Estimation of Tree Diameter at Breast Height (DBH) and Biomass from Allometric Models Using LiDAR Data: A Case of the Lake Broadwater Forest in Southeast Queensland, Australia
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. LiDAR Data Collection
2.3. Field Measurements
2.4. Data Analysis
2.5. DBH Models
2.6. Aboveground Biomass (AGB) Models
2.7. Data Validation (Statistical Analysis)
3. Results
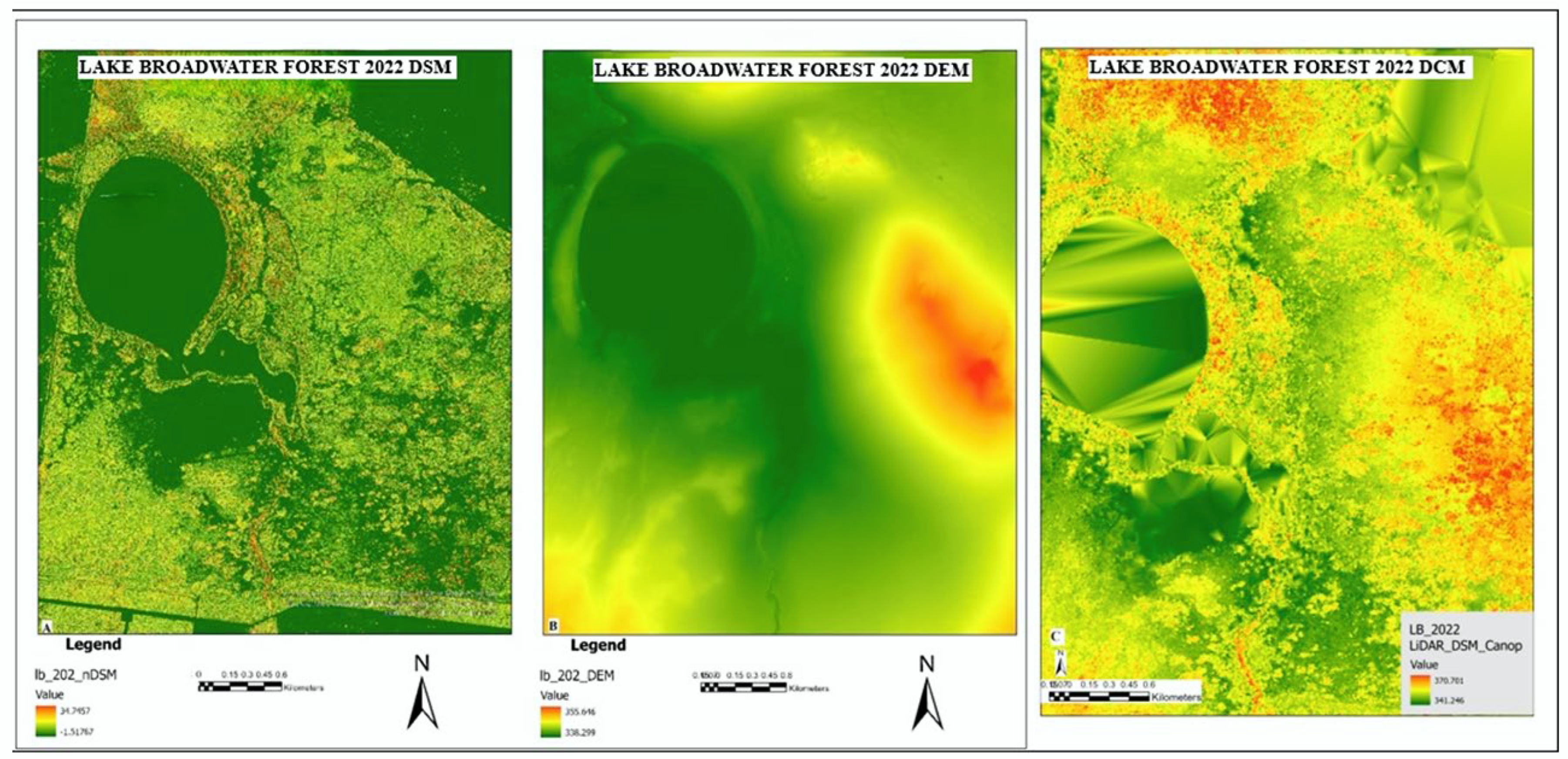
3.1. LiDAR Data Outputs
3.1.1. LiDAR Data Analysis
3.1.2. 2022 LiDAR Forest and Tree Parameters
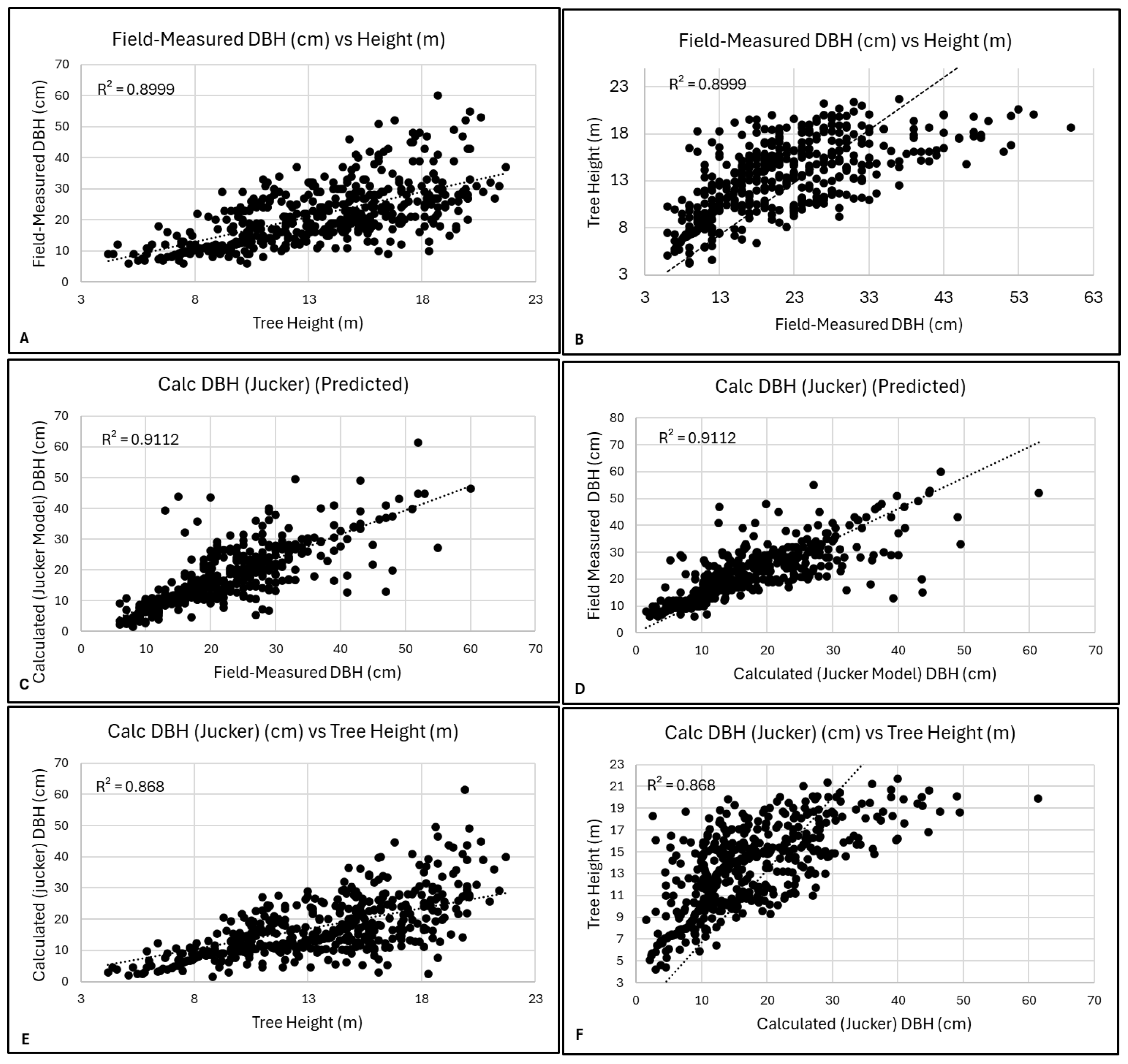
3.2. DBH Calculation Models
3.2.1. Jucker Model for DBH Calculation
3.2.2. Gonzalez-Benecke Equation (1) Model for DBH Calculation
3.2.3. Gonzalez-Benecke Equation (2) Model for DBH Calculation
3.3. Model Validation and Performance
R2 Values for the Jucker and the Gonzalez-Benecke DBH Models
3.4. Aboveground Biomass (AGB)
4. Discussion
4.1. LiDAR Data and Technology
4.2. DBH Estimation Models’ Statistical Metrics
4.2.1. RMSE and MAE
4.2.2. Mean Percentage Bias (MBias)
4.2.3. Mean Absolute Percentage Error (MAPE)
4.2.4. Coefficient of Determination (R2)
4.2.5. DBH Models’ Performance
4.2.6. Graphical Interpretations (Asymptote and Heteroskedasticity)
4.3. ABG and Carbon Sequestration
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AGB | Above-Ground Biomass |
| AGC | Above-ground Carbon |
| AHD | Australian Height Datum |
| ALS | Airborne Laser Scanning |
| Av | Average |
| BGB | Below Ground Biomass |
| C | Carbon |
| CA | Canopy Area |
| CD | Canopy Diameter |
| CD N_S | Canopy Diameter North_South |
| CD E_W | Canopy Diameter East_West |
| CH | Canopy Height |
| CO2 | Carbon Dioxide |
| CV | Canopy Volume |
| D | Diameter |
| DBH | Diameter at Breast Height |
| DCM | Digital Canopy Model |
| DEM | Digital Elevation Model |
| DETSI | Department of Environment, Tourism, Science and Innovation |
| DSM | Digital Surface Model |
| nDSM | Normalized Digital Surface Model |
| ELVIS | Earth Observation and Land-Vectoring Infrastructure System |
| Eu | Eucalyptus |
| GDA | Geocentric Datum of Australia |
| GIS | Geographical Information System |
| GNSS | Global Navigation Satellite System |
| Ha | Hectare |
| HAGL | Height Above Ground Level |
| IPCC | Intergovernmental Panel on Climate Change |
| LAS | LiDAR Aerial Survey |
| LiDAR | Light Detection and Ranging |
| MBias | Mean Percentage Bias |
| MAE | Mean Absolute Error |
| MAPE | Mean Absolute Percentage Error |
| Mg | Mega Gram |
| R2 | Coefficient of Determination |
| RMSE | Root Mean Square Error |
| SOC | Soil Organic Carbon |
| TPH | Trees per Hectare |
References
- Xu, D.; Wang, H.; Xu, W.; Luan, Z.; Xu, X. LiDAR applications to estimate forest biomass at individual tree scale: Opportunities, challenges and future perspectives. Forests 2021, 12, 550. [Google Scholar] [CrossRef]
- Lu, D.; Chen, Q.; Wang, G.; Liu, L.; Li, G.; Moran, E. A survey of remote sensing-based aboveground biomass estimation methods in forest ecosystems. Int. J. Digit. Earth 2016, 9, 63–105. [Google Scholar] [CrossRef]
- Riano, D.; Chuvieco, E.; Condés, S.; González-Matesanz, J.; Ustin, S.L. Generation of crown bulk density for Pinus sylvestris L. from lidar. Remote Sens. Environ. 2004, 92, 345–352. [Google Scholar] [CrossRef]
- Fauzi, N.; Hambali, K.; Nawawi, S.; Busu, I.; Yew, S. Biomass and carbon stock estimation along different altitudinal gradients in tropical forest of Gunung Basor, Kelantan, Malaysia. Malay. Nat. J. 2017, 69, 57–62. [Google Scholar]
- Gubena, A.F.; Soromessa, T. Variations in forest carbon stocks along environmental gradients in egdu forest of oromia region, ethiopia: Implications for sustainable forest management. Am. J. Environ. Prot. 2016, 12, 1–8. [Google Scholar]
- Joshi, R.; Singh, H. Carbon sequestration potential of disturbed and non-disturbed forest ecosystem: A tool for mitigating climate change. Afr. J. Environ. Sci. Technol. 2020, 14, 385–393. [Google Scholar] [CrossRef]
- Fawzy, S.; Osman, A.I.; Doran, J.; Rooney, D.W. Strategies for mitigation of climate change: A review. Environ. Chem. Lett. 2020, 18, 2069–2094. [Google Scholar] [CrossRef]
- McRoberts, R.E.; Næsset, E.; Gobakken, T.; Bollandsås, O.M. Indirect and direct estimation of forest biomass change using forest inventory and airborne laser scanning data. Remote Sens. Environ. 2015, 164, 36–42. [Google Scholar] [CrossRef]
- Gonzalez-Benecke, C.; Gezan, S.A.; Samuelson, L.J.; Cropper, W.P.; Leduc, D.J.; Martin, T.A. Estimating Pinus palustris tree diameter and stem volume from tree height, crown area and stand-level parameters. J. For. Res. 2014, 25, 43–52. [Google Scholar] [CrossRef]
- West, P. Biomass. In Tree and Forest Measurement; Springer: Berlin/Heidelberg, Germany, 2015; pp. 53–70. [Google Scholar]
- Torre-Tojal, L.; Bastarrika, A.; Boyano, A.; Lopez-Guede, J.M.; Grana, M. Above-ground biomass estimation from LiDAR data using random forest algorithms. J. Comput. Sci. 2022, 58, 101517. [Google Scholar] [CrossRef]
- Fonseka, C. Extracting dendrometric parameters of urban trees using remotely sensed data for quantifying their ecological services in Valls Hage. Master’s Thesis, University of Gävle, Gävle, Sweden, 2023. [Google Scholar]
- Walker, S.M.; Murray, L.; Tepe, T. Allometric Equation Evaluation Guidance Document; Winrock International: Little Rock, AR, USA, 2016. [Google Scholar]
- Galán, C.O.; Rodríguez-Pérez, J.R.; Torres, J.M.; Nieto, P.G. Analysis of the influence of forest environments on the accuracy of GPS measurements by using genetic algorithms. Math. Comput. Model. 2011, 54, 1829–1834. [Google Scholar] [CrossRef]
- Mukuralinda, A.; Kuyah, S.; Ruzibiza, M.; Ndoli, A.; Nabahungu, N.L.; Muthuri, C. Allometric equations, wood density and partitioning of aboveground biomass in the arboretum of Ruhande, Rwanda. Trees For. People 2021, 3, 100050. [Google Scholar] [CrossRef]
- Khan, M.N.I.; Islam, M.R.; Rahman, A.; Azad, M.S.; Mollick, A.S.; Kamruzzaman, M.; Sadath, M.N.; Feroz, S.; Rakkibu, M.G.; Knohl, A. Allometric relationships of stand level carbon stocks to basal area, tree height and wood density of nine tree species in Bangladesh. Glob. Ecol. Conserv. 2020, 22, e01025. [Google Scholar] [CrossRef]
- Delcourt, C.J.F.; Veraverbeke, S. Allometric equations and wood density parameters for estimating aboveground and woody debris biomass in Cajander larch (Larix cajanderi) forests of northeast Siberia. Biogeosciences 2022, 19, 4499–4520. [Google Scholar] [CrossRef]
- Torgersen, C.E.; Le Pichon, C.; Fullerton, A.H.; Dugdale, S.J.; Duda, J.J.; Giovannini, F.; Tales, É.; Belliard, J.; Branco, P.; Bergeron, N.E.; et al. Riverscape approaches in practice: Perspectives and applications. Biol. Rev. 2022, 97, 481–504. [Google Scholar] [CrossRef] [PubMed]
- Ferraz, A.; Saatchi, S.; Mallet, C.; Meyer, V. Lidar detection of individual tree size in tropical forests. Remote Sens. Environ. 2016, 183, 318–333. [Google Scholar] [CrossRef]
- Gonzalez de Tanago, J.; Lau, A.; Bartholomeus, H.; Herold, M.; Avitabile, V.; Raumonen, P.; Martius, C.; Goodman, R.C.; Disney, M.; Manuri, S.; et al. Estimation of above-ground biomass of large tropical trees with terrestrial LiDAR. Methods Ecol. Evol. 2018, 9, 223–234. [Google Scholar] [CrossRef]
- Caccamo, G.; Iqbal, I.; Osborn, J.; Bi, H.; Arkley, K.; Melville, G.; Aurik, D.; Stone, C. Comparing yield estimates derived from LiDAR and aerial photogrammetric point-cloud data with cut-to-length harvester data in a Pinus radiata plantation in Tasmania. Aust. For. 2018, 81, 131–141. [Google Scholar] [CrossRef]
- Pu, R. Mapping tree species using advanced remote sensing technologies: A state-of-the-art review and perspective. J. Remote Sens. 2021, 2021, 9812624. [Google Scholar] [CrossRef]
- Yermo, M.; Laso, R.; Lorenzo, O.G.; Pena, T.F.; Cabaleiro, J.C.; Rivera, F.F.; Vilariño, D.L. Powerline detection and characterization in general-purpose airborne LiDAR surveys. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2024, 17, 10137–10157. [Google Scholar] [CrossRef]
- Terryn, L.; Calders, K.; Bartholomeus, H.; Bartolo, R.E.; Brede, B.; D’hont, B.; Disney, M.; Herold, M.; Lau, A.; Shenkin, A.; et al. Quantifying tropical forest structure through terrestrial and UAV laser scanning fusion in Australian rainforests. Remote Sens. Environ. 2022, 271, 112912. [Google Scholar] [CrossRef]
- Baccini, A.; Goetz, S.; Walker, W.; Laporte, N.; Sun, M.; Sulla-Menashe, D.; Hackler, J.; Beck, P.; Dubayah, R.; Friedl, M.; et al. Estimated carbon dioxide emissions from tropical deforestation improved by carbon-density maps. Nat. Clim. Change 2012, 2, 182–185. [Google Scholar] [CrossRef]
- Avitabile, V.; Herold, M.; Heuvelink, G.B.; Lewis, S.L.; Phillips, O.L.; Asner, G.P.; Armston, J.; Ashton, P.S.; Banin, L.; Bayol, N.; et al. An integrated pan-tropical biomass map using multiple reference datasets. Glob. Change Biol. 2016, 22, 1406–1420. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Chevallier, F.; Gomez, C.; Guanter, L.; Hicke, J.A.; Huete, A.R.; Ichii, K.; Ni, W.; Pang, Y.; Rahman, A.F.; et al. Remote sensing of the terrestrial carbon cycle: A review of advances over 50 years. Remote Sens. Environ. 2019, 233, 111383. [Google Scholar] [CrossRef]
- Santoro, M.; Cartus, O.; Carvalhais, N.; Rozendaal, D.M.; Avitabile, V.; Araza, A.; De Bruin, S.; Herold, M.; Quegan, S.; Rodríguez-Veiga, P.; et al. The global forest above-ground biomass pool for 2010 estimated from high-resolution satellite observations. Earth Syst. Sci. Data 2021, 13, 3927–3950. [Google Scholar] [CrossRef]
- Jurjević, L.; Liang, X.; Gašparović, M.; Balenović, I. Is field-measured tree height as reliable as believed–Part II, A comparison study of tree height estimates from conventional field measurement and low-cost close-range remote sensing in a deciduous forest. ISPRS J. Photogramm. Remote Sens. 2020, 169, 227–241. [Google Scholar] [CrossRef]
- Liao, K.; Li, Y.; Zou, B.; Li, D.; Lu, D. Examining the role of UAV Lidar data in improving tree volume calculation accuracy. Remote Sens. 2022, 14, 4410. [Google Scholar] [CrossRef]
- Shao, T.; Qu, Y.; Du, J. A low-cost integrated sensor for measuring tree diameter at breast height (DBH). Comput. Electron. Agric. 2022, 199, 107140. [Google Scholar] [CrossRef]
- Cao, L.; Gao, S.; Li, P.; Yun, T.; Shen, X.; Ruan, H. Aboveground biomass estimation of individual trees in a coastal planted forest using full-waveform airborne laser scanning data. Remote Sens. 2016, 8, 729. [Google Scholar] [CrossRef]
- Cao, A.; Esteban, M.; Onuki, M.; Takagi, H.; Thao, N.D.; Tsuchiya, N. Future of Asian Deltaic Megacities under sea level rise and land subsidence: Current adaptation pathways for Tokyo, Jakarta, Manila, and Ho Chi Minh City. Curr. Opin. Environ. Sustain. 2021, 50, 87–97. [Google Scholar] [CrossRef]
- Ahmad, A.; Gilani, H.; Ahmad, S.R. Forest aboveground biomass estimation and mapping through high-resolution optical satellite imagery—A literature review. Forests 2021, 12, 914. [Google Scholar] [CrossRef]
- Coops, N.C.; Tompalski, P.; Goodbody, T.R.; Queinnec, M.; Luther, J.E.; Bolton, D.K.; White, J.C.; Wulder, M.A.; van Lier, O.R.; Hermosilla, T. Modelling lidar-derived estimates of forest attributes over space and time: A review of approaches and future trends. Remote Sens. Environ. 2021, 260, 112477. [Google Scholar] [CrossRef]
- Willkens, D.S.; Liu, J.; Alathamneh, S. A case study of integrating terrestrial laser scanning (TLS) and building information modeling (BIM) in heritage bridge documentation: The Edmund Pettus Bridge. Buildings 2024, 14, 1940. [Google Scholar] [CrossRef]
- Bi, H.; Fox, J.C.; Li, Y.; Lei, Y.; Pang, Y. Evaluation of nonlinear equations for predicting diameter from tree height. Can. J. For. Res. 2012, 42, 789–806. [Google Scholar] [CrossRef]
- Gonzalez-Benecke, C.A.; Fernández, M.P.; Gayoso, J.; Pincheira, M.; Wightman, M.G. Using Tree Height, Crown Area and Stand-Level Parameters to Estimate Tree Diameter, Volume, and Biomass of Pinus radiata, Eucalyptus globulus and Eucalyptus nitens. Forests 2022, 13, 2043. [Google Scholar] [CrossRef]
- Jucker, T.; Caspersen, J.; Chave, J.; Antin, C.; Barbier, N.; Bongers, F.; Dalponte, M.; van Ewijk, K.Y.; Forrester, D.I.; Haeni, M.; et al. Allometric equations for integrating remote sensing imagery into forest monitoring programmes. Glob. Change Biol. 2017, 23, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Department of the Environment, T.; Science and Innovation. Lake Broadwater Tourist Park. 2024. Available online: https://www.desi.qld.gov.au/ (accessed on 26 April 2024).
- Howard, M.; Pearl, H.; McDonald, B.; Shimizu, Y.; Srivastava, S.K.; Shapcott, A. The Conservation of Biodiverse and Threatened Dry Rainforest Plant Communities Is Vital in a Changing Climate. Conservation 2024, 4, 657–684. [Google Scholar] [CrossRef]
- Collette, J.C.; Emery, N.J. A new project investigating the floral phenology and seed biology of threatened ecological communities in northwest NSW. Australas. Plant Conserv. J. Aust. Netw. Plant Conserv. 2020, 29, 21–25. [Google Scholar] [CrossRef]
- Ahmad, F.; Goparaju, L.; Qayum, A. Natural resource mapping using Landsat and LIDAR towards identifying digital elevation, digital surface and canopy height models. Int. J. Environ. Sci. Nat. Res. 2017, 2, 555580. [Google Scholar]
- Nam, V.T.; Van Kuijk, M.; Anten, N.P. Allometric equations for aboveground and belowground biomass estimations in an evergreen forest in Vietnam. PLoS ONE 2016, 11, e0156827. [Google Scholar] [CrossRef] [PubMed]
- Balenović, I.; Simic Milas, A.; Marjanović, H. A comparison of stand-level volume estimates from image-based canopy height models of different spatial resolutions. Remote Sens. 2017, 9, 205. [Google Scholar] [CrossRef]
- Tristan, G. Calculate Vegetation Biomass from LiDAR Data in Python. 2024. Available online: https://www.neonscience.org/resources/learning-hub/tutorials/calc-biomass-py (accessed on 24 July 2024).
- Pucha-Cofrep, F.; Fries, A. ArcGIS Pro Manual; Franz Pucha Cofrep: Senftenberg, Germany, 2025; ISBN 9942510400. [Google Scholar]
- Baskerville, G. Use of logarithmic regression in the estimation of plant biomass. Can. J. For. Res. 1972, 2, 49–53. [Google Scholar] [CrossRef]
- Chave, J.; Andalo, C.; Brown, S.; Cairns, M.A.; Chambers, J.Q.; Eamus, D.; Fölster, H.; Fromard, F.; Higuchi, N.; Kira, T.; et al. Tree allometry and improved estimation of carbon stocks and balance in tropical forests. Oecologia 2005, 145, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Dawkins, H. Estimating total volume of some Caribbean trees. Caribb. For. 1961, 22, 62–63. [Google Scholar]
- Chave, J.; Réjou-Méchain, M.; Búrquez, A.; Chidumayo, E.; Colgan, M.S.; Delitti, W.B.; Duque, A.; Eid, T.; Fearnside, P.M.; Goodman, R.C.; et al. Improved allometric models to estimate the aboveground biomass of tropical trees. Glob. Change Biol. 2014, 20, 3177–3190. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Li, Z.; Shen, B.; Wang, X.; Xu, D.; Yan, R.; Yan, Y.; Xin, X.; Xiao, J.; Li, M.; et al. Spatial patterns and driving factors of aboveground and belowground biomass over the eastern Eurasian steppe. Sci. Total Environ. 2022, 803, 149700. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.; Shakhela, R.; Jat, J. Growth, biomass production and CO2 sequestration of some important multipurpose trees under rainfed condition. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 1943–1950. [Google Scholar] [CrossRef]
- Boomiraj, K.; Jagadeeswaran, R.; Karthik, S.; Poornima, R.; Jothimani, S.; Sudhagar, R.J. Assessing the Carbon Sequestration Potential of Coconut Plantation in Vellore District of Tamil Nadu, India. Int. J. Environ. Clim. Change 2020, 10, 618–624. [Google Scholar] [CrossRef]
- Khokher, F.K.; Khan, A.U.; Bargali, H.S.; Ahmed, S.; Gobato, R.; Zaman, S.; Mitra, A. Stored carbon in dominant mangrove species in Indian Sundarbans: A situation analysis with two different methods. Parana. J. Sci. Educ. 2023, 9, 1–9. [Google Scholar]
- Lawal, A.; Ayeni, O.; Ayanniyi, O. Carbon-dioxide sequestration potential of trees species in Nigerian Tertiary Institutions: A case study of Ondo State, Nigeria. World News Nat. Sci. 2023, 50, 247–257. [Google Scholar]
- Dada, A.D.; Matthew, O.J.; Odiwe, A.I. Nexus between carbon stock, biomass, and CO2 emission of woody species composition: Evidence from Ise-Ekiti Forest Reserve, Southwestern Nigeria. Carbon. Res. 2024, 3, 40. [Google Scholar] [CrossRef]
- Ahmed, M.; Miah, M.; Abdullah, H.; Hossain, M.; Rubayet, M. Measurement of Sequestered Standing Carbon Stock in Major Agroforestry Tree Species. Can. J. Agric. Crops 2020, 5, 153–159. [Google Scholar]
- Konishi, S.; Kitagawa, G. Information Criteria and Statistical Modeling; Springer Science & Business Media: New York, NY, USA, 2008. [Google Scholar]
- Li, N.; Ho, C.P.; Xue, J.; Lim, L.W.; Chen, G.; Fu, Y.H.; Lee, L.Y.T. A progress review on solid-state LiDAR and nanophotonics-based LiDAR sensors. Laser Photonics Rev. 2022, 16, 2100511. [Google Scholar] [CrossRef]
- Sharma, M.; Garg, R.D.; Badenko, V.; Fedotov, A.; Min, L.; Yao, A. Potential of airborne LiDAR data for terrain parameters extraction. Quat. Int. 2021, 575, 317–327. [Google Scholar] [CrossRef]
- West, P. Modelling maximum stem basal area growth rates of individual trees of Eucalyptus pilularis Smith. For. Sci. 2021, 67, 633–636. [Google Scholar] [CrossRef]
- Hodson, T.O. Root mean square error (RMSE) or mean absolute error (MAE): When to use them or not. Geosci. Model. Dev. Discuss. 2022, 15, 5481–5487. [Google Scholar] [CrossRef]
- Zhang, Z. A law of likelihood for composite hypotheses. arXiv 2009, arXiv:0901.0463. [Google Scholar]
- De Myttenaere, A.; Golden, B.; Le Grand, B.; Rossi, F. Mean absolute percentage error for regression models. Neurocomputing 2016, 192, 38–48. [Google Scholar] [CrossRef]
- Hyndman, R.; Athanasopoulos, G. Forecasting: Principles and Practice; OTexts: Melbourne, Australia, 2018. [Google Scholar]
- Makridakis, S.; Wheelwright, S.C.; Hyndman, R.J. Forecasting Methods and Applications; John Wiley & Sons: Hoboken, NJ, USA, 2008. [Google Scholar]
- Tofallis, C. A better measure of relative prediction accuracy for model selection and model estimation. J. Oper. Res. Soc. 2015, 66, 1352–1362. [Google Scholar] [CrossRef]
- Burnham, K.P.; Anderson, D.R.; Huyvaert, K.P. AIC model selection and multimodel inference in behavioral ecology: Some background, observations, and comparisons. Behav. Ecol. Sociobiol. 2011, 65, 23–35. [Google Scholar] [CrossRef]
- Chatterjee, S.; Simonoff, J.S. Handbook of Regression Analysis with Applications in R.; John Wiley & Sons: Hoboken, NJ, USA, 2020. [Google Scholar]
- Montgomery, D.C.; Peck, E.A.; Vining, G.G. Introduction to Linear Regression Analysis; John Wiley & Sons: Hoboken, NJ, USA, 2021. [Google Scholar]
- Dormann, C.F.; Elith, J.; Bacher, S.; Buchmann, C.; Carl, G.; Carré, G.; Marquéz, J.R.G.; Gruber, B.; Lafourcade, B.; Leitão, P.J.; et al. Collinearity: A review of methods to deal with it and a simulation study evaluating their performance. Ecography 2013, 36, 27–46. [Google Scholar] [CrossRef]
- King, D.A. Linking tree form, allocation and growth with an allometrically explicit model. Ecol. Model. 2005, 185, 77–91. [Google Scholar] [CrossRef]
- King, D.A.; Clark, D.A. Allometry of emergent tree species from saplings to above-canopy adults in a Costa Rican rain forest. J. Trop. Ecol. 2011, 27, 573–579. [Google Scholar] [CrossRef]
- Duncanson, L.; Rourke, O.; Dubayah, R. Small sample sizes yield biased allometric equations in temperate forests. Sci. Rep. 2015, 5, 17153. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Zhou, X.; Liu, Z.; Li, P.; Liu, C.; Huang, H.; Tang, J.; Zhang, C.; Zou, Z.; Xie, B.; et al. Dynamic carbon allocation trade-off: A robust approach to model tree biomass allometry. Methods Ecol. Evol. 2024, 15, 886–899. [Google Scholar] [CrossRef]
- Demol, M.; Aguilar-Amuchastegui, N.; Bernotaite, G.; Disney, M.; Duncanson, L.; Elmendorp, E.; Espejo, A.; Furey, A.; Hancock, S.; Hansen, J.; et al. Multi-scale lidar measurements suggest miombo woodlands contain substantially more carbon than thought. Commun. Earth Environ. 2024, 5, 366. [Google Scholar] [CrossRef]
- Peng, Y.; Fornara, D.A.; Yue, K.; Peng, X.; Peng, C.; Wu, Q.; Ni, X.; Liao, S.; Yang, Y.; Wu, F.; et al. Globally limited individual and combined effects of multiple global change factors on allometric biomass partitioning. Glob. Ecol. Biogeogr. 2022, 31, 454–469. [Google Scholar] [CrossRef]
- Thornton, C.; Elledge, A. Heavy grazing of buffel grass pasture in the Brigalow Belt bioregion of Queensland, Australia, more than tripled runoff and exports of total suspended solids compared to conservative grazing. Mar. Pollut. Bull. 2021, 171, 112704. [Google Scholar] [CrossRef] [PubMed]
- Dalal, R.; Chan, K.Y.; So, H.B.; Carroll, C.; Rose, C.W.; Greene, R.; Murphy, B. Global Degradation of Soil and Water Resources; Springer: Singapore, 2022. [Google Scholar]
- Carroll, C.; Rose, C.W.; Greene, R.; Murphy, B.; Dalal, R.; Chan, K.Y.; So, H.B. Issues and Challenges in the Sustainable Use of Soil and Water Resources in Australian Agricultural Lands. In Global Degradation of Soil and Water Resources: Regional Assessment and Strategies; Springer Nature: Singapore, 2022; pp. 537–564. [Google Scholar]
- Muir, K. Submission to the Independent Review of the Environment Protection and Biodiversity Conservation Act; Local Government NSW: Sydney, Australia, 1999. [Google Scholar]
- Macintosh, A. Australia’s National Environmental Legislation: A Response to Early. J. Int. Wildl. Law Policy 2009, 12, 166–179. [Google Scholar] [CrossRef]
- Ngugi, M.R.; Johnson, R.W.; McDonald, W.J. Restoration of ecosystems for biodiversity and carbon sequestration: Simulating growth dynamics of brigalow vegetation communities in Australia. Ecol. Model. 2011, 222, 785–794. [Google Scholar] [CrossRef]
- Yang, S.; Sterck, F.J.; Sass-Klaassen, U.; Cornelissen, J.H.C.; van Logtestijn, R.S.; Hefting, M.; Goudzwaard, L.; Zuo, J.; Poorter, L. Stem trait spectra underpin multiple functions of temperate tree species. Front. Plant Sci. 2022, 13, 769551. [Google Scholar] [CrossRef] [PubMed]
- MacFarlane, D.W. Functional relationships between branch and stem wood density for temperate tree species in North America. Front. For. Glob. Change 2020, 3, 63. [Google Scholar] [CrossRef]
- Slik, J.F.; Paoli, G.; McGuire, K.; Amaral, I.; Barroso, J.; Bastian, M.; Blanc, L.; Bongers, F.; Boundja, P.; Clark, C.; et al. Large trees drive forest aboveground biomass variation in moist lowland forests across the tropics. Glob. Ecol. Biogeogr. 2013, 22, 1261–1271. [Google Scholar] [CrossRef]
- Bastin, J.-F.; Barbier, N.; Réjou-Méchain, M.; Fayolle, A.; Gourlet-Fleury, S.; Maniatis, D.; De Haulleville, T.; Baya, F.; Beeckman, H.; Beina, D.; et al. Seeing Central African forests through their largest trees. Sci. Rep. 2015, 5, 13156. [Google Scholar] [CrossRef] [PubMed]
- Feldpausch, T.R.; Banin, L.; Phillips, O.L.; Baker, T.R.; Lewis, S.L.; Quesada, C.A.; Affum-Baffoe, K.; Arets, E.J.; Berry, N.J.; Bird, M.; et al. Height-diameter allometry of tropical forest trees. Biogeosciences 2011, 8, 1081–1106. [Google Scholar] [CrossRef]
- Fournier, M.; Stokes, A.; Coutand, C.; Fourcaud, T.; Moulia, B. Tree biomechanics and growth strategies in the context of forest functional ecology. In Ecology and Biomechanics: A Mechanical Approach to the Ecology of Animals and Plants; CRC Press: Boca Raton, FL, USA, 2006; pp. 1–34. [Google Scholar]
- Zianis, D.; Mencuccini, M. On simplifying allometric analyses of forest biomass. For. Ecol. Manag. 2004, 187, 311–332. [Google Scholar] [CrossRef]
- Ismail, M.J.; Poudel, T.R.; Ali, A.; Dong, L. Incorporating stand parameters in nonlinear height-diameter mixed-effects model for uneven-aged Larix gmelinii forests. Front. For. Glob. Change 2025, 7, 1491648. [Google Scholar] [CrossRef]
- Dutcă, I.; McRoberts, R.E.; Næsset, E.; Blujdea, V.N. Accommodating heteroscedasticity in allometric biomass models. For. Ecol. Manag. 2022, 505, 119865. [Google Scholar] [CrossRef]
- Pekár, S.; Brabec, M. Marginal models via GLS: A convenient yet neglected tool for the analysis of correlated data in the behavioural sciences. Ethology 2016, 122, 621–631. [Google Scholar] [CrossRef]
- Brown, S.; Sathaye, J.; Cannell, M.; Kauppi, P.E. Mitigation of carbon emissions to the atmosphere by forest management. Commonw. For. Rev. 1996, 96, 80–91. [Google Scholar]
- Houghton, R.; Davidson, E.; Woodwell, G. Missing sinks, feedbacks, and understanding the role of terrestrial ecosystems in the global carbon balance. Glob. Biogeochem. Cycles 1998, 12, 25–34. [Google Scholar] [CrossRef]
- Houghton, R. Aboveground forest biomass and the global carbon balance. Glob. Change Biol. 2005, 11, 945–958. [Google Scholar] [CrossRef]
- Canadell, J.G.; Raupach, M.R. Managing forests for climate change mitigation. Science 2008, 320, 1456–1457. [Google Scholar] [CrossRef] [PubMed]
- Alemu, B. The role of forest and soil carbon sequestrations on climate change mitigation. Res. J. Agr. Environ. Manage 2014, 3, 492–505. [Google Scholar]
- Grassi, G.; House, J.; Dentener, F.; Federici, S.; den Elzen, M.; Penman, J. The key role of forests in meeting climate targets requires science for credible mitigation. Nat. Clim. Change 2017, 7, 220–226. [Google Scholar] [CrossRef]
- Sadatshojaei, E.; Wood, D.A.; Rahimpour, M.R. Potential and challenges of carbon sequestration in soils. In Applied Soil Chemistry; Wiley-Scrivener: Hoboken, NJ, USA, 2021; pp. 1–21. [Google Scholar]
- Khan, I.A.; Khan, S.M.; Jahangir, S.; Ali, S.; Tulindinova, G.K. Carbon Storage and Dynamics in Different Agroforestry Systems. In Agroforestry; Wiley: Hoboken, NJ, USA, 2024; pp. 345–374. [Google Scholar]
- Zhao, J.; Liu, D.; Cao, Y.; Zhang, L.; Peng, H.; Wang, K.; Xie, H.; Wang, C. An integrated remote sensing and model approach for assessing forest carbon fluxes in China. Sci. Total Environ. 2022, 811, 152480. [Google Scholar] [CrossRef] [PubMed]
- Lokuge, N.; Anders, S. Carbon-credit systems in agriculture: A review of literature. Sch. Public. Policy Publ. 2022, 15. [Google Scholar] [CrossRef]
- Geroe, S. Regulatory Support for Biosequestration Projects in Australia: A Useful Model for Transition to Net-Zero Emissions? Sriwij. Law Rev. 2022, 6, 1–23. [Google Scholar] [CrossRef]
- Hemming, P.; Armistead, A.; Venketasubramanian, S. An Environmental Fig Leaf? Restoring Integrity to the Emissions Reduction Fund; The Australia Institute: Griffith, Australia, 2022. [Google Scholar]
- Chakraborti, J.; Mondal, S.; Palit, D. Green Approaches to Mitigate Climate Change Issues in Indian Subcontinent. In Ecosystem Management: Climate Change and Sustainability; Wiley: Hoboken, NJ, USA, 2024; pp. 79–114. [Google Scholar]
- Figueredo, A. A review of research on farmers’ perspectives and attitudes towards carbon farming as a climate change mitigation strategy. In Urban and Rural Reports; Department of Urban and Rural Development, Swedish University of Agricultural Sciences: Uppsala, Sweden, 2024. [Google Scholar]
- Shockley, J.; Snell, W. Carbon Markets 101; Economic and Policy Update (21); University of Kentucky: Lexington, KY, USA, 2021; Volume 21, Available online: https://agecon.ca.uky.edu/sites/agecon.ca.uky.edu/files/Carbon%20Markets%20101.pdf (accessed on 28 January 2025).
- Aiken, J.D. Growing Climate Solutions Act of 2021; 2021. Available online: https://digitalcommons.unl.edu/cgi/viewcontent.cgi?article=2114&context=agecon_cornhusker (accessed on 15 June 2025).






| Plot Number | Plot Size (ha) | Number of Trees | Trees/ha−1 | Av Tree H (m) | CD (m) | CD N_S (m) | CD E_W (m) | CA (m2) | CV (m3) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.13 | 38 | 302 | 17.69 | 3.71 | 4.02 | 4.24 | 12.88 | 28.06 |
| 2 | 0.13 | 65 | 517 | 8.05 | 3.88 | 4.17 | 4.48 | 14.10 | 43.71 |
| 3 | 0.13 | 74 | 589 | 13.63 | 3.61 | 3.45 | 4.57 | 12.52 | 37.70 |
| 4 | 0.13 | 52 | 414 | 14.37 | 3.48 | 3.43 | 4.38 | 11.00 | 23.60 |
| 5 | 0.13 | 89 | 708 | 10.54 | 3.10 | 3.25 | 3.89 | 9.25 | 22.83 |
| 6 | 0.13 | 45 | 358 | 16.45 | 3.61 | 3.98 | 4.08 | 12.17 | 25.48 |
| 7 | 0.13 | 65 | 517 | 11.94 | 3.59 | 3.86 | 4.29 | 12.40 | 34.39 |
| 8 | 0.13 | 46 | 366 | 9.43 | 3.27 | 3.36 | 3.96 | 10.02 | 22.19 |
| 9 | 0.13 | 62 | 493 | 12.85 | 3.49 | 3.91 | 3.97 | 11.26 | 22.32 |
| 10 | 0.13 | 39 | 310 | 15.29 | 3.74 | 3.89 | 4.62 | 13.40 | 33.28 |
| 11 | 0.13 | 70 | 557 | 10.99 | 3.25 | 3.33 | 3.89 | 10.02 | 22.62 |
| 12 | 0.13 | 56 | 446 | 15.47 | 3.98 | 4.51 | 4.53 | 15.19 | 40.24 |
| 13 | 0.13 | 68 | 541 | 11.28 | 3.52 | 3.64 | 4.24 | 11.1 | 23.93 |
| 14 | 0.13 | 55 | 438 | 17.29 | 3.83 | 4.14 | 4.50 | 13.34 | 30.66 |
| 15 | 0.13 | 74 | 589 | 9.85 | 3.64 | 4.19 | 4.12 | 12.72 | 30.05 |
| 16 | 0.13 | 69 | 549 | 14.59 | 2.99 | 3.22 | 3.58 | 8.55 | 13.90 |
| 17 | 0.13 | 60 | 477 | 11.29 | 3.55 | 3.99 | 3.98 | 11.64 | 25.37 |
| 18 | 0.13 | 55 | 438 | 20.70 | 4.09 | 4.42 | 4.84 | 16.28 | 45.51 |
| 19 | 0.13 | 79 | 628 | 11.65 | 3.45 | 3.57 | 4.21 | 10.96 | 26.01 |
| 22 | 0.13 | 66 | 525 | 17.16 | 3.77 | 3.93 | 4.55 | 13.88 | 38.20 |
| Average | 0.13 | 61 | 488 | 13.60 | 3.60 | 3.80 | 4.20 | 12.20 | 30.00 |
| Model | Plot | 2024 | 2022 | |
|---|---|---|---|---|
| Av Field Measured Diameter (cm) | Average Calculated Diameter (cm) | Residuals (cm) | ||
| Jucker Model | 4 | 22 | 13 | 9 |
| 5 | 24 | 13 | 11 | |
| 6 | 26 | 22 | 4 | |
| 7 | 19 | 18 | 1 | |
| 11 | 17 | 14 | 2 | |
| 15 | 13 | 10 | 3 | |
| 22 | 29 | 23 | 6 | |
| 24 | 27 | 14 | 12 | |
| Gonzalez-Benecke Model 1 | 4 | 22 | 14 | 8 |
| 5 | 24 | 11 | 13 | |
| 6 | 26 | 15 | 11 | |
| 7 | 19 | 12 | 7 | |
| 11 | 17 | 11 | 6 | |
| 15 | 13 | 10 | 3 | |
| 22 | 29 | 16 | 13 | |
| 24 | 27 | 17 | 10 | |
| Gonzalez-Benecke Model 2 | 4 | 22 | 15 | 7 |
| 5 | 24 | 12 | 12 | |
| 6 | 26 | 17 | 9 | |
| 7 | 19 | 15 | 4 | |
| 11 | 17 | 12 | 5 | |
| 15 | 13 | 12 | 1 | |
| 22 | 29 | 19 | 10 | |
| 24 | 27 | 15 | 12 | |
| Model | RMSE (Plot 6) | RMSE (All Plots) | PBias % (Plots 6) | PBias % (All Plots) | MAE (Plot 6) | MAE (All Plots) | MAPE (Plot 6) | MAPE (All Plots) |
|---|---|---|---|---|---|---|---|---|
| Jucker DBH | 8.60 | 8.6 | −13.54 | −21.94 | 6 | 6 | 13.63 | 22.05 |
| Gonzalez-Benecke DBH 1 | 13.24 | 9.29 | −40.35 | −26.05 | 12 | 6 | 35.13 | 24.56 |
| Gonzalez-Benecke DBH 2 | 12.36 | 8.93 | −33.18 | −24.92 | 8 | 6 | 30.99 | 24.26 |
| Tree Species | Plots | Total Biomass AGB + BGB (AGB × 1.2)/ha (kg) | Total Carbon (TC) 50%BM/ha (kg) | Total CO2 (TC × 3.67)/ha (kg) | |||
|---|---|---|---|---|---|---|---|
| 2022 | 2024 | 2022 | 2024 | 2022 | 2024 | ||
| Eucalyptus | 5 | 88,716 | 194,623 | 32,159 | 70,551 | 118,025 | 258,921 |
| 6 | 295,048 | 211,688 | 106,955 | 76,737 | 392,525 | 281,624 | |
| White cypress pine | 4 | 68,652 | 425,883 | 24,886 | 154,383 | 91,333 | 566,584 |
| 7 | 157,302 | 145,303 | 57,022 | 52,672 | 209,271 | 193,308 | |
| 11 | 109,950 | 92,392 | 39,857 | 33,492 | 146,275 | 122,915 | |
| 15 | 124,030 | 64,728 | 44,961 | 23,464 | 165,007 | 86,113 | |
| Acacia harpophylla | 22 | 385,781 | 385,781 | 139,846 | 139,846 | 513,233 | 513,233 |
| 24 | 217,649 | 471,730 | 78,898 | 171,002 | 289,555 | 627,577 | |
| Average | 180,891 | 249,016 | 65,573 | 90,268 | 240,653 | 331,284 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhebhe, Z.M.; Liu, X.; Zhang, Z.; Paudyal, D.R. Estimation of Tree Diameter at Breast Height (DBH) and Biomass from Allometric Models Using LiDAR Data: A Case of the Lake Broadwater Forest in Southeast Queensland, Australia. Remote Sens. 2025, 17, 2523. https://doi.org/10.3390/rs17142523
Bhebhe ZM, Liu X, Zhang Z, Paudyal DR. Estimation of Tree Diameter at Breast Height (DBH) and Biomass from Allometric Models Using LiDAR Data: A Case of the Lake Broadwater Forest in Southeast Queensland, Australia. Remote Sensing. 2025; 17(14):2523. https://doi.org/10.3390/rs17142523
Chicago/Turabian StyleBhebhe, Zibonele Mhlaba, Xiaoye Liu, Zhenyu Zhang, and Dev Raj Paudyal. 2025. "Estimation of Tree Diameter at Breast Height (DBH) and Biomass from Allometric Models Using LiDAR Data: A Case of the Lake Broadwater Forest in Southeast Queensland, Australia" Remote Sensing 17, no. 14: 2523. https://doi.org/10.3390/rs17142523
APA StyleBhebhe, Z. M., Liu, X., Zhang, Z., & Paudyal, D. R. (2025). Estimation of Tree Diameter at Breast Height (DBH) and Biomass from Allometric Models Using LiDAR Data: A Case of the Lake Broadwater Forest in Southeast Queensland, Australia. Remote Sensing, 17(14), 2523. https://doi.org/10.3390/rs17142523








