRef]
- Cong, Y.; Khanna, S.; Meng, C.; Liu, P.; Rozi, E.; He, Y.; Burke, M.; Lobell, D.B.; Ermon, S. Satmae: Pre-training transformers for temporal and multi-spectral satellite imagery. Adv. Neural Inf. Process Syst. 2022, 35, 197–211. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).