Sensor-Based Yield Prediction in Durum Wheat Under Semi-Arid Conditions Using Machine Learning Across Zadoks Growth Stages
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Experimental Design and Meteorological Data
2.1.2. Soil Properties of Experimental Area
2.2. Methods
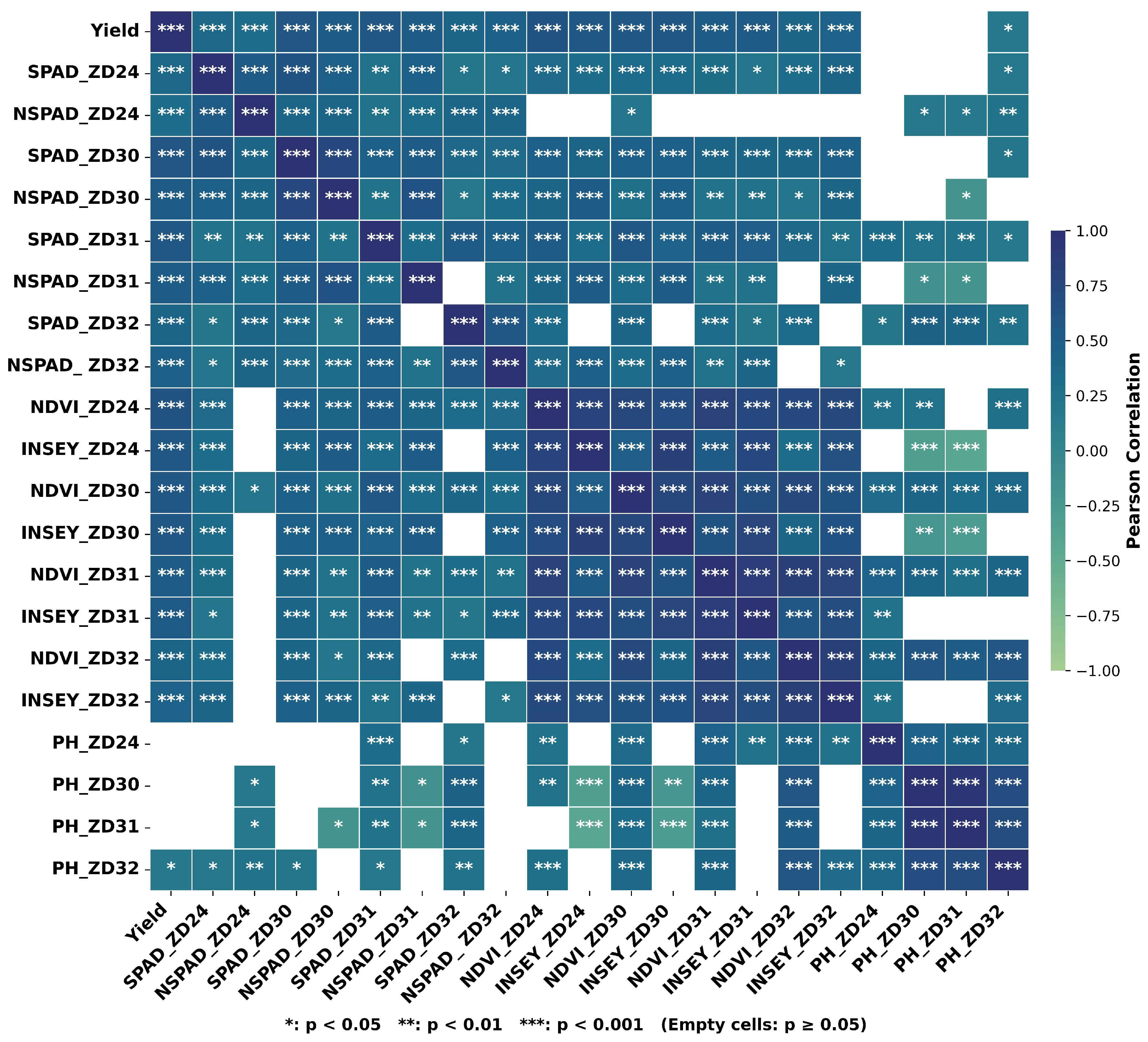
2.2.1. Obtaining Plant Indices (NDVI and INSEY) Using an Optical Sensor (GreenSeeker)
2.2.2. Chlorophyll (SPAD) Measurements
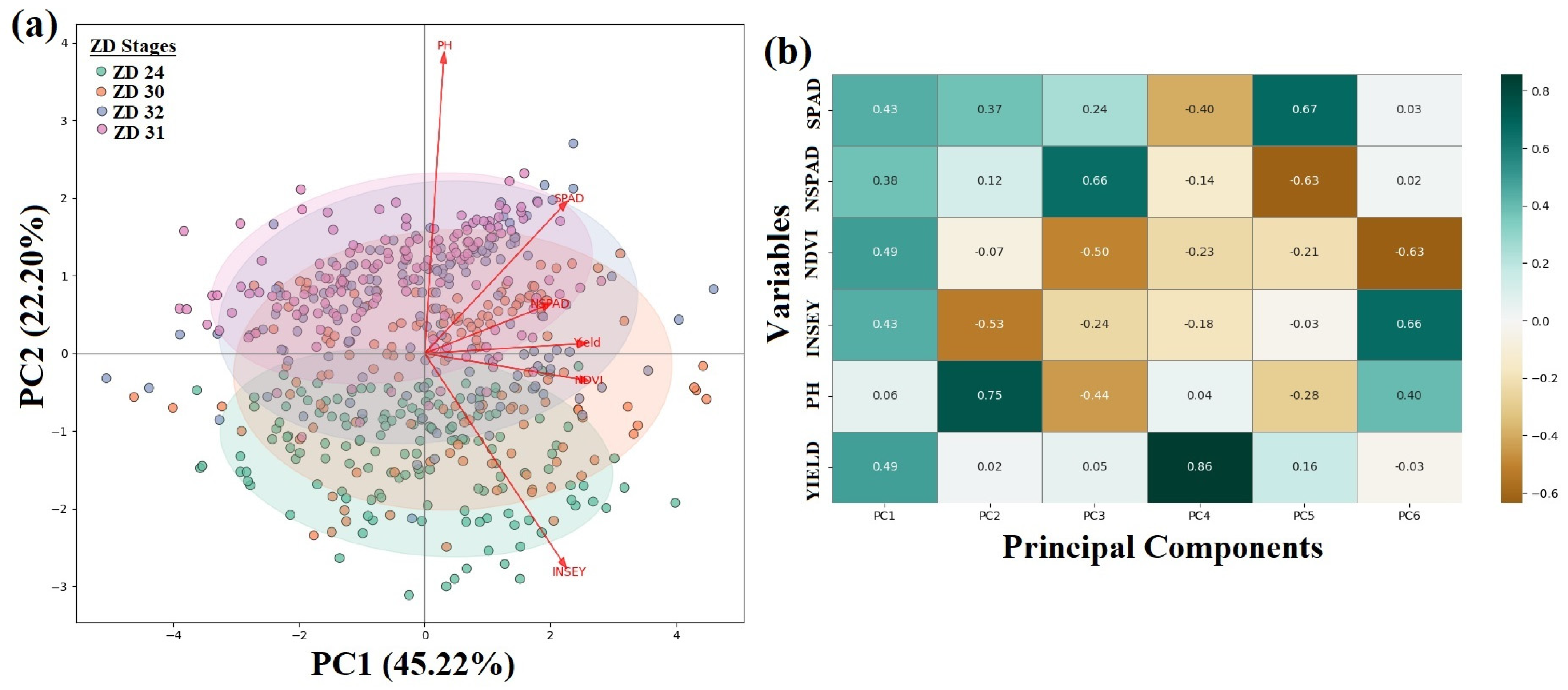
2.2.3. Principal Component Analysis (PCA)
2.2.4. SHAP (SHapley Additive exPlanations)
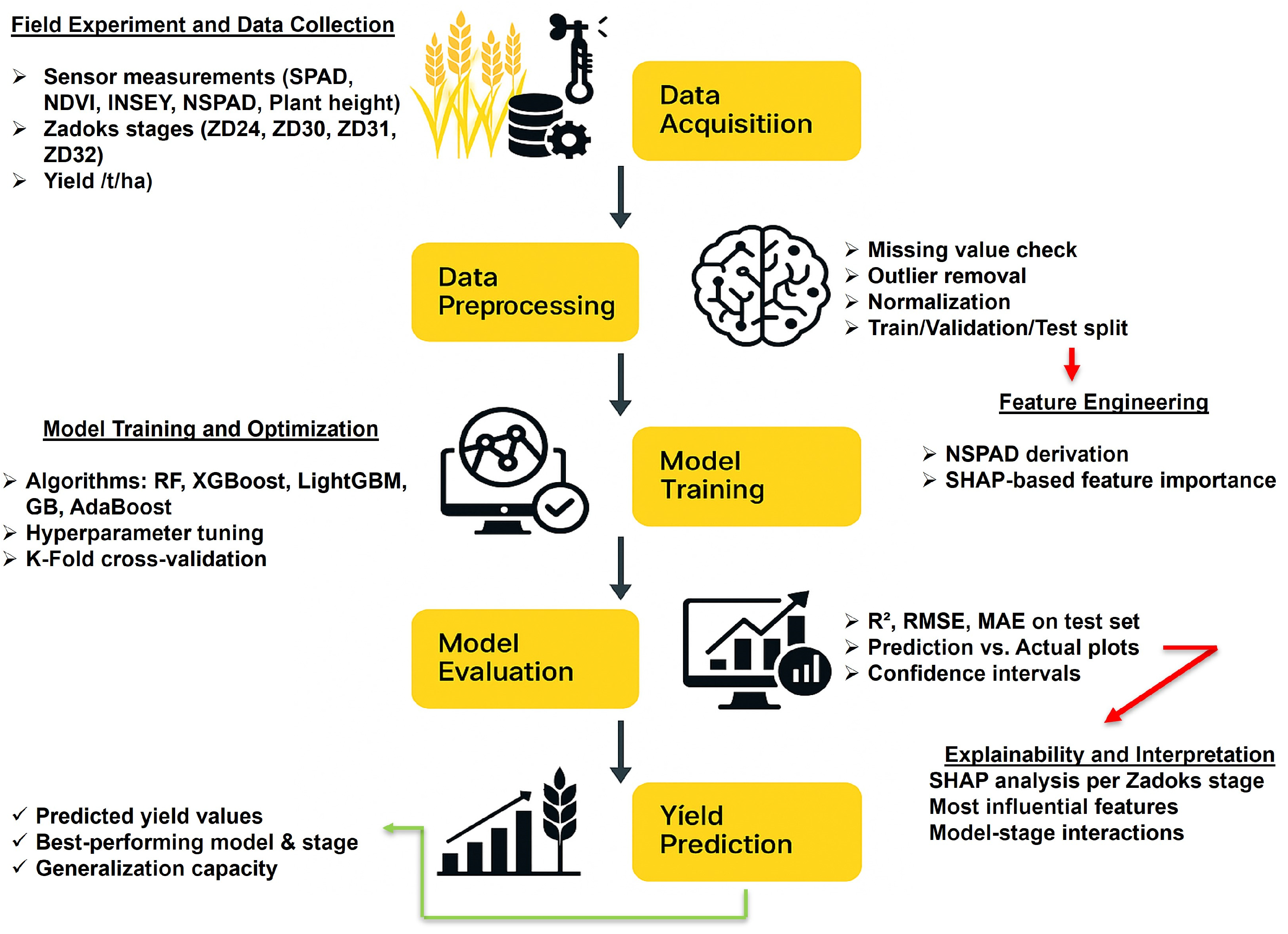
2.3. Architecture of Algorithms
2.3.1. Algorithms
2.3.2. Data Preprocessing and Exploratory Data Analysis (EDA)
2.3.3. Data Analysis
2.3.4. Performance Evaluation Metrics
3. Results
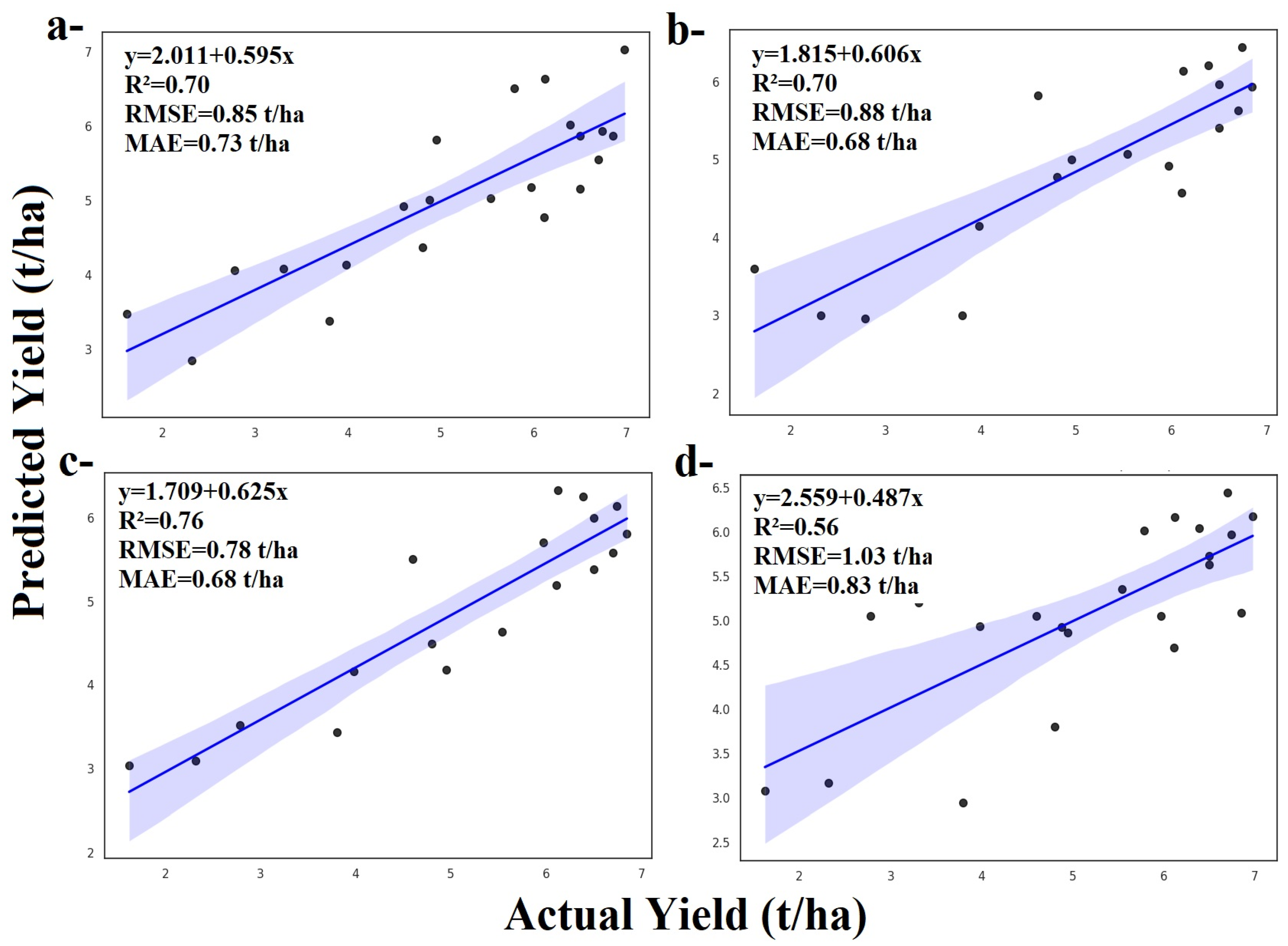
3.1. Yield Prediction by ZD Stages Using Machine Learning Algorithms
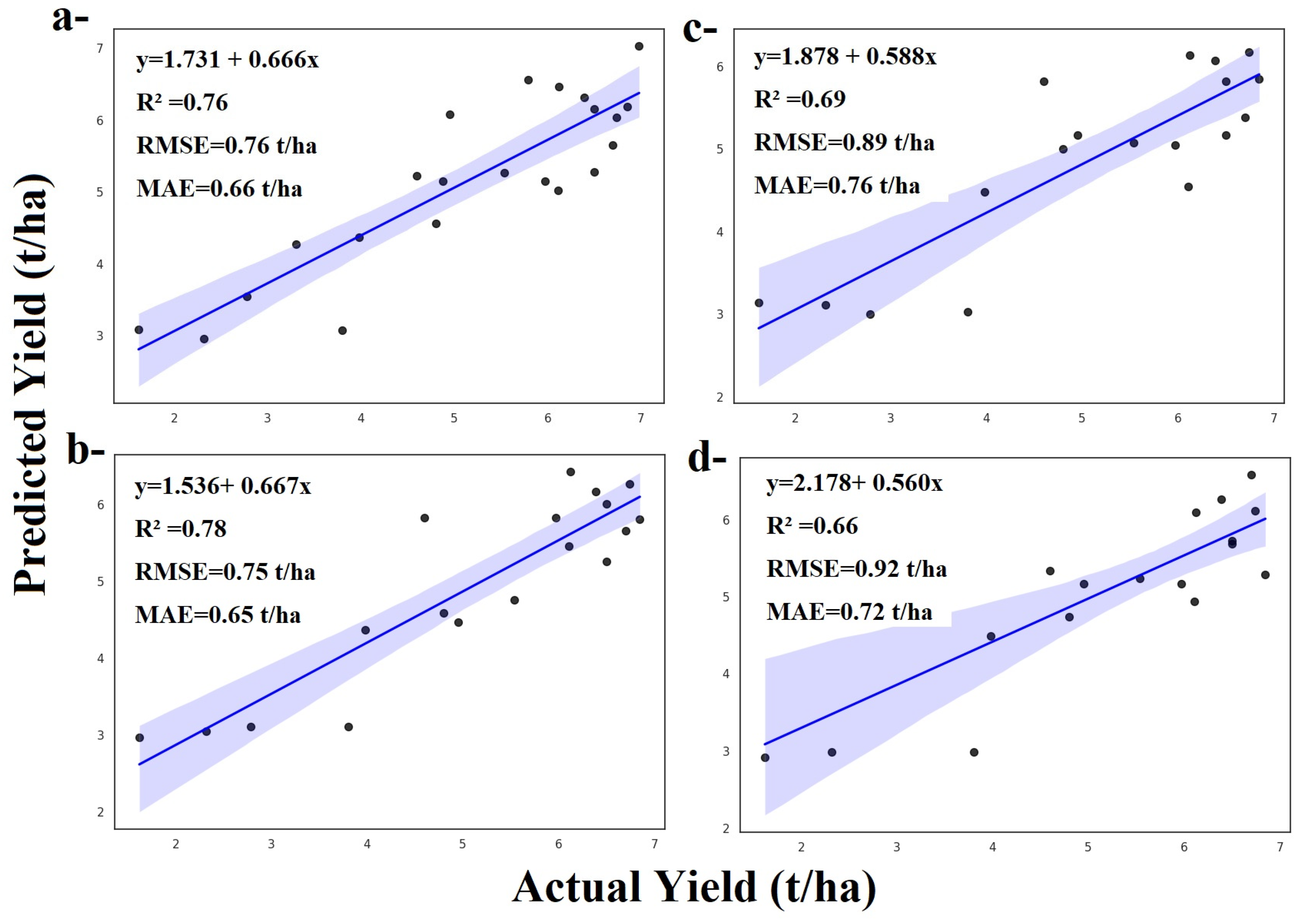
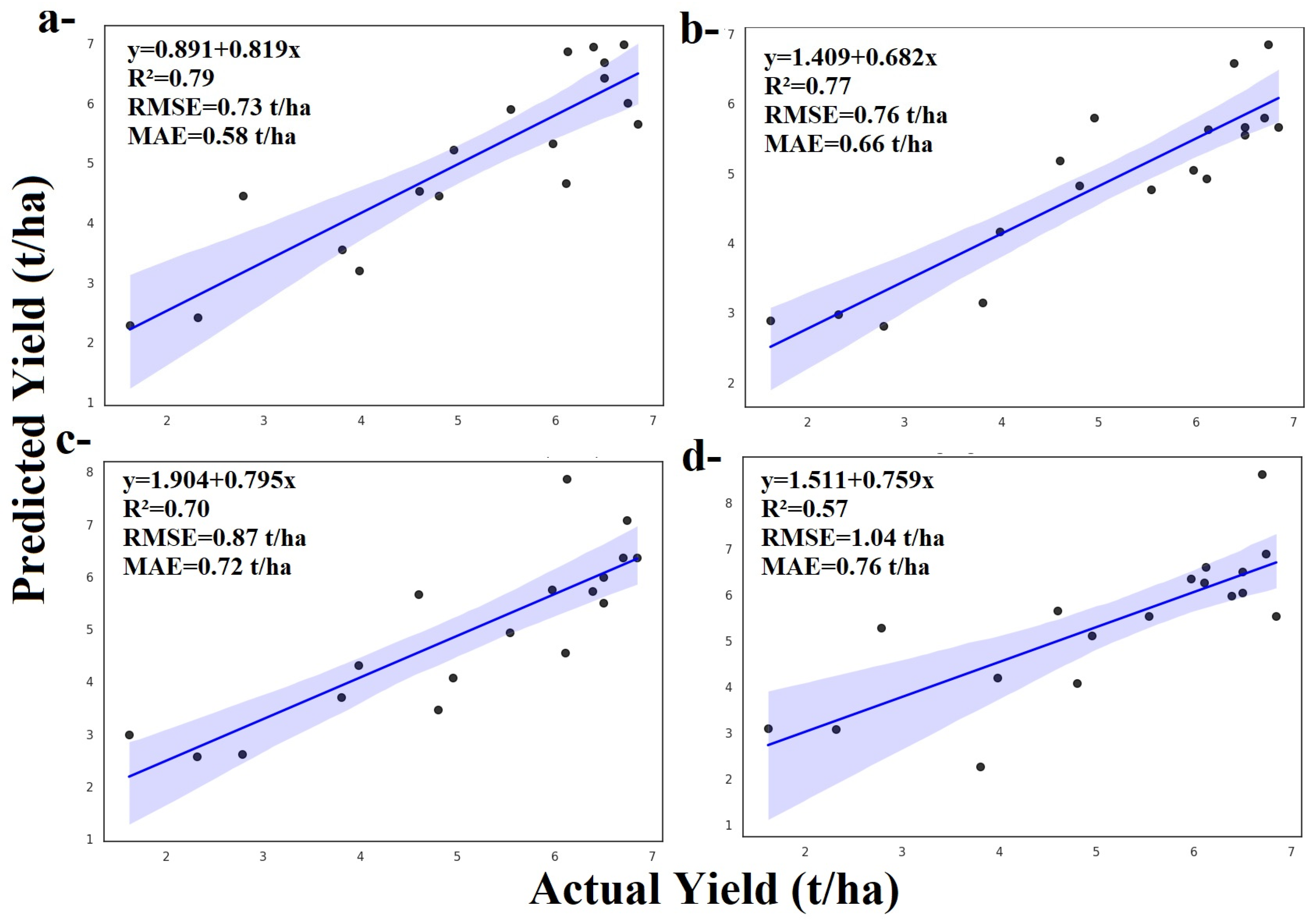
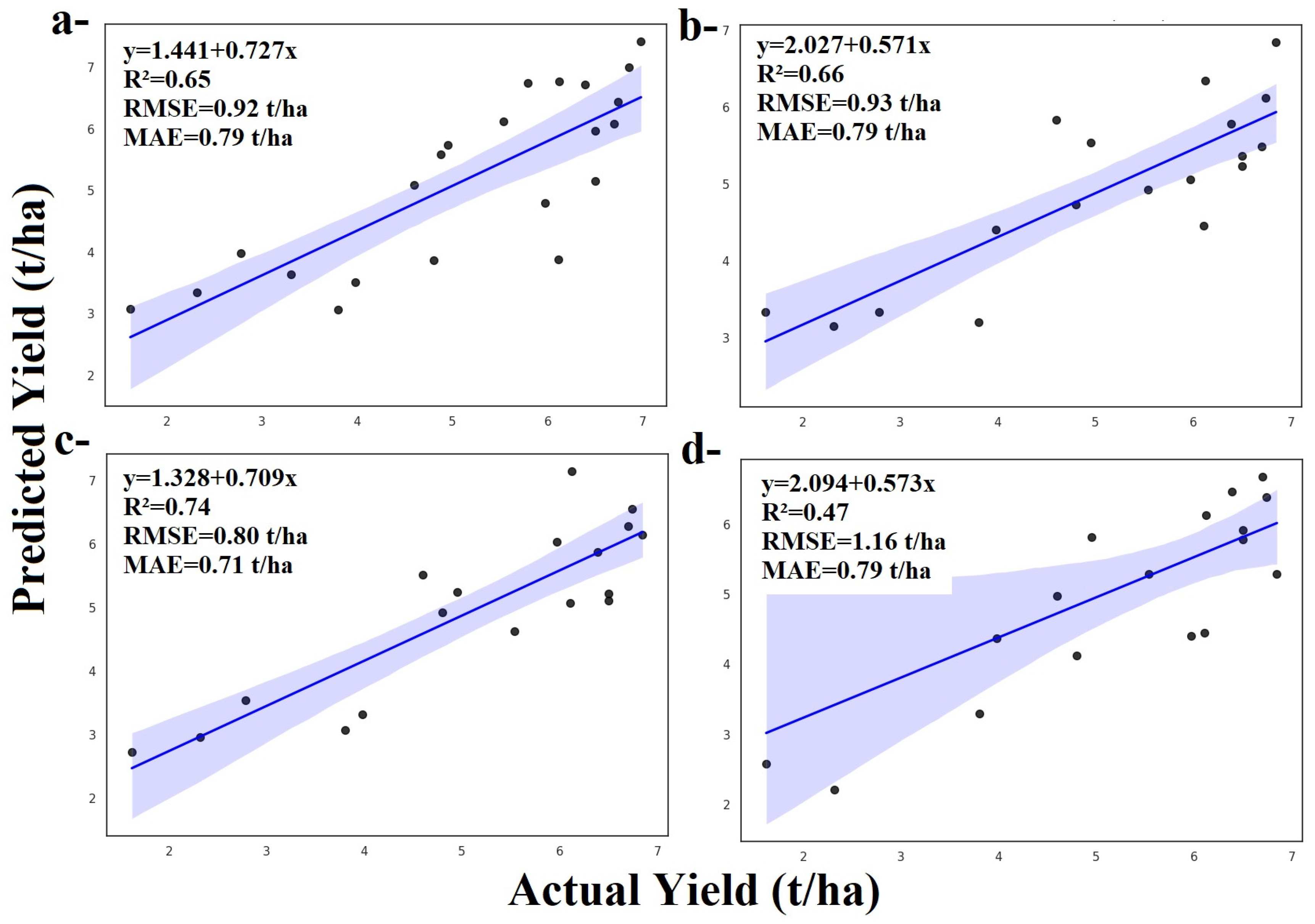
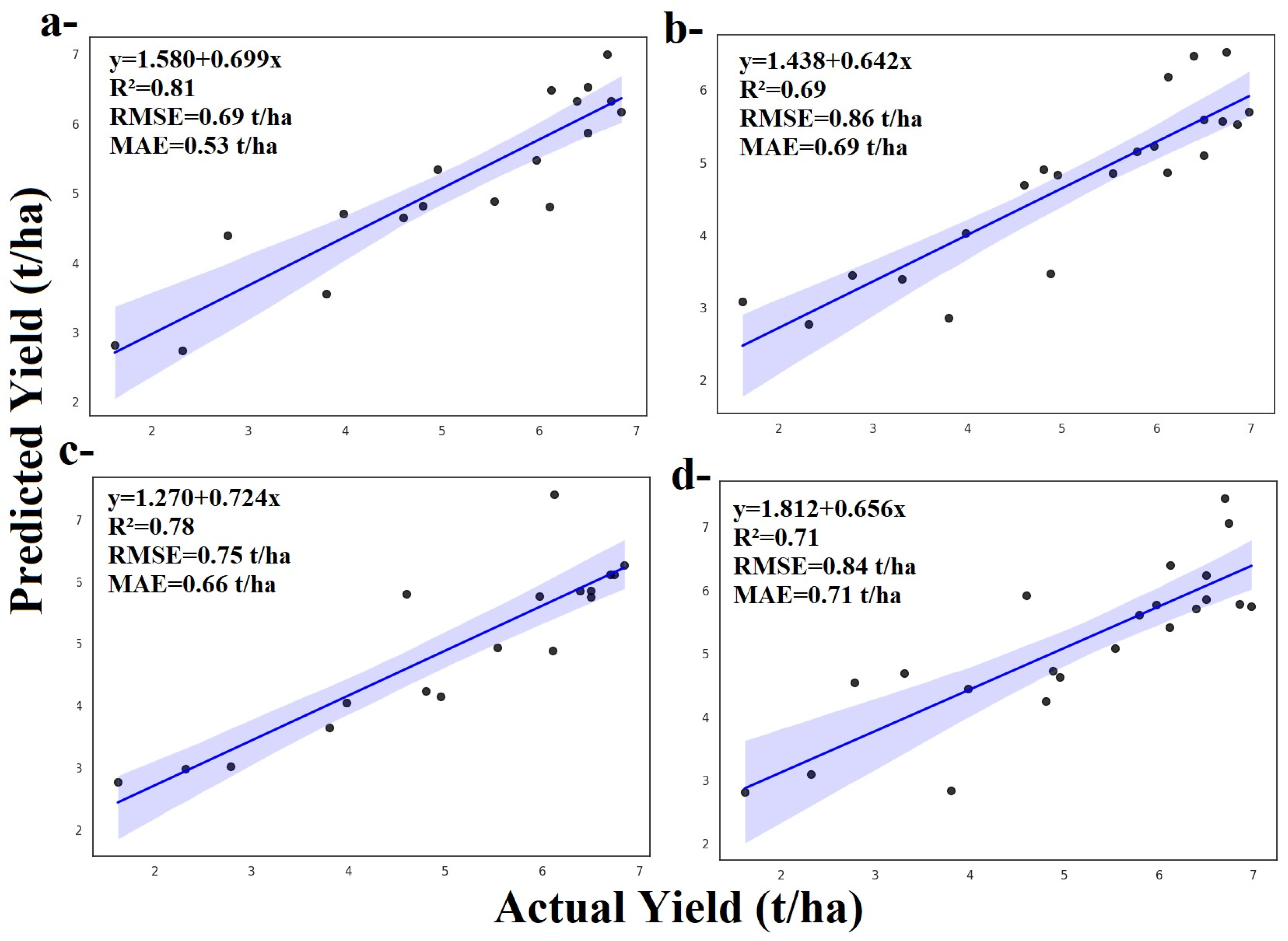
3.1.1. Random Forest and ZD Stages
3.1.2. Adaboost and ZD Stages
3.1.3. Gradient Boosting and ZD Stages
3.1.4. LightGBM and ZD Stages
3.1.5. XGBoost and ZD Stages
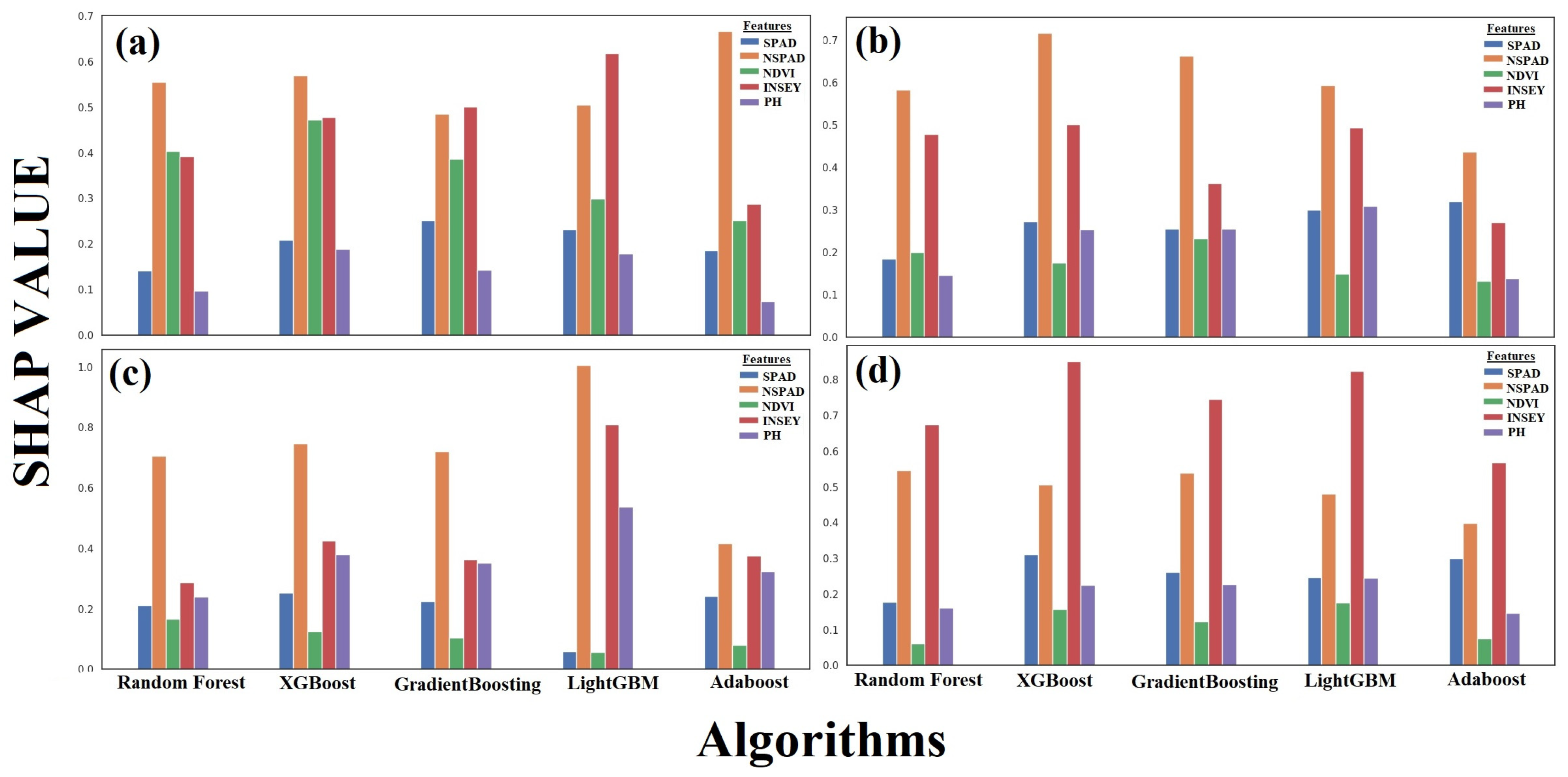
3.1.6. SHAP Analysis Interpretation Across ZD Stages
3.2. Comparision of Algorithms and ZD Periods
3.2.1. Model Performances
3.2.2. Zadoks Stage-Specific Observations
3.2.3. Feature Contribution and SHAP Values
4. Discussion
4.1. Limitations and Contributions of the Study
4.1.1. Limitations
4.1.2. Contributions
4.2. Rationale for Algorithm Selection and Structural Superiority
4.3. The Influence of Zadoks Growth Stages
4.4. The Effect of Regional Variability and Model Robustness
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jabed, M.A.; Murad, M.A.A. Crop yield prediction in agriculture: A comprehensive review of machine learning and deep learning approaches, with insights for future research and sustainability. Heliyon 2024, 10, e40836. [Google Scholar] [CrossRef] [PubMed]
- TUIK. Türkiye İstatistik Kurumu Bitkisel Üretim İstatistikleri Turkish Statistical Institute. Turkish Statistical Institute—Agricultural Production Statistics. 2023. Available online: https://data.tuik.gov.tr (accessed on 5 June 2025).
- Wang, X.; Zhang, J.; Xun, L.; Wang, J.; Wu, Z.; Henchiri, M.; Zhang, S.; Zhang, S.; Bai, Y.; Yang, S.; et al. Evaluating the effectiveness of machine learning and deep learning models combined time-series satellite data for multiple crop types classification over a large-scale region. Remote Sens. 2022, 14, 2341. [Google Scholar] [CrossRef]
- Raza, A.; Shahid, M.A.; Zaman, M.; Miao, Y.; Huang, Y.; Safdar, M.; Maqbool, S.; Muhammad, N.E. Improving Wheat Yield Prediction with Multi-Source Remote Sensing Data and Machine Learning in Arid Regions. Remote Sens. 2025, 17, 774. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Z.; Feng, L.; Du, Q.; Runge, T. Combining multi-source data and machine learning approaches to predict winter wheat yield in the conterminous United States. Remote Sens. 2020, 12, 1232. [Google Scholar] [CrossRef]
- Silvestri, N.; Ercolini, L.; Grossi, N.; Ruggeri, M. Integrating NDVI and agronomic data to optimize the variable-rate nitrogen fertilization. Precis. Agric. 2024, 25, 2554–2572. [Google Scholar] [CrossRef]
- Fava, F.; Colombo, R.; Bocchi, S.; Meroni, M.; Sitzia, M.; Fois, N.; Zucca, C. Identification of hyperspectral vegetation indices for Mediterranean pasture characterization. Int. J. Appl. Earth Obs. Geoinf. 2009, 11, 233–243. [Google Scholar] [CrossRef]
- Üstündağ, B.B.; Aktaş, H. Evaluation of NSPAD and NDVI as indicators of nitrogen status in wheat under Mediterranean conditions. Agric. Water Manag. 2017, 189, 54–62. [Google Scholar] [CrossRef]
- Savasli, E.; Onder, O.; Cekic, C.; Kalayci, H.M.; Dayioglu, R.; Karaduman, Y.; Gezgin, S. Calibration optimization for sensor-based in-season nitrogen management of rainfed winter wheat in central Anatolian conditions. KSU J. Agric. Nat. 2021, 24, 130–140. [Google Scholar] [CrossRef]
- Savaşlı, E.; Karaduman, Y.; Önder, O.; Özen, D.; Dayıoğlu, R.; Ateş, Ö.; Özdemir, S. Estimating technological quality parameters of bread wheat using sensor-based normalized difference vegetation index. J. Cereal. Sci. 2022, 107, 103535. [Google Scholar] [CrossRef]
- Lu, J.; Li, J.; Fu, H.; Tang, X.; Liu, Z.; Chen, H.; Sun, Y.; Ning, X. Deep learning for multi-source data-driven crop yield prediction in northeast China. Agriculture 2023, 14, 794. [Google Scholar] [CrossRef]
- Joshi, A.; Pradhan, B.; Gite, S.; Chakraborty, S. Remote-sensing data and deep-learning techniques in crop mapping and yield prediction: A systematic review. Remote Sens. 2023, 15, 2014. [Google Scholar] [CrossRef]
- Zhao, Y.; Han, S.; Meng, Y.; Feng, H.; Li, Z.; Chen, J.; Song, X.; Zhu, Y.; Yang, G. Transfer-learning-based approach for yield prediction of winter wheat from planet data and SAFY Model. Remote Sens. 2022, 14, 5474. [Google Scholar] [CrossRef]
- Wang, W.; Cheng, Y.; Ren, Y.; Zhang, Z.; Geng, H. Prediction of chlorophyll content in multi-temporal winter wheat based on multispectral and machine learning. Front. Plant Sci. 2022, 13, 896408. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Yang, F.; Du, J. Yield forecasting for winter wheat using time series NDVI from HJ satellite. Trans. Chin. Soc. Agric. Eng. 2013, 29, 124–131. [Google Scholar]
- Hussein, E.E.; Zerouali, B.; Bailek, N.; Derdour, A.; Ghoneim, S.S.; Santos, C.A.G.; Hashim, M.A. Harnessing Explainable AI for Sustainable Agriculture: SHAP-Based Feature Selection in Multi-Model Evaluation of Irrigation Water Quality Indices. Water 2024, 17, 59. [Google Scholar] [CrossRef]
- Bouyoucos, G.J. A recalibration of the hydrometer method for making mechanical analysis of soils. Agron. J. 1951, 43, 434–438. [Google Scholar] [CrossRef]
- Mclean, E.O. Soil pH and lime requirement. In Methods of Soil Analysis, Part 2, 2nd ed.; Page, A.L., Ed.; ASA and SSSA: Salt Lake City, UT, USA, 1982; pp. 199–224. [Google Scholar]
- Nelson, D.W.; Sommers, L.E. Total carbon, organic carbon, and organic matter. Methods Soil Anal. Part 3 Chem. Methods 1996, 5, 961–1010. [Google Scholar]
- Tüzüner, A. Toprak ve su Analiz Laboratuvarları el Kitabı; Tarım Orman ve Köyişleri Bakanlığı, Köy Hizmetleri Genel Müdürlüğü: Ankara, Turky, 1990. [Google Scholar]
- Olsen, S.R. Estimation of available phosphorus in soils by extraction with sodium bicarbonate. U.S. Dep. Agric. Circ. 1954, 939, 1–19. [Google Scholar]
- Bremner, J.M. Total nitrogen. In Methods of Soil Analysis; Black, C.A., Ed.; ASA and SSSA: Salt Lake City, UT, USA, 1965; pp. 1149–1178. [Google Scholar]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A decimal code for the growth stages of cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- Raun, W.R.; Solie, J.B.; Johnson, G.V.; Stone, M.L.; Mullen, R.W.; Freeman, K.W.; Lukina, E.V. Improving nitrogen use efficiency in cereal grain production with optical sensing and variable rate application. Agron. J. 2002, 94, 815–820. [Google Scholar] [CrossRef]
- Mullen, R.W.; Freeman, K.W.; Raun, W.R.; Johnson, G.V.; Stone, M.L.; Solie, J.B. Identifying an in-season response index and the potential to increase wheat yield with nitrogen. Agron. J. 2003, 95, 347–351. [Google Scholar] [CrossRef]
- Peñuelas, J.; Gamon, J.A.; Griffin, K.L.; Field, C.B. Assessing community type, plant biomass, pigment composition, and photosynthetic efficiency of aquatic vegetation from spectral reflectance. Remote Sens. Environ. 1993, 46, 110–118. [Google Scholar] [CrossRef]
- Garrido-Lestache, E.; López-Bellido, R.J.; López-Bellido, L. Effect of N rate, timing and splitting and N type on bread-making quality in hard red spring wheat under rainfed Mediterranean conditions. Field Crops Res. 2004, 85, 213–236. [Google Scholar] [CrossRef]
- Varvel, G.E.; Schepers, J.S.; Francis, D.D. Ability for in-season correction of nitrogen deficiency in corn using chlorophyll meters. Soil Sci. Soc. Am. J. 1997, 61, 1233–1239. [Google Scholar] [CrossRef]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, S.M.; Lee, S.I. A unified approach to interpreting model predictions. Adv. Neural Inf. Process Syst. 2017, 30, 4765–4774. [Google Scholar]
- Friedman, J.H. Greedy Function Approximation: A Gradient Boosting Machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, 2nd ed.; Springer: New York, NY, USA, 2009; pp. 154–196. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Duchesnay, É. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Frost, J. Regression Analysis: An Intuitive Guide for Using and Interpreting Linear Models; Life Course Research and Social Policies; Springer: London, UK, 2019. [Google Scholar]
- Freedman, D.A. Statistical Models: Theory and Practice; Cambridge University Press: Cambridge, UK, 2009. [Google Scholar]
- Chai, T.; Draxler, R.R. Root mean square error (RMSE) or mean absolute error (MAE)?—Arguments against avoiding RMSE in the literature. Geosci. Model Dev. 2014, 7, 1247–1250. [Google Scholar] [CrossRef]
- Willmott, C.J.; Matsuura, K. Advantages of the mean absolute error (MAE) over the root mean square error (RMSE) in assessing average model performance. Clim. Res. 2005, 30, 79–82. [Google Scholar] [CrossRef]
- Kyratzis, A.C.; Skarlatos, D.P.; Menexes, G.C.; Vamvakousis, V.F.; Katsiotis, A. Assessment of vegetation indices derived by UAV imagery for durum wheat phenotyping under a water limited and heat stressed Mediterranean environment. Front. Plant Sci. 2017, 8, 1114. [Google Scholar] [CrossRef]
- Ji, R.; Min, J.; Wang, Y.; Cheng, H.; Zhang, H.; Shi, W. In-season yield prediction of cabbage with a hand-held active canopy sensor. Sensors 2017, 17, 2287. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, T.K.; Roy, D.K.; Kang, Y.S.; Jun, S.R.; Park, J.W.; Ryu, C.S. Ensemble of machine learning algorithms for rice grain yield prediction using UAV-based remote sensing. J. Biosyst. Eng. 2024, 49, 1–19. [Google Scholar] [CrossRef]
- Ruan, R.; Lin, L.; Li, Q.; He, T. Integration of remote sensing and machine learning to improve crop yield prediction in heterogeneous environments. Precis. Agric. 2024, 25, 389–408. [Google Scholar]
- Cao, J.; Zhang, Z.; Tao, F.; Zhang, L.; Luo, Y.; Zhang, J.; Han, J.; Xie, J. Integrating multi-source data for rice yield prediction across China using machine learning and deep learning approaches. Agric. For. Meteorol. 2021, 297, 108275. [Google Scholar] [CrossRef]
- Liu, X.; Wu, L.; Zhang, F.; Huang, G.; Yan, F.; Bai, W. Splitting and length of years for improving tree-based models to predict reference crop evapotranspiration in the humid regions of China. Water 2021, 13, 3478. [Google Scholar] [CrossRef]
- Yang, S.; Li, L.; Fei, S.; Yang, M.; Tao, Z.; Meng, Y.; Xiao, Y. Wheat yield prediction using machine learning method based on UAV remote sensing data. Drones 2024, 8, 284. [Google Scholar] [CrossRef]
- Kılıç, M. Modeling of Land Use/Land Cover Change and Its Effects on Soil Properties: The Case of Besni District, Adiyaman. Ph.D. Thesis, Harran University, Şanlıurfa, Turkey, 2023. [Google Scholar]
- Meng, L.; Liu, H.; Ustin, S.L.; Zhang, X. Predicting maize yield at the plot scale of different fertilizer systems by multi-source data and machine learning methods. Remote Sens. 2021, 13, 3760. [Google Scholar] [CrossRef]
- Hu, T.; Zhang, X.; Bohrer, G.; Liu, Y.; Zhou, Y.; Martin, J.; Yang, L.; Zhao, K. Crop yield prediction via explainable AI and interpretable machine learning: Dangers of black box models for evaluating climate change impacts on crop yield. Agric. For. Meteorol. 2023, 336, 109458. [Google Scholar] [CrossRef]
- Zhou, X.; Kono, Y.; Win, A.; Matsui, T.; Tanaka, T.S. Predicting within-field variability in grain yield and protein content of winter wheat using UAV-based multispectral imagery and machine learning approaches. Plant Prod. Sci. 2021, 24, 137–151. [Google Scholar] [CrossRef]
- Minolta, K. Chlorophyll Meter SPAD-502; Instruction Manual. Minolta: Osaka, Japan, 1989. [Google Scholar]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Liu, T.Y. Lightgbm: A highly efficient gradient boosting decision tree. Adv. Neural Inf. Process Syst. 2017, 30, 3149–3157. [Google Scholar]
- Kumar, C.; Dhillon, J.; Huang, Y.; Reddy, K. Explainable machine learning models for corn yield prediction using UAV multispectral data. Comput. Electron. Agric. 2025, 231, 109990. [Google Scholar] [CrossRef]
- Piekutowska, M.; Niedbała, G. Review of Methods and Models for Potato Yield Prediction. Agriculture 2025, 15, 367. [Google Scholar] [CrossRef]
- Van Klompenburg, T.; Kassahun, A.; Catal, C. Crop yield prediction using machine learning: A systematic literature review. Comput. Electron. Agric. 2020, 177, 105709. [Google Scholar] [CrossRef]
- Abbasi, M.; Váz, P.; Silva, J.; Martins, P. Machine learning approaches for predicting maize biomass yield: Leveraging feature engineering and comprehensive data integration. Sustainability 2025, 17, 256. [Google Scholar] [CrossRef]

| Soil Properties | Units | 2023 | 2024 |
|---|---|---|---|
| pH | 6.95 | 7.52 | |
| EC | dS/cm | 0.89 | 0.92 |
| Saturation | % | 70 | 72 |
| Organic Matter | % | 1.29 | 0.8 |
| Lime | % | 26.6 | 32.1 |
| Available Phosphorus | kg/da | 3.04 | 2.95 |
| Available Potassium | kg/da | 205 | 213 |
| Total Nitrogen | % | 0.086 | 0.091 |
| Dataset | Model | Hyperparameter | Optimal Value | Optimization | Performance Metrics |
|---|---|---|---|---|---|
| %70 Training %15 Testing %15 Validation | Random Forest | Number of Estimators | 100, 300, 500, 700 | 5-fold cross validation GridSearchCV 95% CI | R2 RMSE MAE |
| Max Depth | 3, 5, 7 | ||||
| Min Samples per Leaf | 1, 2, 3 | ||||
| min_samples_split | 2, 4, 6 | ||||
| max_features | ‘auto’, ‘sqrt’ | ||||
| Gradient Boosting | Number of Estimators | 200 | |||
| Max Depth | 6 | ||||
| Learning Rate | 0.1 | ||||
| min_samples_split | 2 | ||||
| min_samples_leaf | 1,2 | ||||
| AdaBoost | Number of Estimators | 100 | |||
| Learning Rate | 0.1 | ||||
| Loss Function | ‘linear’, ‘square’, ‘exponential | ||||
| LightGBM | Boosting Type | gbdt | |||
| Number of Estimators | 200 | ||||
| Learning Rate | 0.1 | ||||
| Max Depth | 5 | ||||
| num_leaves | 31 | ||||
| XGBoost | Number of Estimators | 100, 200 | |||
| Learning Rate | 0.1 | ||||
| Max Depth | 8 | ||||
| subsample | 0.9 | ||||
| colsample_bytree | 1 |
| Metrics | Explanations | Formulations |
|---|---|---|
| R2 | Ranges from 0 to 1, with values closer to 1 indicating that the model explains a larger proportion of the variance in the data [36,37]. | |
| RMSE | A sensitive indicator for measuring the magnitude of forecast errors, was calculated as the square root of the mean square of the squares of the forecast errors [38]. | |
| MAE | A commonly used metric for evaluating the accuracy of regression models [39]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rufaioğlu, S.B.; Bilgili, A.V.; Savaşlı, E.; Özberk, İ.; Aydemir, S.; Ismael, A.M.; Kaya, Y.; Matos-Carvalho, J.P. Sensor-Based Yield Prediction in Durum Wheat Under Semi-Arid Conditions Using Machine Learning Across Zadoks Growth Stages. Remote Sens. 2025, 17, 2416. https://doi.org/10.3390/rs17142416
Rufaioğlu SB, Bilgili AV, Savaşlı E, Özberk İ, Aydemir S, Ismael AM, Kaya Y, Matos-Carvalho JP. Sensor-Based Yield Prediction in Durum Wheat Under Semi-Arid Conditions Using Machine Learning Across Zadoks Growth Stages. Remote Sensing. 2025; 17(14):2416. https://doi.org/10.3390/rs17142416
Chicago/Turabian StyleRufaioğlu, Süreyya Betül, Ali Volkan Bilgili, Erdinç Savaşlı, İrfan Özberk, Salih Aydemir, Amjad Mohamed Ismael, Yunus Kaya, and João P. Matos-Carvalho. 2025. "Sensor-Based Yield Prediction in Durum Wheat Under Semi-Arid Conditions Using Machine Learning Across Zadoks Growth Stages" Remote Sensing 17, no. 14: 2416. https://doi.org/10.3390/rs17142416
APA StyleRufaioğlu, S. B., Bilgili, A. V., Savaşlı, E., Özberk, İ., Aydemir, S., Ismael, A. M., Kaya, Y., & Matos-Carvalho, J. P. (2025). Sensor-Based Yield Prediction in Durum Wheat Under Semi-Arid Conditions Using Machine Learning Across Zadoks Growth Stages. Remote Sensing, 17(14), 2416. https://doi.org/10.3390/rs17142416










