Unmanned Aerial Vehicle-Based RGB Imaging and Lightweight Deep Learning for Downy Mildew Detection in Kimchi Cabbage
Abstract
1. Introduction
2. Materials and Methods
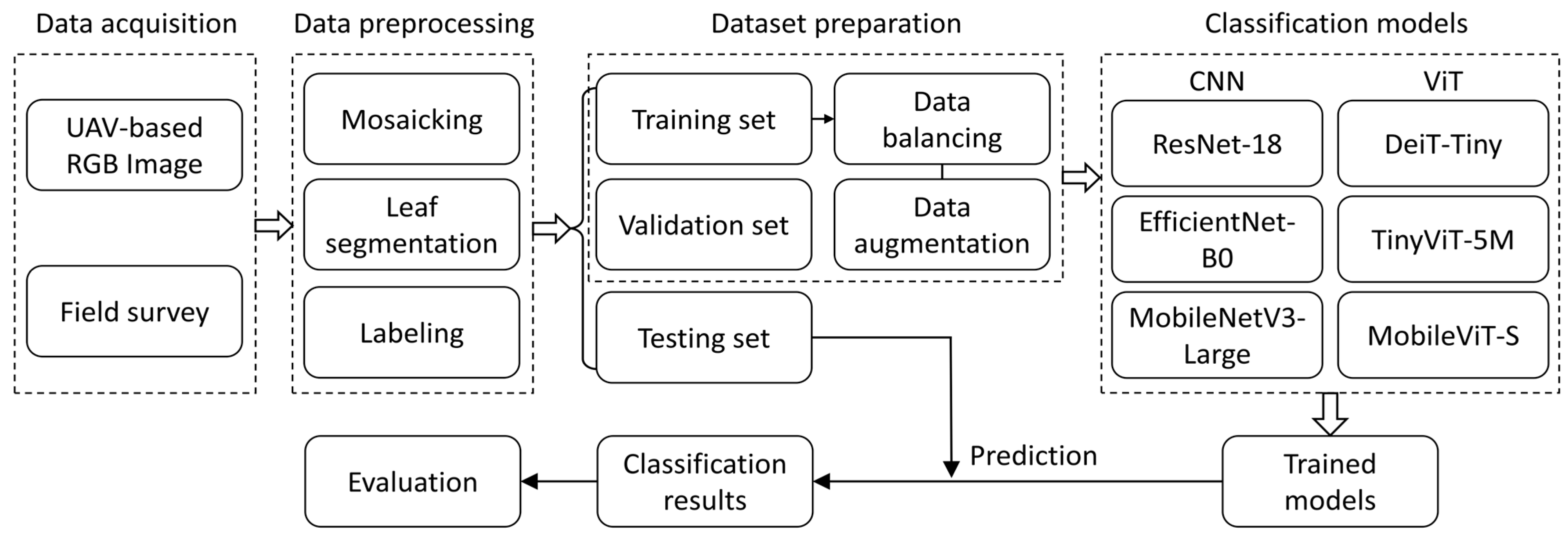
2.1. Overall Workflow
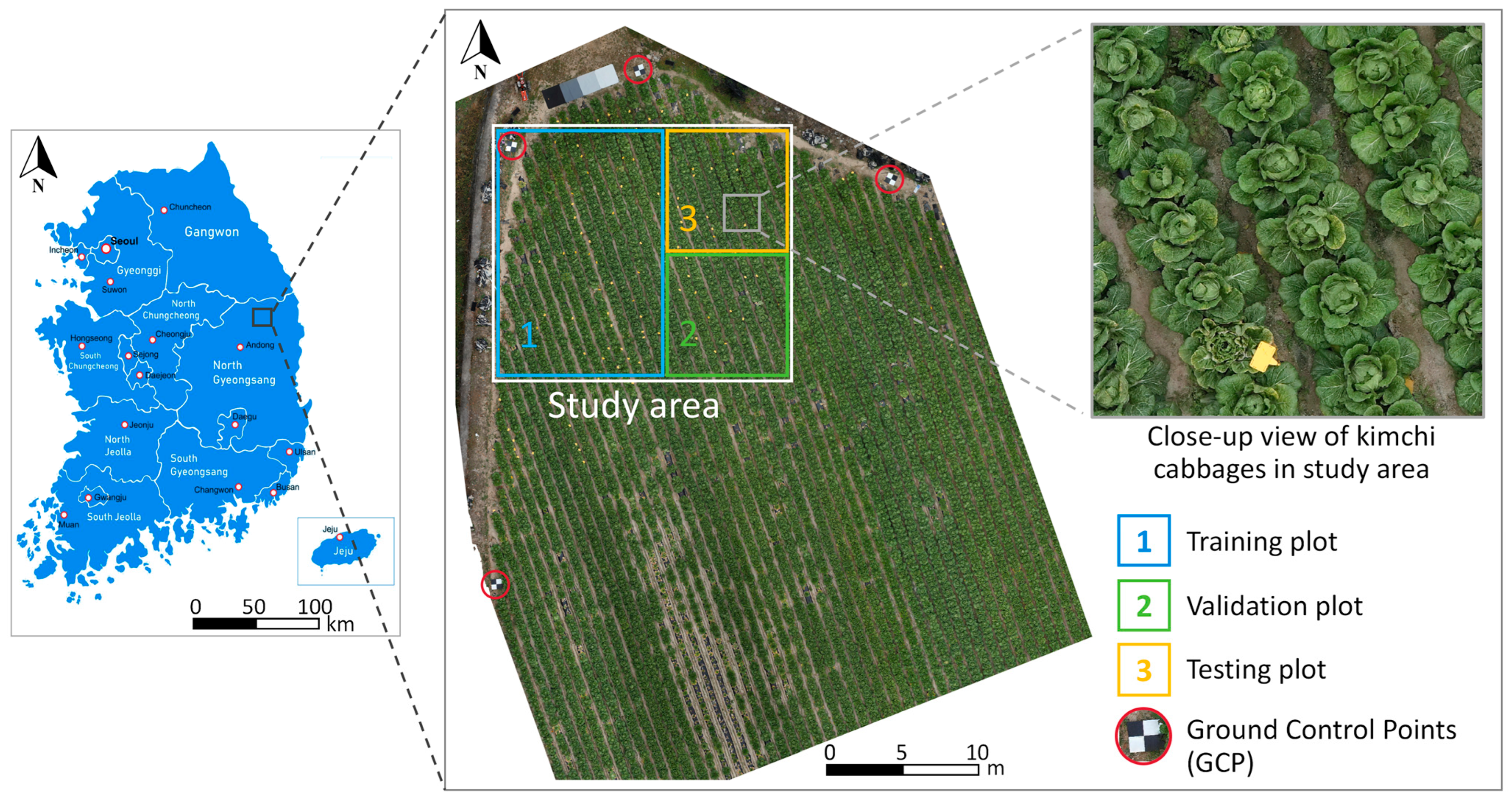
2.2. Study Area and Site Description
2.3. Data Acquisition
2.3.1. UAV-Based Aerial Imaging
2.3.2. Field Survey
2.4. Data Preprocessing
2.4.1. Image Mosaicking
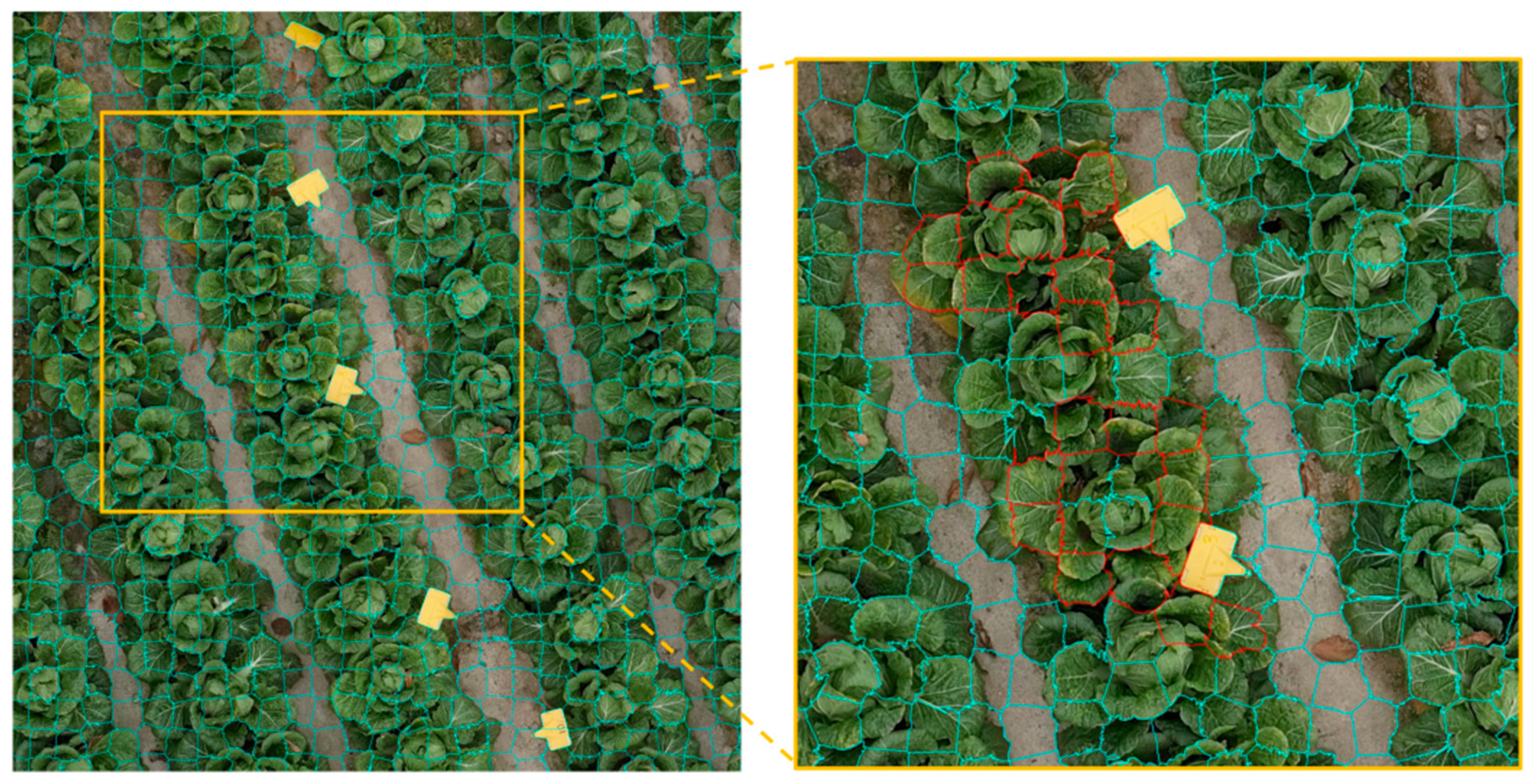
2.4.2. Image Segmentation and Labeling
2.5. Dataset Preparation
2.5.1. Dataset Partitioning
2.5.2. Data Balancing and Augmentation
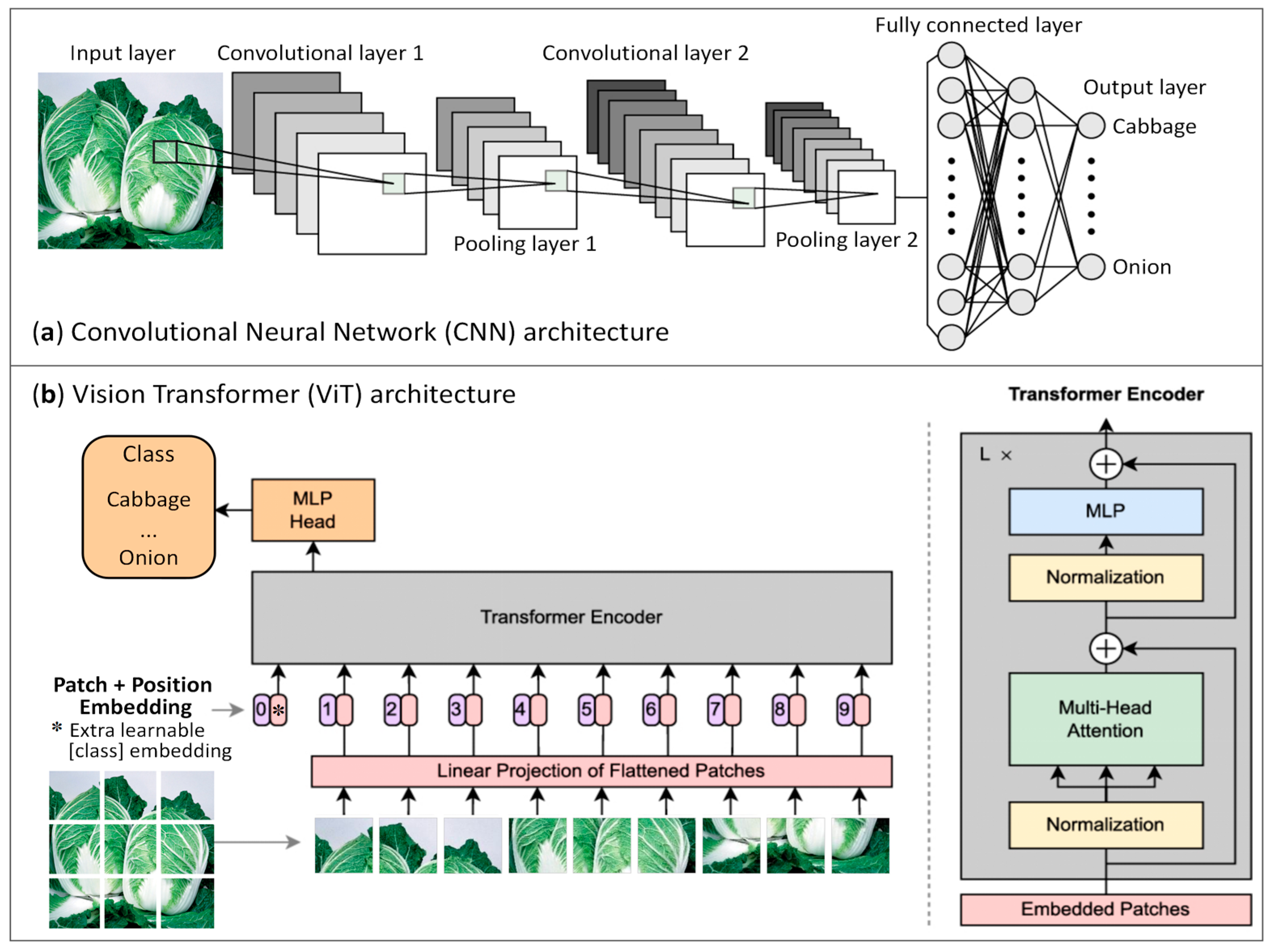
2.6. Lightweight CNN and ViT Models
2.7. Model Training and Experimental Setup
2.8. Model Evaluation
3. Results
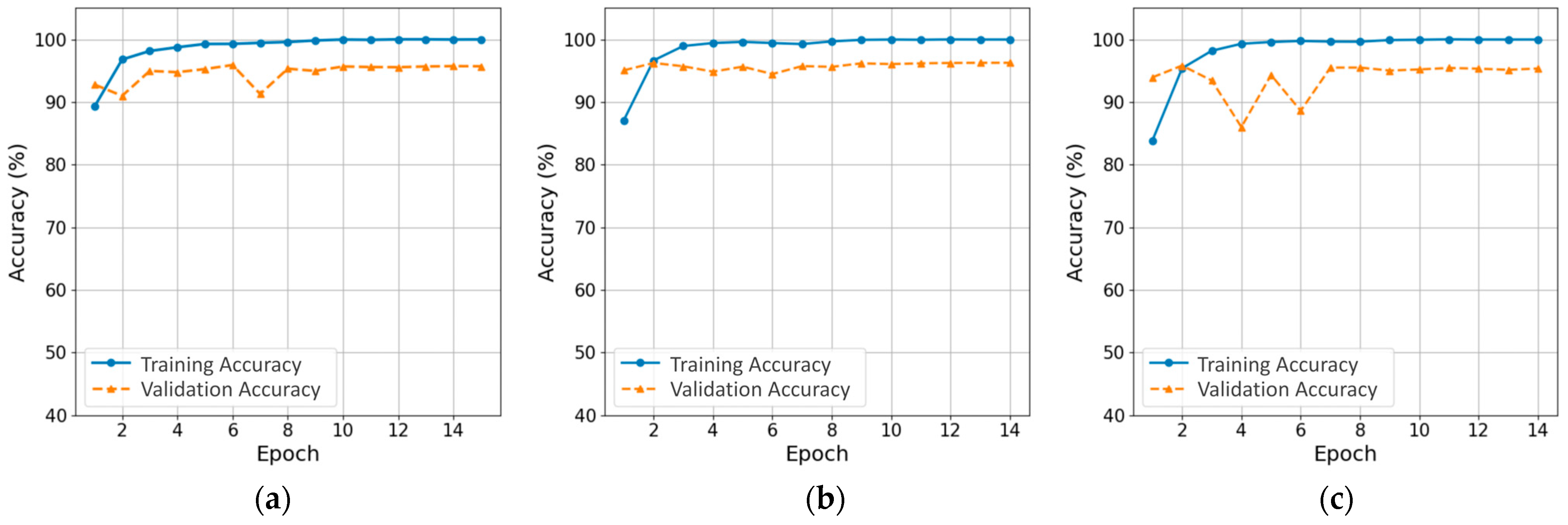
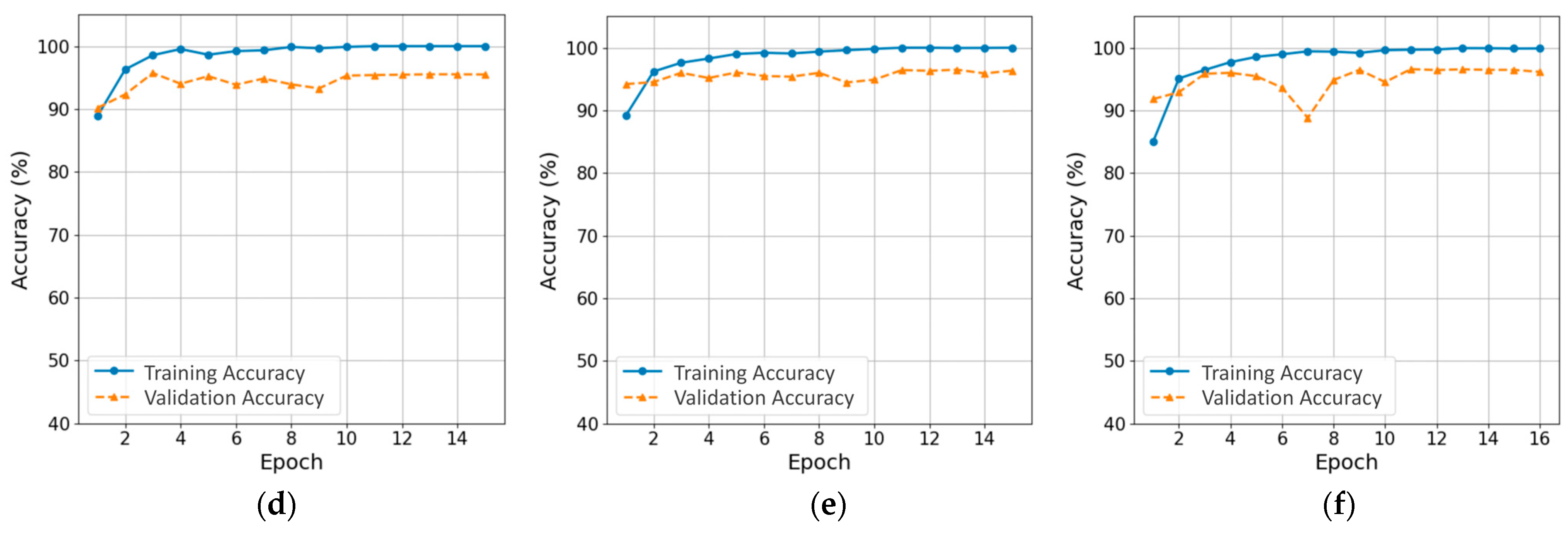
3.1. Training Process and Computational Efficiency of CNN and ViT Models
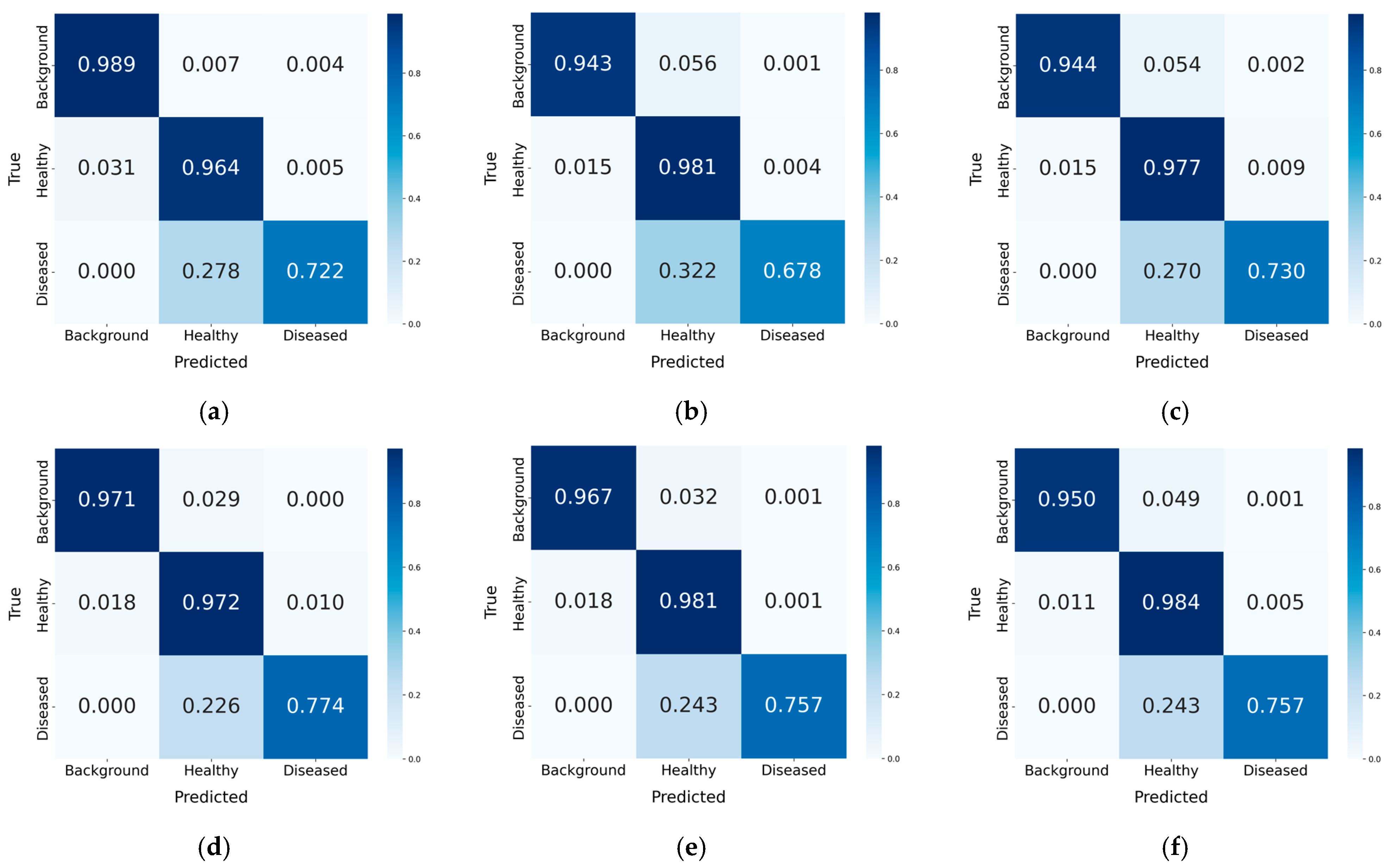
3.2. Classification Performance and Cross-Date Generalization
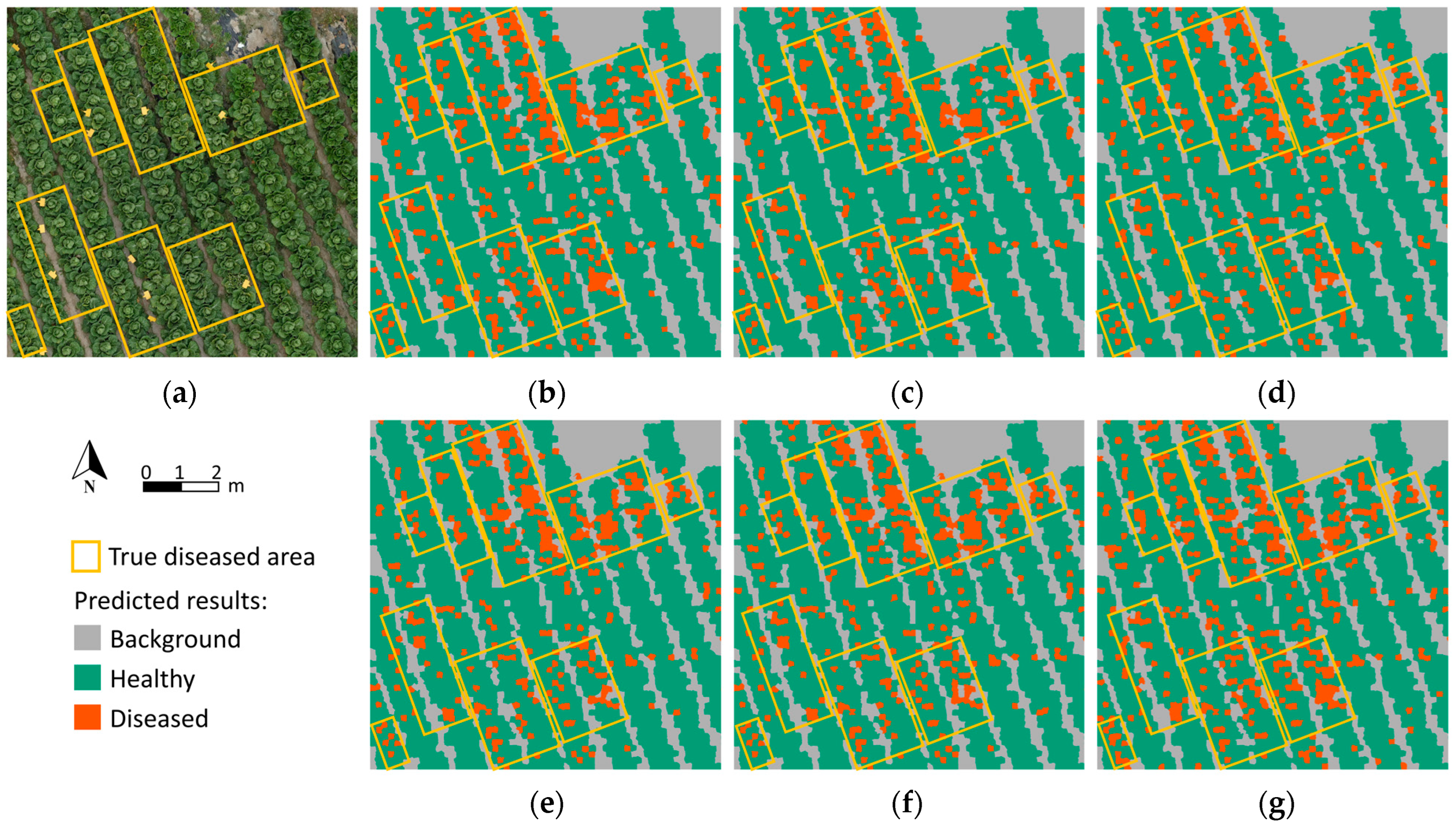
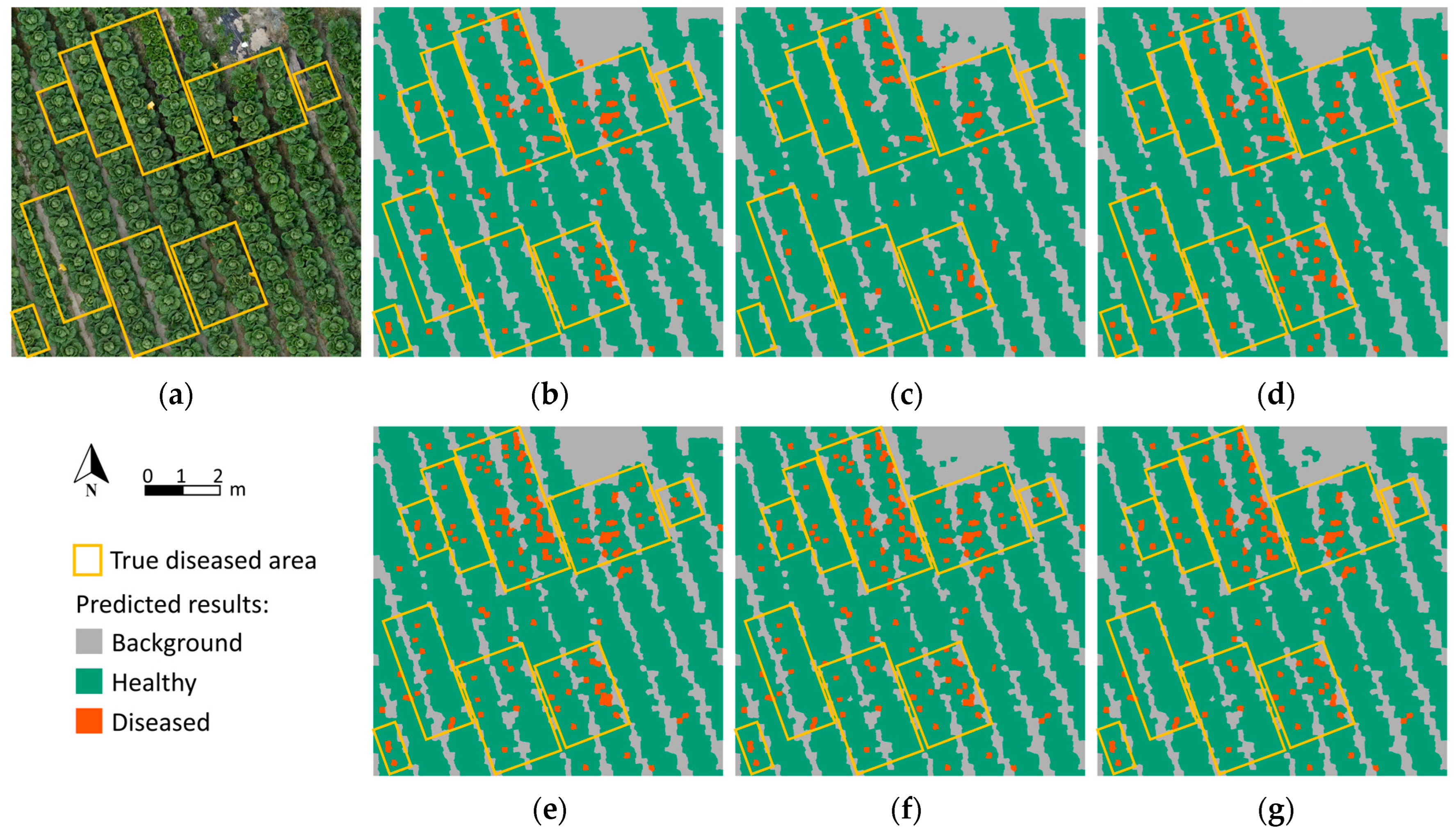
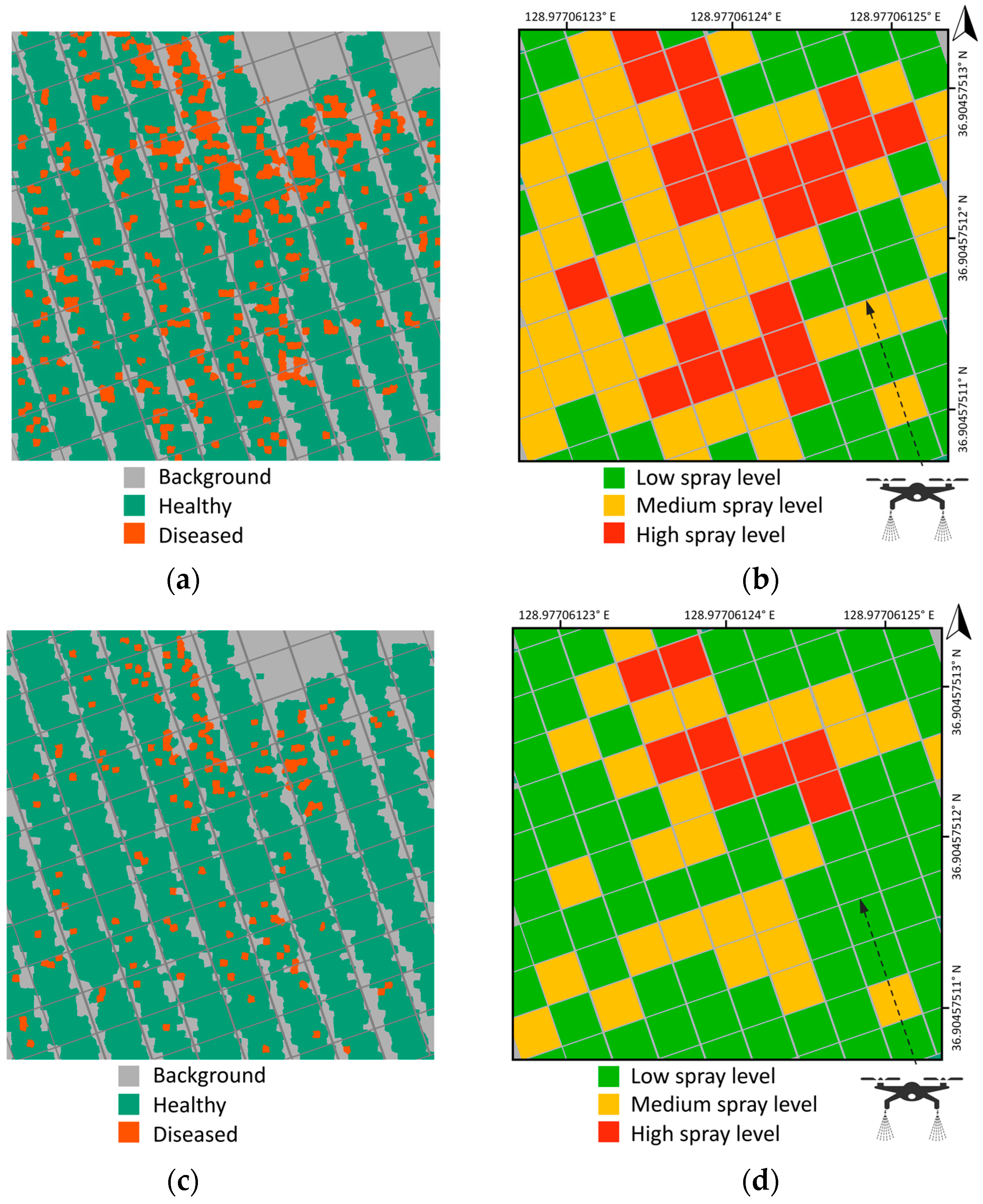
3.3. Classification Visualization and Prescription Map Generation
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Niu, X.; Leung, H.; Williams, P.H. Sources and Nature of Resistance to Downy Mildew and Turnip Mosaic in Chinese Cabbage. J. Am. Soc. Hortic. Sci. 1983, 108, 775–778. [Google Scholar] [CrossRef]
- Kouadio, L.; El Jarroudi, M.; Belabess, Z.; Laasli, S.-E.; Roni, M.Z.K.; Amine, I.D.I.; Mokhtari, N.; Mokrini, F.; Junk, J.; Lahlali, R. A Review on UAV-Based Applications for Plant Disease Detection and Monitoring. Remote Sens. 2023, 15, 4273. [Google Scholar] [CrossRef]
- Barbedo, J. A Review on the Use of Unmanned Aerial Vehicles and Imaging Sensors for Monitoring and Assessing Plant Stresses. Drones 2019, 3, 40. [Google Scholar] [CrossRef]
- Kuswidiyanto, L.W.; Noh, H.-H.; Han, X. Plant Disease Diagnosis Using Deep Learning Based on Aerial Hyperspectral Images: A Review. Remote Sens. 2022, 14, 6031. [Google Scholar] [CrossRef]
- Adão, T.; Hruška, J.; Pádua, L.; Bessa, J.; Peres, E.; Morais, R.; Sousa, J. Hyperspectral Imaging: A Review on UAV-Based Sensors, Data Processing and Applications for Agriculture and Forestry. Remote Sens. 2017, 9, 1110. [Google Scholar] [CrossRef]
- Zhang, N.; Yang, G.; Pan, Y.; Yang, X.; Chen, L.; Zhao, C. A Review of Advanced Technologies and Development for Hyperspectral-Based Plant Disease Detection in the Past Three Decades. Remote Sens. 2020, 12, 3188. [Google Scholar] [CrossRef]
- Kuswidiyanto, L.W.; Wang, P.; Noh, H.H.; Jung, H.Y.; Jung, D.H.; Han, X. Airborne Hyperspectral Imaging for Early Diagnosis of Kimchi Cabbage Downy Mildew Using 3D-ResNet and Leaf Segmentation. Comput. Electron. Agric. 2023, 214, 108312. [Google Scholar] [CrossRef]
- Datta, D.; Mallick, P.K.; Bhoi, A.K.; Ijaz, M.F.; Shafi, J.; Choi, J. Hyperspectral Image Classification: Potentials, Challenges, and Future Directions. Comput. Intell. Neurosci. 2022, 2022, 3854635. [Google Scholar] [CrossRef]
- Bioucas-Dias, J.M.; Plaza, A.; Camps-Valls, G.; Scheunders, P.; Nasrabadi, N.; Chanussot, J. Hyperspectral Remote Sensing Data Analysis and Future Challenges. IEEE Geosci. Remote Sens. Mag. 2013, 1, 6–36. [Google Scholar] [CrossRef]
- Neupane, K.; Baysal-Gurel, F. Automatic Identification and Monitoring of Plant Diseases Using Unmanned Aerial Vehicles: A Review. Remote Sens. 2021, 13, 3841. [Google Scholar] [CrossRef]
- Guo, H.; Cheng, Y.; Liu, J.; Wang, Z. Low-Cost and Precise Traditional Chinese Medicinal Tree Pest and Disease Monitoring Using UAV RGB Image Only. Sci. Rep. 2024, 14, 25562. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Wijewardane, N.K.; Jenkins, S.; Bai, G.; Ge, Y.; Graef, G.L. Early Prediction of Soybean Traits through Color and Texture Features of Canopy RGB Imagery. Sci. Rep. 2019, 9, 14089. [Google Scholar] [CrossRef]
- Pfordt, A.; Paulus, S. A Review on Detection and Differentiation of Maize Diseases and Pests by Imaging Sensors. J. Plant Dis. Prot. 2025, 132, 40. [Google Scholar] [CrossRef]
- Shahi, T.B.; Xu, C.-Y.; Neupane, A.; Guo, W. Recent Advances in Crop Disease Detection Using UAV and Deep Learning Techniques. Remote Sens. 2023, 15, 2450. [Google Scholar] [CrossRef]
- Abade, A.; Ferreira, P.A.; de Barros Vidal, F. Plant Diseases Recognition on Images Using Convolutional Neural Networks: A Systematic Review. Comput. Electron. Agric. 2021, 185, 106125. [Google Scholar] [CrossRef]
- Tugrul, B.; Elfatimi, E.; Eryigit, R. Convolutional Neural Networks in Detection of Plant Leaf Diseases: A Review. Agriculture 2022, 12, 1192. [Google Scholar] [CrossRef]
- Lu, J.; Tan, L.; Jiang, H. Review on Convolutional Neural Network (CNN) Applied to Plant Leaf Disease Classification. Agriculture 2021, 11, 707. [Google Scholar] [CrossRef]
- Bhatt, D.; Patel, C.; Talsania, H.; Patel, J.; Vaghela, R.; Pandya, S.; Modi, K.; Ghayvat, H. CNN Variants for Computer Vision: History, Architecture, Application, Challenges and Future Scope. Electronics 2021, 10, 2470. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image Is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929. [Google Scholar] [CrossRef]
- Mehta, S.; Rastegari, M. MobileViT: Light-Weight, General-Purpose, and Mobile-Friendly Vision Transformer. arXiv 2022, arXiv:2110.02178. [Google Scholar] [CrossRef]
- Wu, K.; Zhang, J.; Peng, H.; Liu, M.; Xiao, B.; Fu, J.; Yuan, L. Tinyvit: Fast pretraining distillation for small vision transformers. In Proceedings of the European Conference on Computer Vision, Tel Aviv, Israel, 23–27 October 2022; pp. 68–85. [Google Scholar] [CrossRef]
- Singh, V.; Misra, A.K. Detection of Plant Leaf Diseases Using Image Segmentation and Soft Computing Techniques. Inf. Process. Agric. 2017, 4, 41–49. [Google Scholar] [CrossRef]
- Nethala, P.; Um, D.; Vemula, N.; Montero, O.F.; Lee, K.; Bhandari, M. Techniques for Canopy to Organ Level Plant Feature Extraction via Remote and Proximal Sensing: A Survey and Experiments. Remote Sens. 2024, 16, 4370. [Google Scholar] [CrossRef]
- Achanta, R.; Shaji, A.; Smith, K.; Lucchi, A.; Fua, P.; Süsstrunk, S. SLIC Superpixels Compared to State-of-the-Art Superpixel Methods. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 2274–2282. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.-S.; Oh, K.-W. Subsampling-Based Acceleration of Simple Linear Iterative Clustering for Superpixel Segmentation. Comput. Vis. Image Underst. 2016, 146, 1–8. [Google Scholar] [CrossRef]
- Tetila, E.C.; Machado, B.B.; Menezes, G.K.; Da Silva Oliveira, A.; Alvarez, M.; Amorim, W.P.; De Souza Belete, N.A.; Da Silva, G.G.; Pistori, H. Automatic Recognition of Soybean Leaf Diseases Using UAV Images and Deep Convolutional Neural Networks. IEEE Geosci. Remote Sens. Lett. 2020, 17, 903–907. [Google Scholar] [CrossRef]
- Karasiak, N.; Dejoux, J.-F.; Monteil, C.; Sheeren, D. Spatial Dependence between Training and Test Sets: Another Pitfall of Classification Accuracy Assessment in Remote Sensing. Mach. Learn. 2022, 111, 2715–2740. [Google Scholar] [CrossRef]
- He, H.; Garcia, E.A. Learning from Imbalanced Data. IEEE Trans. Knowl. Data Eng. 2009, 21, 1263–1284. [Google Scholar] [CrossRef]
- Buda, M.; Maki, A.; Mazurowski, M.A. A Systematic Study of the Class Imbalance Problem in Convolutional Neural Networks. Neural Netw. 2018, 106, 249–259. [Google Scholar] [CrossRef]
- Enkvetchakul, P.; Surinta, O. Effective Data Augmentation and Training Techniques for Improving Deep Learning in Plant Leaf Disease Recognition. Appl. Sci. Eng. Prog. 2021, 15, 3810. [Google Scholar] [CrossRef]
- Owusu-Adjei, M.; Ben Hayfron-Acquah, J.; Frimpong, T.; Abdul-Salaam, G. Imbalanced Class Distribution and Performance Evaluation Metrics: A Systematic Review of Prediction Accuracy for Determining Model Performance in Healthcare Systems. PLoS Digit. Health 2023, 2, e0000290. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 30 June 2016. [Google Scholar]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the International Conference on Machine Learning (ICML), Long Beach, CA, USA, 9–15 June 2019. [Google Scholar]
- Howard, A.; Pang, R.; Adam, H.; Le, Q.; Sandler, M.; Chen, B.; Wang, W.; Chen, L.C.; Tan, M.; Chu, G.; et al. Searching for MobileNetV3. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 1314–1324. [Google Scholar]
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; Jegou, H. Training data-efficient image transformers & distillation through attention. arXiv 2020, arXiv:2012.12877. [Google Scholar]
- Wang, C.-H.; Huang, K.-Y.; Yao, Y.; Chen, J.-C.; Shuai, H.-H.; Cheng, W.-H. Lightweight Deep Learning: An Overview. IEEE Consum. Electron. Mag. 2024, 13, 51–64. [Google Scholar] [CrossRef]
- Qian, S.; Ning, C.; Hu, Y. MobileNetV3 for Image Classification. In Proceedings of the 2021 IEEE 2nd International Conference on Big Data, Artificial Intelligence and Internet of Things Engineering (ICBAIE), Nanchang, China, 26–28 March 2021; pp. 490–497. [Google Scholar]
- Hoang, V.-T.; Jo, K.-H. Practical Analysis on Architecture of EfficientNet. In Proceedings of the 2021 14th International Conference on Human System Interaction (HSI), Gdańsk, Poland, 8–10 July 2021; pp. 1–4. [Google Scholar]
- Borhani, Y.; Khoramdel, J.; Najafi, E. A Deep Learning Based Approach for Automated Plant Disease Classification Using Vision Transformer. Sci. Rep. 2022, 12, 11554. [Google Scholar] [CrossRef] [PubMed]
- Thakur, P.S.; Chaturvedi, S.; Khanna, P.; Sheorey, T.; Ojha, A. Vision Transformer Meets Convolutional Neural Network for Plant Disease Classification. Ecol. Inform. 2023, 77, 102245. [Google Scholar] [CrossRef]
- Wu, X.; Liu, Y.; Xing, M.; Yang, C.; Hong, S. Image Segmentation for Pest Detection of Crop Leaves by Improvement of Regional Convolutional Neural Network. Sci. Rep. 2024, 14, 24160. [Google Scholar] [CrossRef]
- Lin, X.; Li, C.-T.; Adams, S.; Kouzani, A.Z.; Jiang, R.; He, L.; Hu, Y.; Vernon, M.; Doeven, E.; Webb, L.; et al. Self-Supervised Leaf Segmentation under Complex Lighting Conditions. Pattern Recognit. 2023, 135, 109021. [Google Scholar] [CrossRef]
- Javidan, S.M.; Banakar, A.; Rahnama, K.; Vakilian, K.A.; Ampatzidis, Y. Feature Engineering to Identify Plant Diseases Using Image Processing and Artificial Intelligence: A Comprehensive Review. Smart Agric. Technol. 2024, 8, 100480. [Google Scholar] [CrossRef]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- Wang, C.; Du, P.; Wu, H.; Li, J.; Zhao, C.; Zhu, H. A Cucumber Leaf Disease Severity Classification Method Based on the Fusion of DeepLabV3+ and U-Net. Comput. Electron. Agric. 2021, 189, 106373. [Google Scholar] [CrossRef]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using Deep Learning for Image-Based Plant Disease Detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef]
- Lu, Y.; Chen, D.; Olaniyi, E.; Huang, Y. Generative Adversarial Networks (GANs) for Image Augmentation in Agriculture: A Systematic Review. Comput. Electron. Agric. 2022, 200, 107208. [Google Scholar] [CrossRef]
- Wang, Y.; Yao, Q.; Kwok, J.T.; Ni, L.M. Generalizing from a Few Examples. ACM Comput. Surv. 2021, 53, 63. [Google Scholar] [CrossRef]
- Shin, H.-C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef] [PubMed]
- Al Sahili, Z.; Awad, M. The Power of Transfer Learning in Agricultural Applications: AgriNet. Front. Plant Sci. 2022, 13, 992700. [Google Scholar] [CrossRef] [PubMed]
- Weersink, A.; Fraser, E.; Pannell, D.; Duncan, E.; Rotz, S. Opportunities and Challenges for Big Data in Agricultural and Environmental Analysis. Annu. Rev. Resour. Econ. 2018, 10, 19–37. [Google Scholar] [CrossRef]
- Luo, S.; Wen, S.; Zhang, L.; Lan, Y.; Chen, X. Extraction of crop canopy features and decision-making for variable spraying based on unmanned aerial vehicle LiDAR data. Comput. Electron. Agric. 2024, 224, 109197. [Google Scholar] [CrossRef]
- Ahmad, N.; Asif, H.M.S.; Saleem, G.; Younus, M.U.; Anwar, S.; Anjum, M.R. Leaf Image-Based Plant Disease Identification Using Color and Texture Features. Wirel. Pers. Commun. 2021, 121, 1139–1168. [Google Scholar] [CrossRef]
- Ban, S.; Tian, M.; Hu, D.; Xu, M.; Yuan, T.; Zheng, X.; Li, L.; Wei, S. Evaluation and Early Detection of Downy Mildew of Lettuce Using Hyperspectral Imagery. Agriculture 2025, 15, 444. [Google Scholar] [CrossRef]
- Ahmad, U.; Nasirahmadi, A.; Hensel, O.; Marino, S. Technology and Data Fusion Methods to Enhance Site-Specific Crop Monitoring. Agronomy 2022, 12, 555. [Google Scholar] [CrossRef]

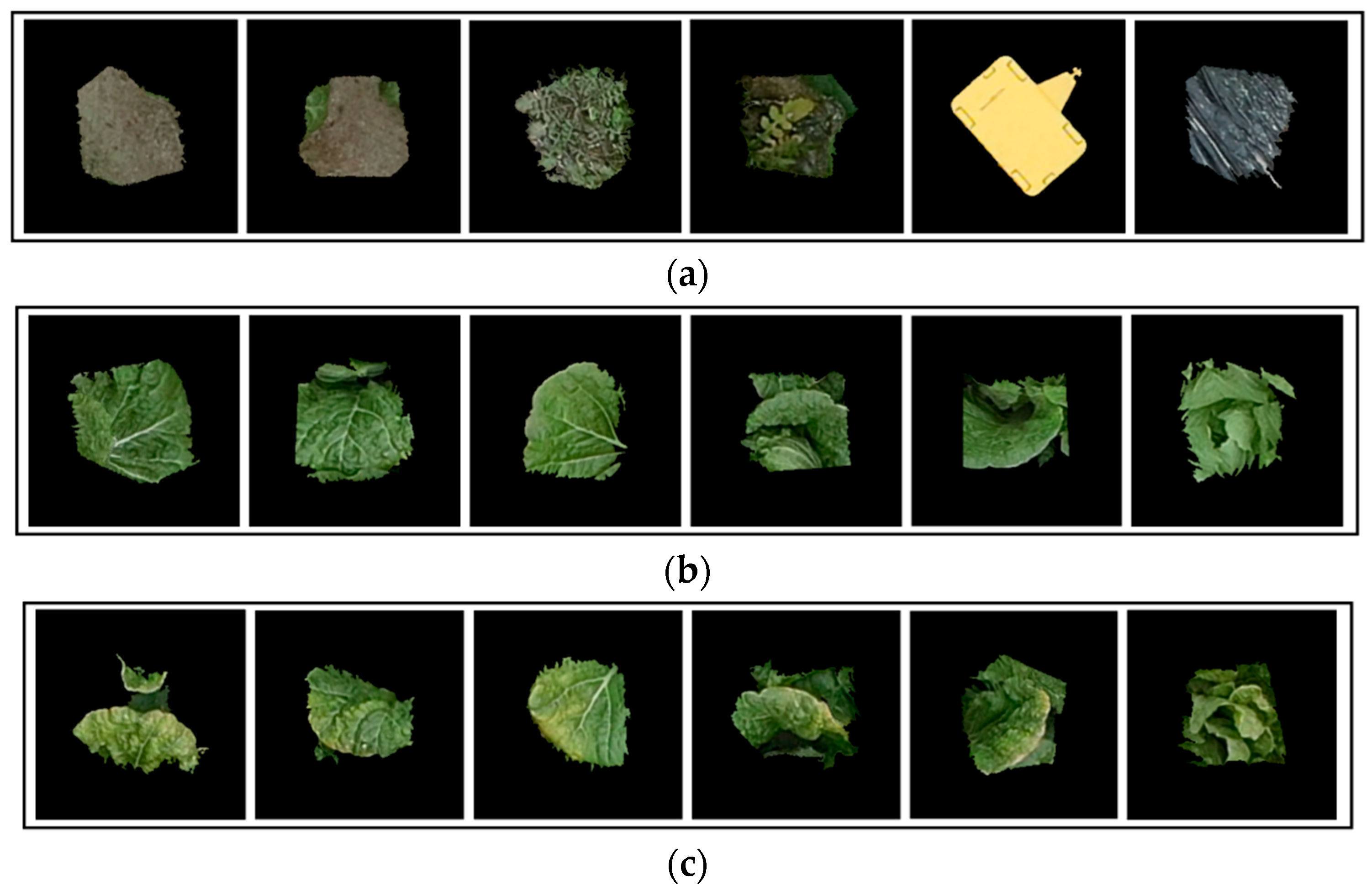




| Class | Original Training Set | Balanced Training Set | Validation Set | Testing Set (25 October) | Testing Set (18 October) |
|---|---|---|---|---|---|
| Background | 1520 | 1520 | 767 | 860 | 819 |
| Healthy | 6742 | 3040 | 2367 | 2211 | 2458 |
| Diseased | 610 | 1520 | 258 | 321 | 115 |
| Total | 8872 | 6080 | 3392 | 3392 | 3392 |
| Model | Type | Parameters (M) | FLOPs 1 (G) | Input Size (Pixels) | Structural Features |
|---|---|---|---|---|---|
| ResNet-18 | CNN | 11.7 | 1.8 | 224 × 224 | Residual blocks, 3 × 3 conv, and skip connections |
| EfficientNet-B0 | CNN | 5.3 | 0.39 | 224 × 224 | MBConv 2, SE 3, and compound scaling |
| MobileNetV3-Large | CNN | 5.4 | 0.22 | 224 × 224 | Inverted residuals, SE, and h-swish 4 |
| DeiT-Tiny | ViT | 5.7 | 1.3 | 224 × 224 | Patch embedding, transformer blocks, class token, and distillation |
| TinyViT-5M | ViT (Hybrid) 5 | 5.4 | 1.3 | 224 × 224 | Local convolution, hierarchical transformer, and window attention |
| MobileViT-S | ViT (Hybrid) | 5.6 | 1.1 | 224 × 224 | Convolution blocks, transformer blocks, and local–global fusion |
| Model | Epochs | Training Time (s) | Inference Time (s) | Test Accuracy | Precision (Diseased) | Recall (Diseased) | F1-Score (Diseased) | Macro F1-Score |
|---|---|---|---|---|---|---|---|---|
| ResNet-18 | 15 | 111.70 | 1.53 | 0.936 | 0.658 | 0.897 | 0.759 | 0.896 |
| EfficientNet-B0 | 14 | 176.79 | 2.09 | 0.941 | 0.704 | 0.882 | 0.783 | 0.904 |
| MobileNetV3-Large | 14 | 105.83 | 1.25 | 0.941 | 0.710 | 0.826 | 0.764 | 0.899 |
| DeiT-Tiny | 15 | 140.83 | 1.78 | 0.948 | 0.731 | 0.882 | 0.799 | 0.913 |
| TinyViT-5M | 15 | 225.15 | 2.59 | 0.947 | 0.704 | 0.903 | 0.791 | 0.911 |
| MobileViT-S | 16 | 326.15 | 3.49 | 0.946 | 0.691 | 0.931 | 0.793 | 0.912 |
| Model | Inference Time (s) | Test Accuracy | Precision (Diseased) | Recall (Diseased) | F1-Score (Diseased) | Macro F1-Score |
|---|---|---|---|---|---|---|
| ResNet-18 | 1.34 | 0.962 | 0.847 | 0.722 | 0.779 | 0.901 |
| EfficientNet-B0 | 2.12 | 0.961 | 0.867 | 0.678 | 0.761 | 0.895 |
| MobileNetV3-Large | 1.42 | 0.960 | 0.785 | 0.730 | 0.757 | 0.893 |
| DeiT-Tiny | 1.88 | 0.965 | 0.781 | 0.774 | 0.777 | 0.904 |
| TinyViT-5M | 2.64 | 0.970 | 0.927 | 0.757 | 0.829 | 0.918 |
| MobileViT-S | 3.57 | 0.968 | 0.870 | 0.757 | 0.809 | 0.915 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lyu, Y.; Han, X.; Wang, P.; Shin, J.-Y.; Ju, M.-W. Unmanned Aerial Vehicle-Based RGB Imaging and Lightweight Deep Learning for Downy Mildew Detection in Kimchi Cabbage. Remote Sens. 2025, 17, 2388. https://doi.org/10.3390/rs17142388
Lyu Y, Han X, Wang P, Shin J-Y, Ju M-W. Unmanned Aerial Vehicle-Based RGB Imaging and Lightweight Deep Learning for Downy Mildew Detection in Kimchi Cabbage. Remote Sensing. 2025; 17(14):2388. https://doi.org/10.3390/rs17142388
Chicago/Turabian StyleLyu, Yang, Xiongzhe Han, Pingan Wang, Jae-Yeong Shin, and Min-Woong Ju. 2025. "Unmanned Aerial Vehicle-Based RGB Imaging and Lightweight Deep Learning for Downy Mildew Detection in Kimchi Cabbage" Remote Sensing 17, no. 14: 2388. https://doi.org/10.3390/rs17142388
APA StyleLyu, Y., Han, X., Wang, P., Shin, J.-Y., & Ju, M.-W. (2025). Unmanned Aerial Vehicle-Based RGB Imaging and Lightweight Deep Learning for Downy Mildew Detection in Kimchi Cabbage. Remote Sensing, 17(14), 2388. https://doi.org/10.3390/rs17142388










