Abstract
Individual tree segmentation (ITS) from terrestrial laser scanning (TLS) point clouds is foundational for deriving detailed forest structural parameters, crucial for precision forestry, biomass calculation, and carbon accounting. Conventional ITS algorithms often struggle in complex forest stands due to reliance on heuristic rules and manual feature engineering. Deep learning methodologies proffer more efficacious and automated solutions, but their segmentation accuracy is restricted by imprecise center offset predictions, particularly in intricate forest environments. To address this issue, we proposed a deep learning method, SPA-Net, for achieving tree instance segmentation of forest point clouds. Unlike methods heavily reliant on potentially error-prone global offset vector predictions, SPA-Net employs a novel sampling-shifting-grouping paradigm within its sparse geometric proposal (SGP) module to directly generate initial proposal candidates from raw point data, aiming to reduce dependence on the offset branch. Subsequently, an affinity aggregation (AA) module robustly refines these proposals by assessing inter-proposal relationships and merging fragmented segments, effectively mitigating oversegmentation of large or complex trees; integrating with SGP eliminates the postprocessing step of scoring/NMS. SPA-Net was rigorously validated on two different forest datasets. On both BaiMa and Hong-Tes Lake datasets, the approach demonstrated superior performance compared to several contemporary segmentation approaches evaluated under the same conditions. It achieved 95.8% precision, 96.3% recall, and 92.9% coverage on BaiMa dataset, and achieved 92.6% precision, 94.8% recall, and 88.8% coverage on the Hong-Tes Lake dataset. This study provides a robust tool for individual tree analysis, advancing the accuracy of individual tree segmentation in challenging forest environments.
1. Introduction
Forest resources, as integral components of terrestrial ecosystems, play a pivotal role in safeguarding essential ecosystem functions, including biodiversity support, atmospheric regulation, and hydrological cycle homeostasis [1,2]. Consequently, detailed forest structural parameters provide the foundational data indispensable for the sustainable stewardship of these vital resources. Conventional mensuration techniques, however, are notably hampered by substantial costs, inherent temporal limitations, and challenges related to site accessibility [3]. The advent of light detection and ranging (LiDAR) technology, particularly terrestrial laser scanning (TLS), represents a paradigm shift in forestry applications [4,5]. These laser-based methodologies have emerged as powerful instruments for detailed forest assessment. Central to leveraging TLS data [6] for detailed forest structural parameters is the capacity for precise individual tree segmentation (ITS), which is indispensable for the accurate estimation of critical biophysical parameters, such as above-ground biomass, sequestered carbon, and taxonomic classification.
Individual tree segmentation (ITS) methodologies based on LiDAR have advanced into three principal paradigms: the canopy height model (CHM)-based method, the point-based method, and the deep learning-based method [7]. CHM-based methods initially involve rasterizing 3D point clouds into 2D CHM representations, followed by tree apex identification through local maxima detection within the CHM [8,9]. Subsequent image segmentation algorithms are then employed to delineate canopy boundaries and accomplish individual tree segmentation. Hyyppä et al. [10] utilized fixed 3 × 3 search windows to detect local maxima as seed points for subsequent region growing segmentation. Li et al. [11] introduced point cloud density features for the first time to diagnose the accuracy of the initial segmentation based on the traditional CHM segmentation, and further utilized the density and height information to guide the three-dimensional morphological analysis of the canopy point cloud, thus accurately correcting the segmentation errors. CHM-based methods offer computationally efficient crown segmentation in forests but face inherent limitations from 3D to 2D rasterization [12] that cause substantial loss of 3D structural data. The point-based approach is a way to overcome the limitations of the CHM-based approach, which is primarily categorized into clustering algorithms and region growing methods [13,14,15]. Clustering approaches partition points within feature spaces analogous to 3D tree point clouds [16]. Morsdorf et al. [17] employed tree apex identification as initial cluster seeds, utilizing k-means clustering for single-tree segmentation. In contrast to clustering methods, region growing operates through iterative rule-based merging and partitioning of point clusters. These methodologies integrate morphological and ecological priors into geometric criteria, thereby enriching semantic information for enhanced segmentation fidelity [18]. Liu et al. [19] pioneered a trunk growth methodology for individual tree segmentation, demonstrating efficacy in natural forest ecosystems. Wang et al. [20] developed an unsupervised segmentation framework leveraging super voxel morphological features, achieving high precision of segmentation in synthetic forest datasets. While point-based extraction directly processes 3D structural data that can enable superior delineation of understory and subcanopy trees [21], it faces limitations in computational efficiency, accuracy degradation at scale, limited generalizability, and hyperparameter sensitivity during large-scale point cloud processing [22,23,24,25,26].
Building upon recent advancements in point cloud semantic segmentation and inspired by emerging developments in point cloud instance segmentation [27,28], researchers have begun exploring deep learning-based methods to address individual tree delineation [29,30,31]. Luo et al. [32] proposed a novel top-down approach that leverages neural networks to learn geometric feature distributions at crown edges, integrating tree center detection and region growing algorithms to achieve high precision individual tree segmentation in complex urban environments. Hu et al. [33] enhanced the point Transformer architecture to precisely separate ground and non-tree points, subsequently combining watershed algorithms with hierarchical clustering to resolve crown occlusion challenges. Further advancing this domain, Chang et al. [34] present a two-stage approach: semantic segmentation isolates tree points while filtering understory noise, followed by a YOLOv3 detector for trunk localization and multi-level adaptive clustering to effectively address small tree omission and dense canopy occlusion. These innovations mark a transition from isolated algorithmic approaches to model symbiosis, enabling robust tree instance delineation. Notably, while deep learning methods adopt 3D semantic segmentation frameworks [35], tree instance segmentation often relies on classical algorithms that often face undersegmentation and oversegmentation issues, with accuracy heavily relying on point cloud density, parameter settings, and terrain noise. This highlights the critical need for automated, robust, and generalizable ITS methods.
Driven by the conceptual advancement of panoptic segmentation, researchers have increasingly focused on integrating semantic and instance segmentation within 3D point cloud processing frameworks [36,37,38,39]. This bottom-up segmentation paradigm enables deep learning networks to concurrently learn semantic features and instance discriminative representations, offering novel pathways toward fully automated individual tree segmentation [40,41,42,43]. Early studies predominantly adapted indoor scene instance segmentation methodologies. Jiang et al. [44] proposed PointGroup, which employs dual coordinate clustering in both raw and centroid offset spaces, coupled with ScoreNet for the quality assessment of candidate instances. Their innovation lies in leveraging inter-object spatial voids, for instance, grouping, which is a strategy with implications for separating adjacent trees in forest environments. Xiang et al. [45,46] proposed a novel urban landscape panoptic segmentation strategy to experimentally compare different 3D backbone networks and instance segmentation strategies for outdoor mobile mapping point clouds. Their experiments demonstrated that, regardless of the backbone, instance segmentation by clustering embedding features is better than using shifted coordinates. Nevertheless, these approaches primarily target urban tree segmentation, with limited attention to forest environments. Xiang et al. [47] further developed ForAINet, which integrates complementary strategies of 3D centroid prediction and discriminative embeddings. By employing a cylindrical block partitioning strategy to mitigate memory constraints in large-scale scenes, the segmentation back-end achieves an over 85% F1-score for individual trees. Concurrently, Wielgosz et al. [48] introduced the SegmentAnyTree, which enhances point cloud data through stochastic downsampling and employs a sparse convolutional network with a multi-task learning head to predict semantic labels and instance offset vectors. This model attained an mAP50 of 64.0% in Norwegian spruce forests. However, postprocessing steps involving the aggregation of fragmented tree projections risk introducing errors such as instance fragmentation. In contrast, Henrich et al. [49] proposed TreeLearn, which circumvents heuristic block merging by predicting offset vectors from points to tree bases, projecting points to their respective trunks, and clustering complete instances across the entire point cloud.
Despite these advancements, deep learning-based individual tree segmentation faces persistent challenges in point cloud processing: (1) Sparse and irregular point distributions complicate precise boundary delineation between adjacent crowns, particularly under dense canopy overlaps. (2) Overreliance on learnable offset vectors for displacement in center shift is inherently limited by point cloud sparsity, density differences, and morphological diversity across tree species, leading to erroneous point assignments. (3) Fragmentation and oversegmentation persist in clustering algorithms, especially for irregularly shaped or large trees, undermining segmentation fidelity.
To address these limitations, we propose an automated individual tree segmentation network, SPA-Net, designed to overcome inaccurate centroid localization and over-segmentation, and enhance the accuracy of segmentation. Our study has four main contributions: (1) An efficient automated segmentation network based on deep learning for individual tree segmentation from forest point clouds was developed to achieve excellent results in both plantation forests. (2) A sparse geometric proposal (SGP) module is designed that effectively addresses the issues of reliance on offset vectors and the challenges of offset prediction, while accelerating clustering speed, through a three-tier cascaded strategy of medoid sampling, centripetal shrink, and topological connectivity clustering. (3) An affinity aggregation (AA) module is proposed that mitigate over-segmentation and significantly improve large tree segmentation coherence. (4) An SGP + AA approach replaces conventional offset/embedding branches and subsequent scoring/NMS filtering methods.
2. Materials
2.1. Study Area
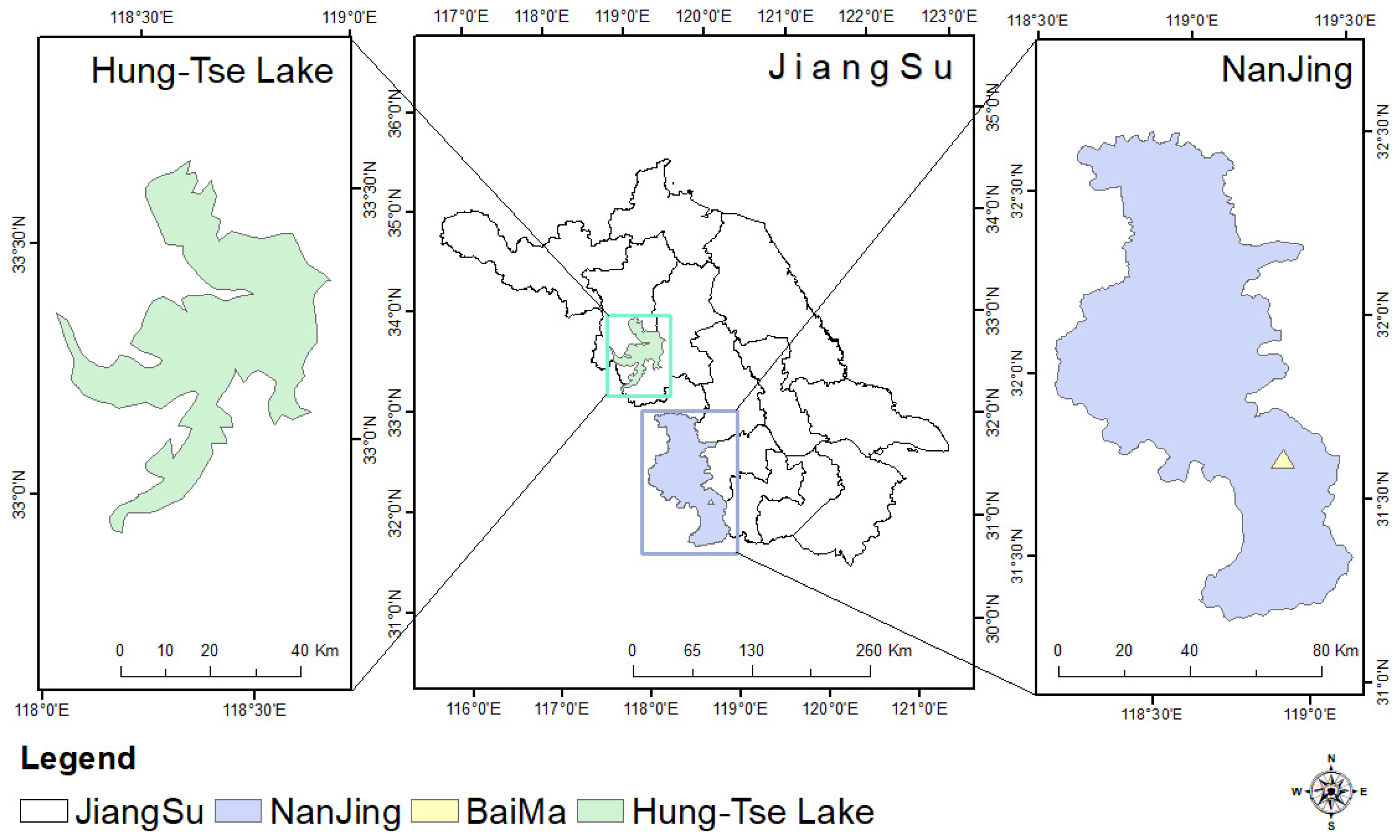
This study used two geographically distinct datasets to encompass diverse tree species and different growth environments. The geographical location is shown in Figure 1. The primary sampling site was the BaiMa Teaching and Research Base of Nanjing Forestry University (31°61′N,119°18′E). The primary tree species in this area are Cherry and Begonia, both of which were seven years old at the time of data collection. The second site was the Hong-Tse Lake wetland in Huaian City, Jiangsu Province, China (31°61′N,119°18′E). The dominant tree species here is Poplar, with ages ranging from 7 to 12 years. Within this plot, the mean measured diameter at breast height (DBH) was 184 mm, and the average tree height was 20.7 m. An overview of the field inventory data from the plots used for validation is provided in Table 1.
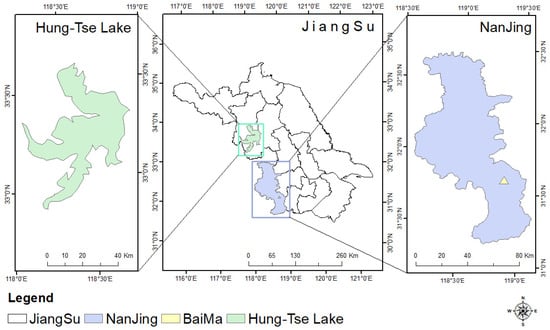
Figure 1.
Geographic location of BaiMa and Hung-Tse Lake in the study area.

Table 1.
The list of data used for validation included density, diameter at breast height (DBH), and tree height.
2.2. Acquisition of Data
Both datasets were acquired using a Trimble TX7 terrestrial laser scanner (TLS). The detailed performance and evaluation metrics of Trimble TX7 are presented in Table 2.

Table 2.
Technical specifications for the Trimble TX7 laser scanner.
Data for the BaiMa dataset were acquired in May 2021. For the Hong-Tes Lake dataset, acquisition occurred in August 2023 under clear weather conditions with light breezes, specifically chosen to minimize the impact of foliage movement on scan quality. Our field measurement setup is shown in Figure 2.

Figure 2.
Field scanning scenarios.
In both study areas, as illustrated in Figure 3, a multi-station scanning strategy was employed to ensure comprehensive capture of the forest 3D structure and to minimize data loss resulting from tree occlusion.
Figure 3.
Distribution of foundations at 12 stations in the poplar sample plot.
Following field acquisition, the point cloud data collected from each individual scan station was coregistered into a unified coordinate system. This process resulted in the generation of complete point cloud datasets containing approximately 150 trees for the BaiMa datasets and 80 trees for the Hong-Tes Lake datasets.
3. Methods
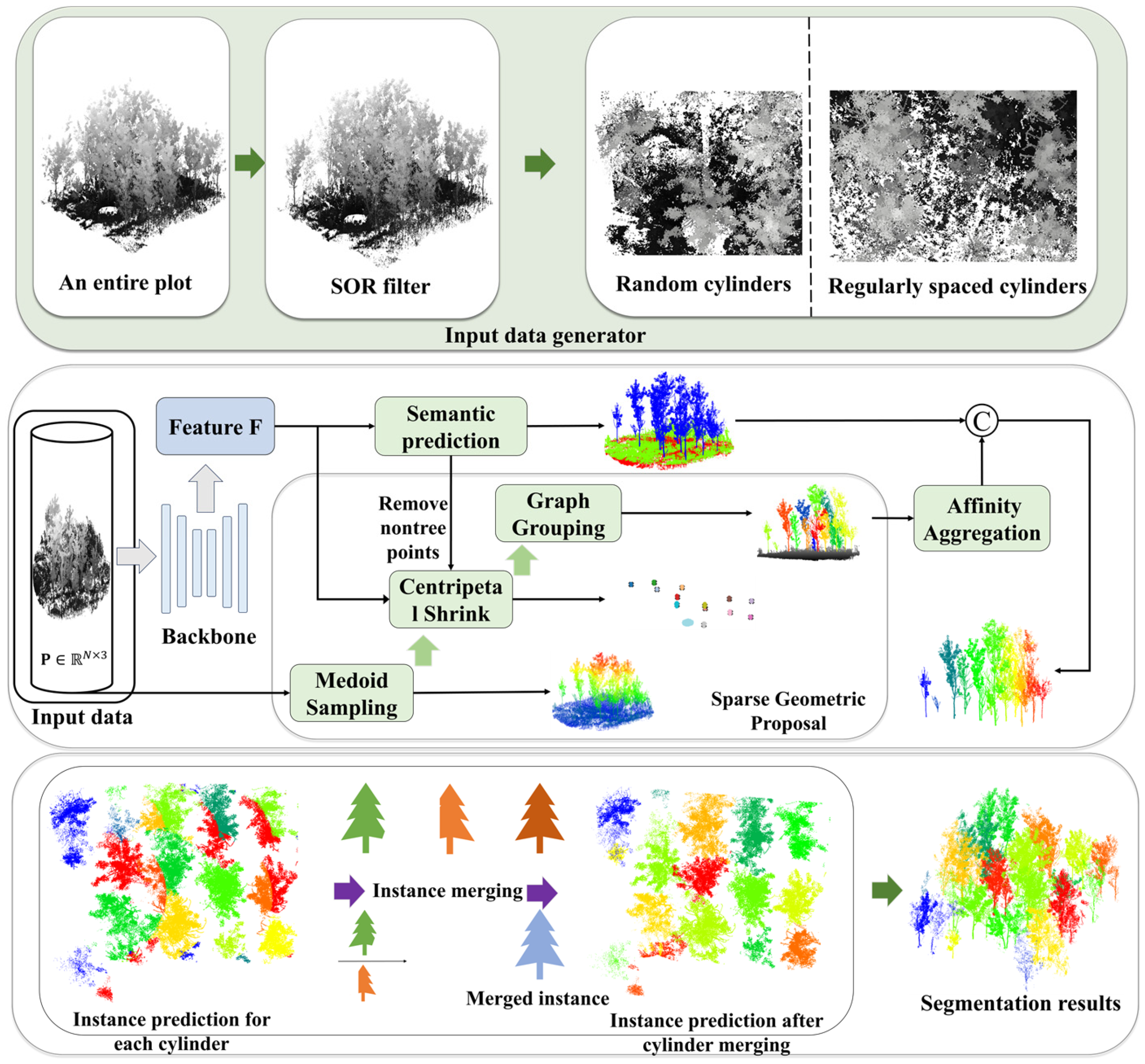
The SPA-Net network for individual tree segmentation initiates processing with an input data generator applying statistical outlier removal (SOR) and cylindrical sampling to large-scale point clouds. These sampled cylinders are subsequently fed into a 3D backbone network for deep per-point feature extraction. A semantic head then performs point-wise classification (tree and non-tree). An instance branch, designed for instance differentiation among tree points, which comprises two key modules: the sparse geometric proposal (SGP) module generates initial instance proposals directly, eschewing traditional offset predictions. Concurrently, the affinity aggregation (AA) module utilizes backbone features to assess inter-proposal relationships, merging fragmented proposals of the same tree and outputting locally consistent instance results per cylinder. Global consistency is achieved via the cylinder merging (CM) module, which integrates results across all cylinders. The CM resolves overlap conflicts and merges cross-boundary fragments, assigning a globally unique instance ID to each tree instance. The final segmentation merges semantic labels and global instance IDs. The architecture of the SPA-Net network is depicted in Figure 4.
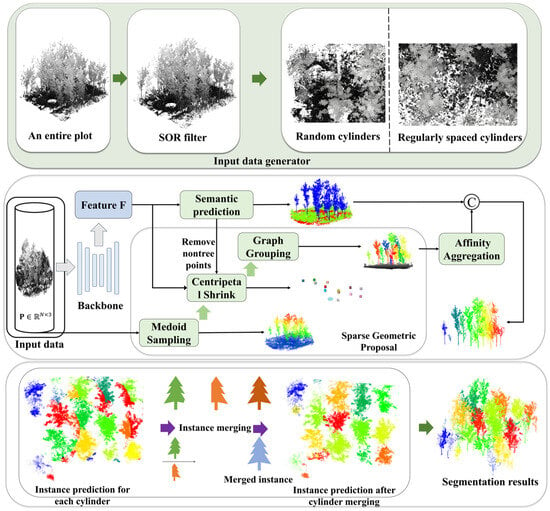
Figure 4.
The flowchart of the SPA-Net network.
3.1. Input Data Generator
To eliminate noise and statistical outliers from the forest point clouds, we utilize the statistical outlier removal (SOR) function within the CloudCompare software (Version: 2.12.4), which serves to eliminate conspicuous noise and statistical outliers, thereby precluding the interference of these invalid points in subsequent analyses and enhancing the overall reliability of the final results. Following this point cloud noise filtration, we implement a cylindrical sampling strategy to generate data subsets suitable for network training and processing. Specifically, a vertical cylindrical volume, defined by a radius of 4 m and centered on a designated point within the point cloud, is extracted to serve as an independent sampling unit. During training, the center point is determined by randomly sampling a training data point, with points belonging to rarer classes given a higher probability of being selected to ensure adequate coverage; specifically, the sampling probability is proportional to the inverse square root of the class frequency. In the testing phase, however, the center points are sampled regularly along a grid defined by the x and y coordinates using a fixed step size to ensure even coverage of the entire point cloud area. Therefore, the specific point is not fixed but is determined either by weighted random sampling during training or by regular grid sampling during testing. All points encompassed within this cylinder constitute a single sample.
3.2. Backbone
Upon acquiring the subsampled point cloud representing a cylindrical subvolume, we employ a 3D U-Net network, implemented using the Minkowski Engine, as the backbone network to extract hierarchical point features [50]. This symmetric encoder–decoder structure, typically enhanced with skip connections, processes the input through multi-resolution sparse volumetric representations. At each hierarchical level, sparse 3D convolutions, operate on the voxelized data to capture contextual relationships across varying spatial extents, ultimately yielding rich point-wise feature vectors F [51]. Finally, the semantic head and instance branch take the fused features F as the input and output semantic label and instance proposals.
3.3. Semantic Branch
The semantic branch performs semantic segmentation by assigning a specific label (tree or non-tree) to each point. The non-tree class encompasses points belonging to the ground or smaller understory vegetation, while the tree class includes all points belonging to actual trees. To achieve this, point features F that are extracted from the U-Net backbone output are input into the semantic segmentation branch that consists of a three-layer multilayer perceptron (MLP) that outputs per-point scores for the tree and non-tree classes. The class with the highest score is assigned as the point’s semantic label. The module is trained under the supervision of a cross-entropy loss function.
3.4. Sparse Geometric Proposal
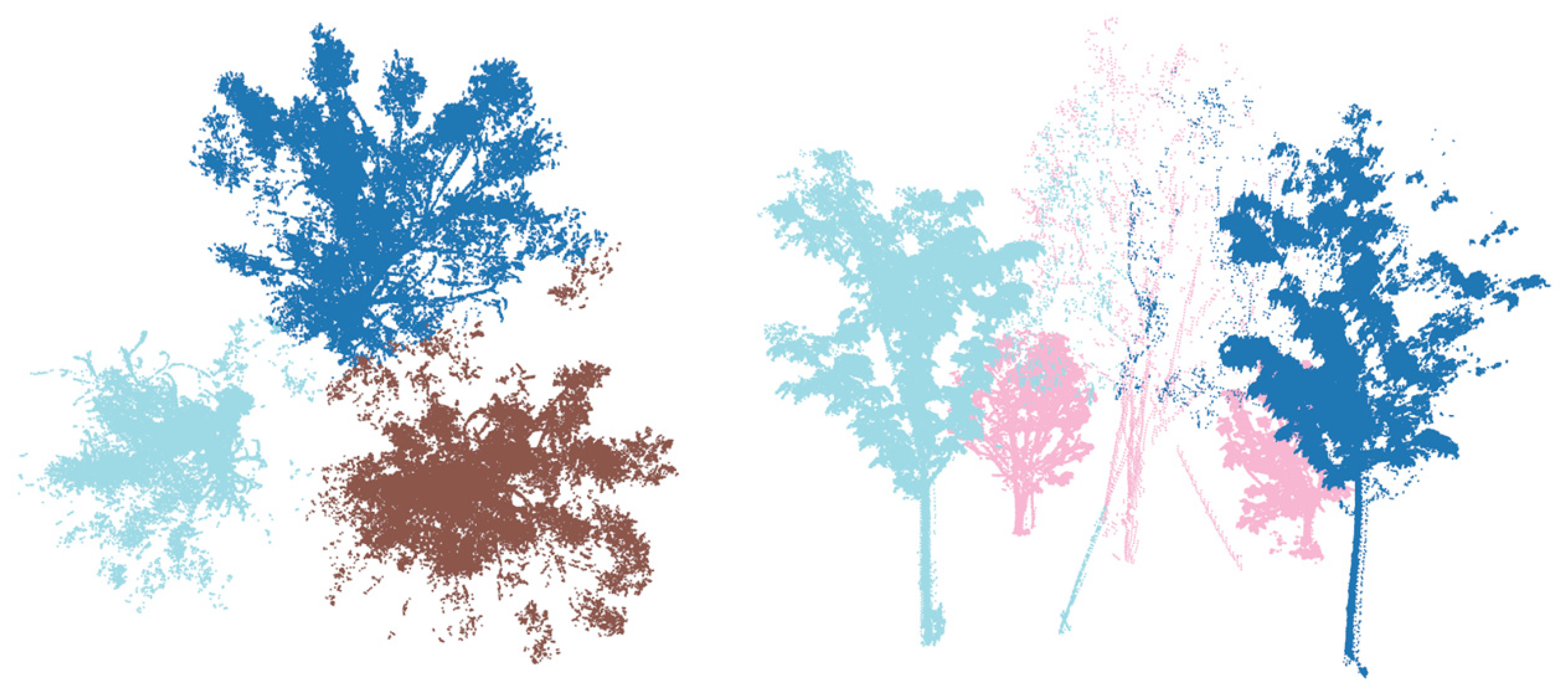
Instance segmentation methods based on 3D center offsets have been widely adopted in previous studies. The core idea is to predict a vector for each point that points toward its instance center, ideally collapsing all points of an instance to a single location. However, accurately predicting these offsets is challenging in sparse LiDAR data. This difficulty is particularly problematic for neighboring trees, as inaccurate offset prediction leads to unreliable estimated centers and erroneous merging of distinct objects. Figure 5 illustrates this incorrect clustering of close instances. To address this critical issue, we propose an SGP module that replaces the problematic offset prediction branch.

Figure 5.
Failure cases in tree clustering due to systematic errors in the space of offset-shifted coordinates.
The overall structure of our proposed SGP module is detailed in Figure 6.
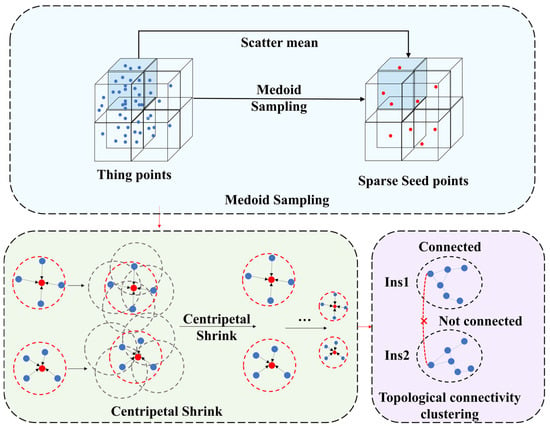
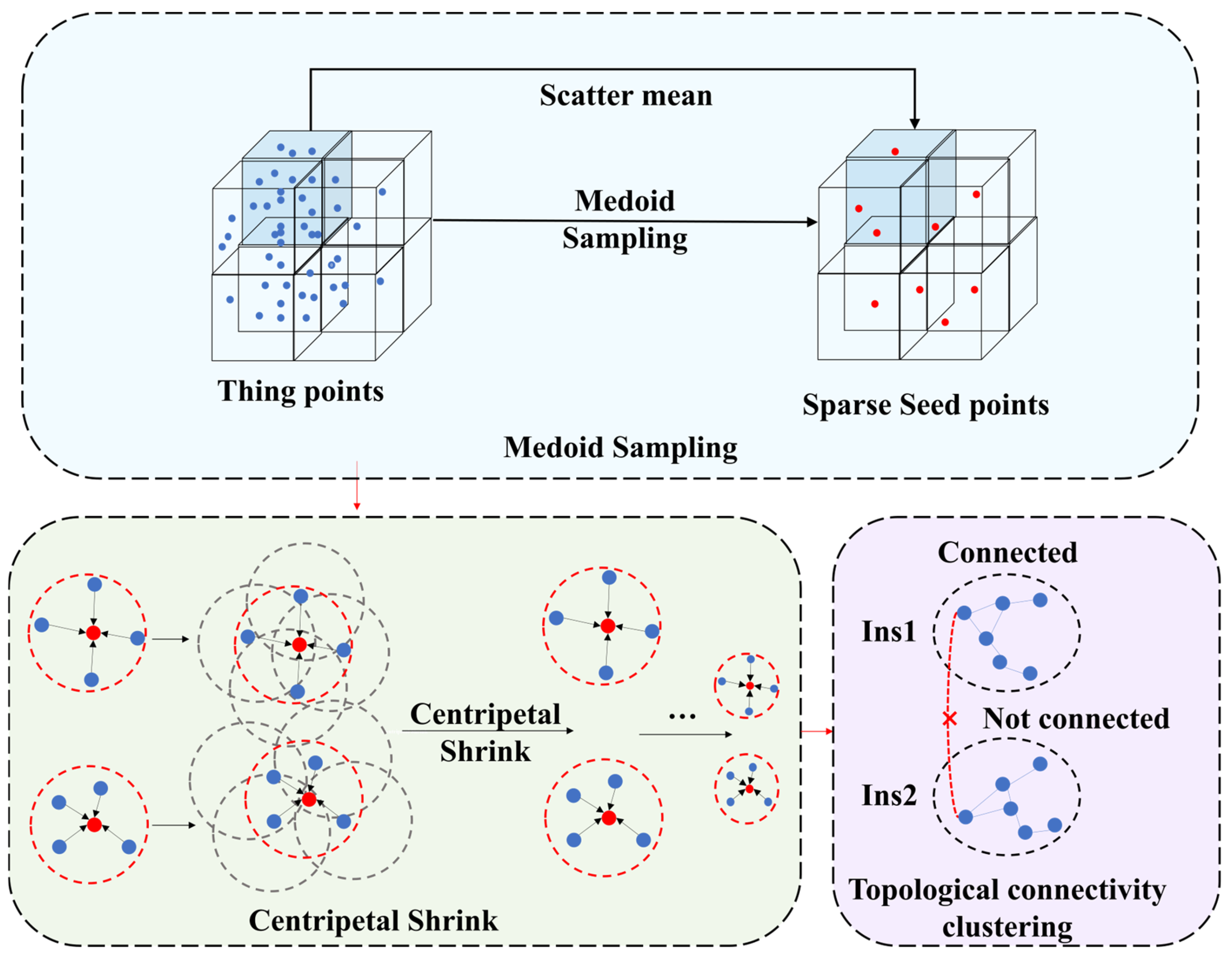
Figure 6.
Flow diagram of the SGP. The red dots represent sparse seed points, and the × represents not connected.
The pseudocode depicting the specific implementation procedure of the SGP module is illustrated as follows (Algorithm 1):
| Algorithm 1. The specific implementation procedure of the SGP module |
| Input: Ptree, Voxel_size, RL, L, RL/2 Output: Instance_IDs |
| // Part 1: Medoid Sampling (MS) |
| S←∅ |
| Voxel_map← Initialize_a_map() for each point p in Ptree do voxel_coord←floor(p.xyz/Voxel_size) Voxel_map[voxel_coord].append(p) end for Point_to_seed_map← Initialize_a_map() |
| for each voxel_coord,points_in_voxel in Voxel_map do Sseed ←Mean(points_in_voxel.xyz) S.append(sseed ) for each point p in points_in_voxel do Point_to_seed_map[p]←Sseed end for end for // Part 2: Centripetal Shrink (CS) Srefined ←S G← Build graph on Srefined with edges connecting points if distance <RL Anorm ← Calculate normalized adjacency matrix from G Define encoding function: f(v)←sign(v)⋅log(1+∣v∣) Define decoding function: f−1(venc )←sign(venc )⋅(exp(∣venc ∣)−1) for i←1 to L do Sencoded ←f(Srefined ) Snext_encoded ←Anorm ⋅Sencoded Srefined ←f−1(Snext_encoded ) end for // Part 3: Topological Connectivity Clustering (TCC) Gfinal ← Build graph on Srefined with edges connecting points if distance <RL/2 Seed_Instance_IDs← Connected_Components_Labeling(Gfinal ) Instance_IDs← Initialize_array_for_all_points_in_Ptree for each original point p in Ptree do Sseed ←Point_to_seed_map[p] Instance_IDs[p]←Seed_Instance_IDs[Sseed ] end for return Instance_IDs |
Raw LiDAR point clouds, with their large volume and spatial heterogeneity that include density variations with sensor distance and between surfaces, complicate analyses and make full dataset processing computationally prohibitive. To address this, we employ a medoid sampling (MS) strategy where the raw cloud is partitioned into uniform voxel blocks, and for each nonempty voxel, we calculate the average that becomes the single seed point representing that voxel of all points contained within it. This approach decouples sampling from point density, yields a spatially balanced set of representative seed points, and inherently links raw points to their seeds via the voxel structure, avoiding explicit proximity searches.
Initial seed points, while providing a sparse representation, may suffer from localization inaccuracies that hinder precise instance delineation. As shown in Figure 6, we proposed the centripetal shrink (CS) module to mitigate this issue, which iteratively refines seed positions toward tree centroids without relying on pre-defined offsets. The CS module takes as input a sparse set of seed points coordinates , where M denotes the quantity of seed points. A graph structure that represents inter-seed point relationships is established and characterized by a pre-computed normalized adjacency matrix, . The matrix delineates connectivity (edges) among seed points (nodes), predicated upon their initial spatial propinquity within a pre-defined radius RL, D represents the corresponding degree matrix. After 10 iterations, the module yields a refined set of seed point coordinates .
The quintessence of the CS module resides in its iterative refinement loop, where the contraction process, driven by neighborhood averaging, is executed entirely within a transformed, encoded domain. This ensures that the encoding directly influences the geometric interpretation of proximity and centrality during each step of the iterative shrinking. We employ an encoding function and its inverse decoding function . These functions operate element-wise on each coordinate of a trivariate vector ; these operations are specified as:
where, is the encoded value of the coordinate component , is the decoded value of the coordinate component, is the exponential function.
The application of transforms the original Euclidean space into a non-linear encoded space where distances and, consequently, the notion of a neighborhood centroid, are rescaled. This logarithmic encoding paradigm compresses substantial coordinate differentials, thereby fostering more stable adjustments across heterogeneous arboreal scales and point cloud densities.
Given initial seed coordinates , the module iteratively updates these positions for = 0 to L − 1. The extant seed point coordinates are initially mapped into the encoded space:
where, is the matrix of 3D coordinates of all M seed points at iteration . is the matrix of encoded 3D coordinates of all seed points at the iteration . All subsequent operations for this iteration occur in this encoded space.
The iterative contraction is achieved by updating each encoded seed point to the average of the encoded coordinates of its connected neighbors in the graph. This operation inherently uses the encoded representations :
Specifically, for each seed point , its new encoded coordinates become the mean of the encoded coordinates of its neighbors:
where, is the 3D encoded coordinates of seed point p after averaging at iteration . is the number of neighbors of seed point in the graph.
Crucially, because this averaging is performed on (the encoded coordinates), the determination of the neighborhood’s geometric center and the subsequent pull on point are directly influenced by the non-linear properties of the encoding function . Large spatial separations in the original space are compressed in the encoded space, modulating their influence during averaging. This results in an adaptive contraction process where the step size and direction are implicitly adjusted by the encoding. The iterative application of this encoded-space averaging causes connected components of the graph to contract towards their respective geometric centers as defined within this non-linear encoded domain.
The newly computed encoded coordinates , representing the contracted positions within the encoded domain, are subsequently transformed back to the primordial coordinate space for the next iteration or final output:
Subsequent to L iterations, the module outputs the ultimately refined seed point coordinates . This strategy of embedding the iterative graph-based contraction entirely within an encoded space imparts a non-linear sensitivity to the displacement adjustment mechanism. This intrinsic coupling ensures that the encoding is not a mere pre/post-processing step but an integral part of how spatial relationships are interpreted and acted upon during each phase of the iterative refinement. It is ensured that this will preserve the capacity for substantial adjustments whilst concurrently augmenting the precision of fine-grained refinements during convergence.
Subsequent to the centripetal shrink (CS), seed points pertaining to the same tree exhibit enhanced spatial compactness. We then employ a topological connectivity clustering (TCC) method that groups the converged seed points based on connectivity and aggregates them into preliminary instance proposals.
To this end, a new connectivity graph is constructed upon . Graph edges connect pairs of points if their Euclidean distance falls below a typically more stringent threshold RL/2 that effectively dictates the spatial granularity of the generated primitive clusters. Subsequently, a connected components labeling (CCL) algorithm [52] is employed to partition this graph. This process assigns all mutually reachable seed points within the graph to the same unique connected component. Each connected component identified is designated as an initial tree instance candidate, denoted , is the total number of proposals. Crucially, all original input points that were initially associated with the seed points constituting a single connected component are then assigned the corresponding unique candidate identifier ID.
3.5. Affinity Aggregation Module
The initial tree instance candidate generated by the SGP module, while locally coherent, may be split into multiple trees for a single real tree instance due to factors such as point cloud sparsity, occlusion, and the complex structure of large trees. In order to restore instance integrity, we design an affinity aggregation (AA) module with the core idea to learn to predict whether any two initial tree instances , should belong to the same final tree instance, and perform a merger based on this prediction.
For each constituent point within a tree instance , given its point set position and point-wise features , we first employ a 1 × 1 convolutional layer to refine that produces new point-wise features . Multi-layer perceptrons (MLPs) are used to enhance by incorporating the position . Finally, the resulting composite representation is aggregated via max-pooling to generate the global instance feature , which can be calculated as follows:
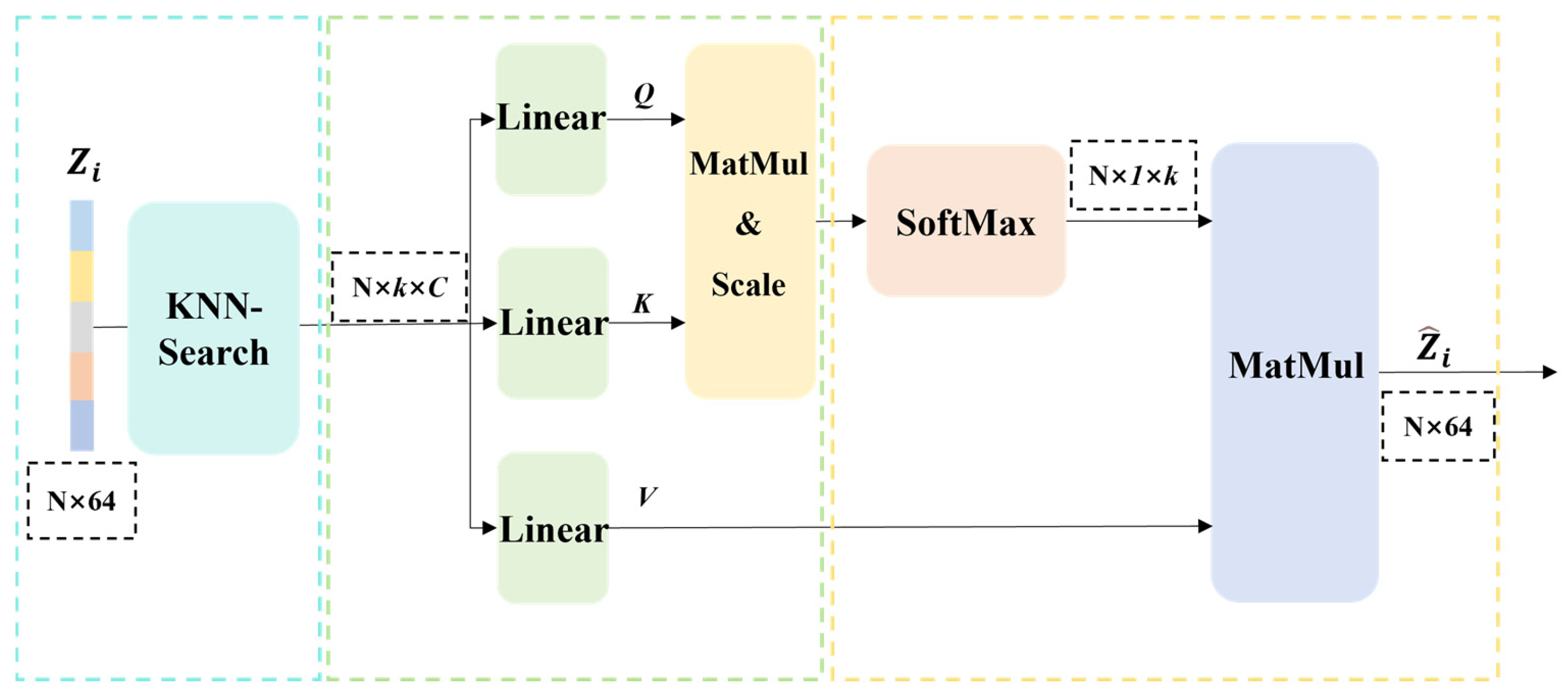
To address the lack of sufficient contextual information in the characteristics of a single instance proposal to judge its relationship with other proposals, we further apply the KNN-transformer [53,54] within the AA module to enhance the interactions among global instance features, as shown in Figure 7, thereby integrating potentially fragmented instances.
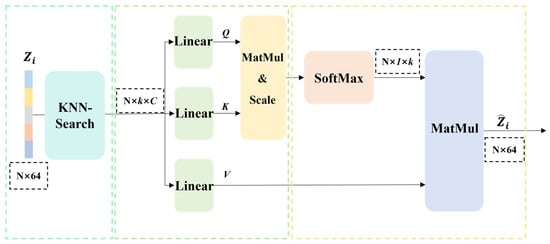
Figure 7.
Flow diagram of the KNN-transformer algorithm.
Pertaining to each instance proposal , its centroid is determined. Predicated upon these centroidal locations, the K-nearest neighbors (KNN) algorithm identifies K spatially proximal proposals . Thereafter, the aggregate instance descriptors corresponding to these adjacent proposals are leveraged for subsequent feature interaction.
To facilitate informative interaction between each instance and its pertinent surrounding counterparts, we leverage the K-nearest neighbors (KNN) algorithm. Based on the spatial coordinates of an instance centroid, we identify, for instance, the K spatially proximal candidate regions among other instances. These identified neighbors collectively constitute the neighborhood, for instance . The criticality of this step resides in its exclusive focus on locally relevant instances, thereby circumventing the computational complexity associated with global calculations.
For each instance candidate region, a global instance feature is acquired. Analogously, for each adjacent candidate region , its corresponding global instance feature is likewise obtained. Subsequently, these global instance features are leveraged for feature interaction.
The global feature is linearly projected as a query vector , while neighborhood features are projected as key vectors and value vectors . Subsequently, the scaled dot-product similarity between the query vector and all key vectors is computed. The resulting similarity scores are then normalized using the Softmax function to yield attention weights. These weights are subsequently employed to compute a weighted sum of the corresponding value vectors , resulting in an enhanced feature vector that integrates the neighborhood context. is calculated as follows:
where * denotes the dot product and = 64 denotes the feature dimension.
Following the acquisition of enhanced features and , we constructed a discriminative network structured as an MLP, which is designed to predict the affinity between any two candidate instances, and , indicating their likelihood of originating from the same ground-truth object. It utilizes the enhanced features of both instances (,), the Euclidean distance between their centroids and the minimum point-to-point distance between them , as input. The resulting scalar value , after applying the Sigmoid activation function, represents the posterior probability P(,) that and originate from the same tree, effectively quantifying this affinity.
The training of this probabilistic predictor is supervised by a binary cross-entropy loss Laff:
where, is set to 1 if and share the same instance ID, otherwise it is set to 0. The instance ID for each proposal is determined by majority voting.
3.6. Cylinder Merging Module
After processing overlapping cylindrical subvolumes, the cylinder merging (CM) module, inspired by Xiang [47], consolidates these local segmentations. It iteratively merges local tree instances into a globally consistent map by evaluating their spatial overlap with existing global instances. If the overlap exceeds a threshold, points are assigned to the existing global ID; otherwise, a new global ID is created. This yields a global instance label map for the final segmentation.
3.7. Evaluation Metrics
Since all of the methods compared in this work achieve a reasonable segmentation into tree and non-tree points, we decided to perform no evaluation of this aspect. Evaluation focused solely on individual tree instance segmentation. In order to evaluate the description accuracy of individual tree instance segmentation, we adopt the evaluation scheme of Xiang [45]. The evaluation process commences by formally defining the ground-truth tree instance set as .
denotes the cardinality of ground-truth instances. Correspondingly, the predicted tree instance set is expressed as .
For each pairwise combination of ground-truth instance and predicted instance , the point-wise intersection over union (IoU) metric is computed through the following formulation:
where , , and , respectively, denote the true positive, false positive, and false negative point sets derived from the spatial correspondence between and . For each ground truth tree, we determine the index of the predicted tree with the highest IoU score:
This pairwise approach ensures that each true tree instance is associated with a predicted instance, even in the presence of mergers or splits. In order to systematically compare the segmentation performance between the different methods, even if the number of trees matched according to a specific criterion varies, the coverage, which is defined as the average of the IoUs between all pairs of true and predicted values, is used as follows:
For a more detailed performance evaluation, we also calculated the average precision, recall, and F1 as follows:
3.8. Implementation Details
For SPA-Net, all experiments were performed using PyTorch 1.12.1 on a workstation equipped with an Intel (Clara, CA, USA) i7-14700K processor, NVIDIA (Clara, CA, USA) GeForce RTX 3090 GPUs (24 GB memory), and 512 GB of RAM. The system ran Ubuntu 22.04 with Python 3.9, CUDA 11.3, and cuDNN 8.2.1. Our PyTorch-based implementation utilized Minkowski Engine 0.5.4 for efficient sparse 3D convolutions. The AdamW algorithm was selected as the optimizer.
4. Results
4.1. Ablation Studies on SGP and AA Module
To understand how each part of our SGP and AA module contributes to its performance, we conducted an ablation study that evaluated the contributions of the instance branch which consists of medoid sampling (MS), centripetal shrink (CS), topological connectivity clustering (TCC), and AA modules, with the results in Table 3.

Table 3.
Ablation experiments on the SGP module and the AA module.
Where, √ indicates that the module is available, × indicates that it has been removed. The full SPA-Net network (MS + CS + TCC + AA) served as the baseline, achieving 95.8% Prec, 96.3% Rec, and 92.9% Cov. The importance of the centripetal shrink (CS) within SGP was tested by comparing the full SGP (MS + CS + TCC) with a version lacking CS (MS + TCC). Removing CS substantially lowered Prec by 2.2%, Rec by 2.5%, and Cov by 4.0% to 86.5%. This shows CS is vital for optimizing initial instance seed locations before TCC grouping.
Effective center shifting thus leads to better clustering by reducing both tree segmentation and incorrect point assignments between trees. In summary, these experiments confirm the benefits of both the CS component within SGP and the AA model.
4.2. Ablation on Clustering Algorithms
In this study, we perform a comprehensive ablation study of clustering algorithms, systematically comparing their performance with established clustering algorithms including MeanShift, DBSCAN and HDBScan [55,56,57]. To adapt these clustering algorithms for instance segmentation tasks, all baseline methods are equipped with an offset prediction head that consist of three consecutive linear layers to estimate per-point offsets towards instance centroids.
As shown in Table 4, we found that guiding standard clustering algorithms like MeanShift or DBScan with predicted offsets improved results, whereas refining point coordinates using center shift prior to clustering was notably more effective.

Table 4.
Ablation on clustering algorithms.
However, our proposed SGP module, which does not rely on separate offset prediction, proved superior. SGP alone reached 95.7% Prec and 90.5% Cov, surpassing the optimized offset-guided approach using HDBScan by 1.7% in Prec and 1% in Cov. This indicates that the SGP integrated grouping process is more potent than the pipeline of offset prediction followed by refinement and standard clustering for generating high-accuracy instance proposals. The final AA module further refines these SGP proposals, enabling the complete SPA-Net network to achieve the overall performance with 95.8% Prec, 96.3% Rec, and 92.9% Cov.
We also conducted a direct quantitative comparison between our SGP module and DS module [55], using the BaiMa dataset. SGP demonstrated superior segmentation accuracy overall, surpassing DS with 1.4% higher Prec and 1.3% higher Cov. Critically, as shown in Table 5, SGP also proved significantly more efficient, executing over thirty times faster than DS.

Table 5.
Ablation experiments on the SGP and DS modules.
Observations suggest that while DS can effectively cluster points for individual instances, potentially boosting Rec under favorable initial offset conditions, its strong grouping mechanism may sometimes introduce boundary imperfections. This can result in compromised Prec and Cov compared to the results achieved by the SGP methodology.
4.3. Comparison with 3D Offset
To validate that our SPA-Net architecture can effectively segment individual trees without relying on traditional 3D center offset prediction, we conducted controlled experiments. These compared four architectural variants: a baseline using learned 3D offsets, our SGP module alone, the offset baseline refined by AA, and the complete SPA-Net. The experimental results are shown in Table 6.

Table 6.
The segmentation results based on the four offset structures.
We compared SGP against the learned 3D offset approach directly, without AA refinement. In this comparison, the SGP module demonstrated superior performance over the baseline 3D offset method. It achieved 1.5% higher Prec and 1.3% higher Cov, albeit with a marginal 0.4% deficit in Rec. This highlights the strength of SGP in producing more accurate initial candidates without relying on direct offset regression. Furthermore, the foundational role of the AA refinement stage was substantiated. Adding AA to the conventional 3D offset architecture substantially improved performance over the offset-only baseline, most notably increasing Cov by 2.2% and Rec by 0.7%.
Finally, the synergistic integration of SGP and AA within the full SPA-Net architecture culminated in the highest performance across all metrics. Crucially, this complete SGP plus AA configuration significantly outperformed the 3D offset baseline. SPA-Net exhibited clear superiority, surpassing the refined offset approach by 3.6% in Cov, 1.9% in Prec, and 1.2% in Rec. This decisively validates our method that the SPA-Net network, using SGP for proposals followed by AA for refinement, represents a more effective paradigm than even 3d offset-based approaches.
4.4. Comparison with the Existing Network
In order to compare with advanced ITS methods, we conducted a systematic performance evaluation comparing our SPA-Net method against state-of-the-art techniques on the BaiMa Plantation Forest Dataset and the Hung-tse Lake dataset. The results confirm the effectiveness of SPA-Net. On the BaiMa dataset, our method achieved 95.8% Prec, 96.3% Rec, 92.9% Cov, and a 96.0% F1.
The corresponding metrics in the Hung-Tse Lake dataset were 92.6% Prec, 94.8% Rec, 88.8% Cov, and 93.7% F1. The result of the comparison on different datasets for individual tree segmentation is shown in Table 7.

Table 7.
Comparison of different datasets for individual tree segmentation performance metrics.
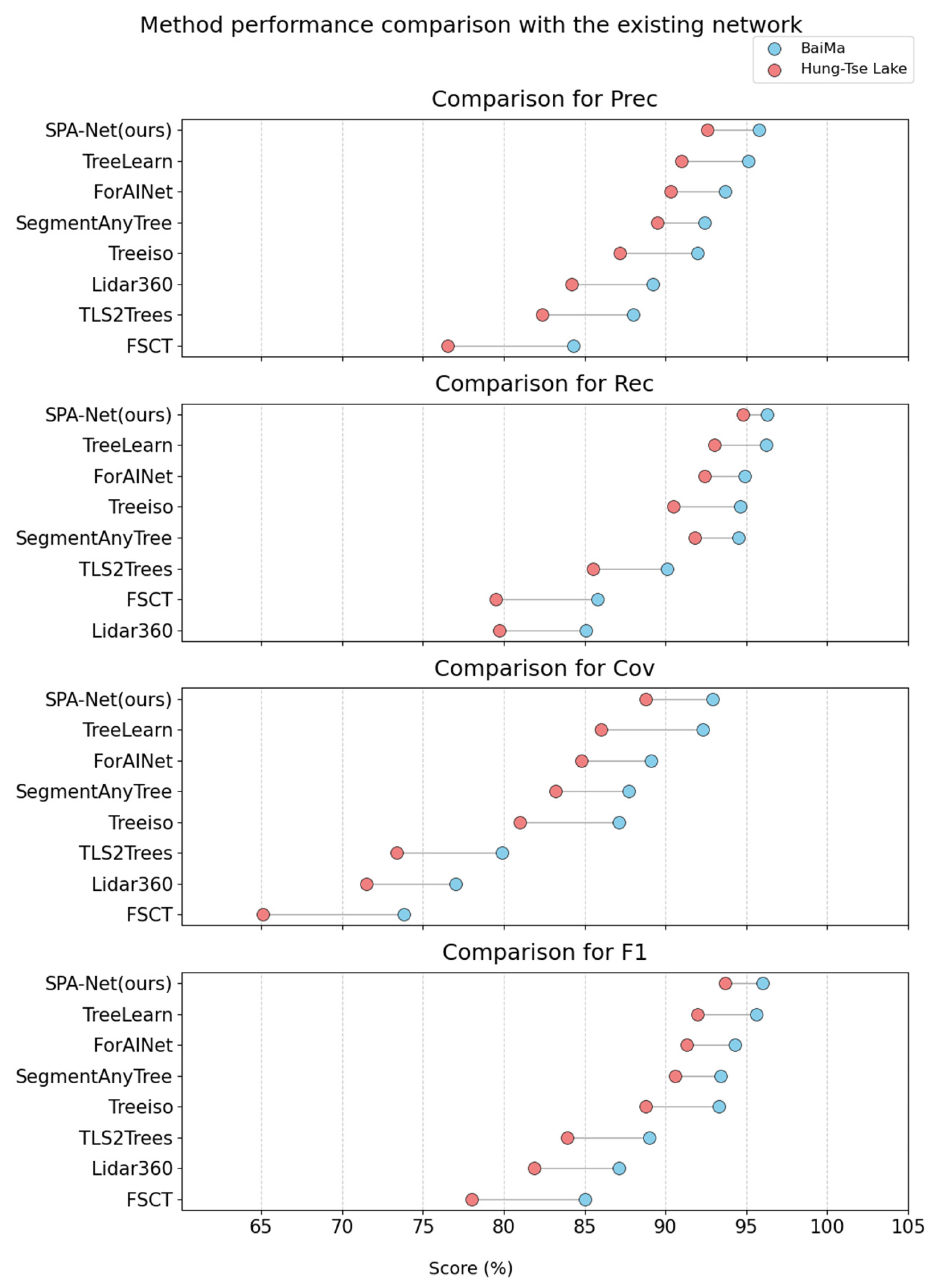
Evaluation on the BaiMa dataset demonstrates that SPA-Net consistently surpasses the strongest baseline TreeLearn, as shown in Figure 8, across all reported metrics. Specifically, our method yielded improvements of 0.7% in Prec, 0.1% in Rec, 0.6% in Cov, and 0.4% in F1 over TreeLearn. Performance levels attained by the leading methodologies on the relatively sparse BaiMa dataset were exceptionally high, with the top five methods exceeding 93% F1.
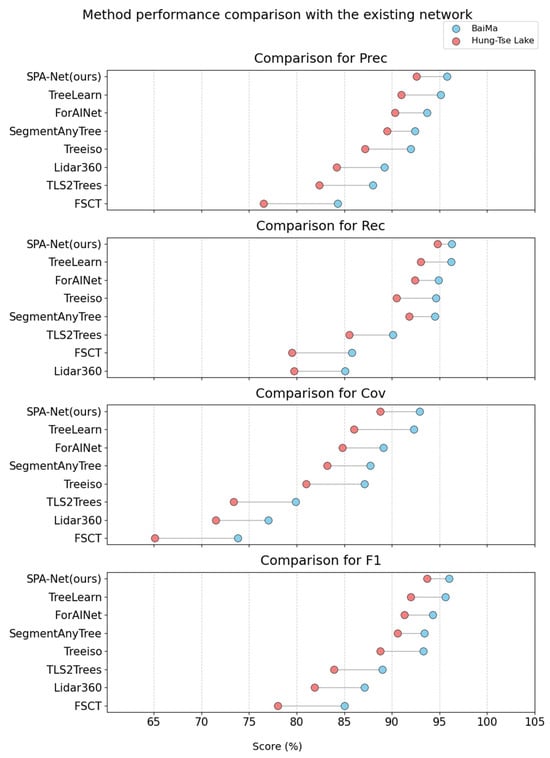
Figure 8.
Method performance comparison with the existing network.
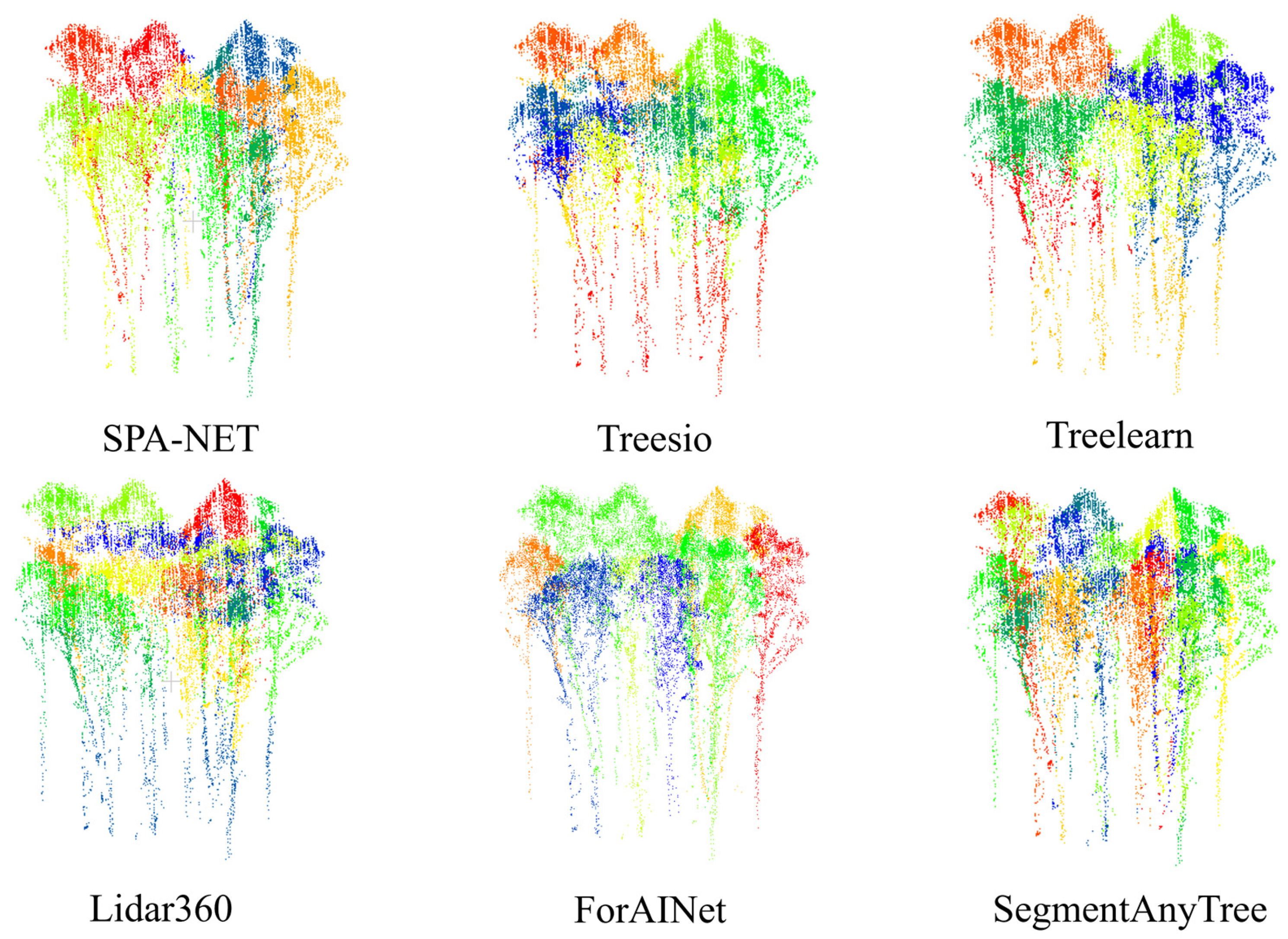
Consistent with advancements in point cloud analysis, deep learning approaches [58,59,60] significantly outperformed methods predominantly based on algorithms or rules when applied to the BaiMa dataset. This highlights the advantage of learned feature representations for robust segmentation, even in sparse point clouds. The segmentation results of different methods are shown in Figure 9.

Figure 9.
Method performance comparison with the existing network. (a–c) from the BaiMa dataset, (d) from the Hong-Tes Lake dataset.
To provide a cross-domain validation of our architecture, we benchmarked SPA-Net against PointGroup, an influential method developed for large-scale indoor scenes like ScanNet v2 and S3DIS. Given that the structured geometry of these indoor environments differs significantly from that of complex outdoor forests, this experiment tests the robustness of our forestry-focused approach against a leading model from a different application domain. The results are presented in Table 8.

Table 8.
Comparison with the PointGroup model.
This performance of SPA-Net significantly outperforms the classic PointGroup method, which only reached a 92.3% F1 score. PointGroup’s limitations are evident because it was originally designed for structured indoor scenes; challenges in forestry applications, such as irregular tree crowns and point cloud sparsity, hinder its ability to predict offset vectors, resulting in lower Prec (91.5%) and Cov (85.8%).
5. Discussion
5.1. Performance on Plantation Forest and Wetland Forest
The empirical results from the BaiMa Plantation Forest and Hung-Tse Lake TLS datasets substantiate the effectiveness of SPA-Net design choices across differing environmental contexts. On the BaiMa dataset, as shown in Table 7, potentially characterized by more uniform tree structures typical of plantations, SPA-Net achieved 95.8% Prec, 96.3% Rec, 92.9% Cov, and a 96.0% F1. This performance level, surpassing a suite of contemporary ITS methods [25], suggests that SPA-Net’s offset-free proposal SGP module and AA module are adept at handling conditions even where tree delineation might be considered relatively straightforward due to stand regularity. In contrast, the Hung-Tse Lake dataset, which presents greater structural complexity, specific challenges of wetland species, and denser understory, still saw SPA-Net achieve robust results, again leading the compared methods. The consistent leading performance across these two distinct TLS environments demonstrates the adaptability of the SPA-Net network.
Despite SPA-Net’s excellent performance, its accuracy is still influenced by forest complexity. The model’s accuracy decreased when moving from the relatively uniform BaiMa plantation forest to the more complex Hung-Tse Lake wetland forest. This indicates that extreme crown overlap and stand density remain a challenge. A potential failure case, particularly for large or structurally complex trees, occurs when the Affinity Aggregation (AA) module fails to merge all fragments belonging to the same tree. This situation can arise in cases of severe occlusion or unusual tree morphology. This implies that while factors like crown morphology and stand density, known to influence segmentation accuracy, do affect absolute performance [29], as shown in Figure 9, architectural features of SPA-Net provide a more consistent ability to resolve individual trees compared to other state-of-the-art deep learning and traditional approaches in these specific TLS-scanned environments.
5.2. Comparison with Existing Methods
The SPA-Net network, introduced in this study, offers a distinct approach to individual tree segmentation (ITS) when compared to other contemporary deep learning networks. Our method, which is validated by the ablation studies, was that an SGP module could achieve superior accuracy by moving beyond the direct 3D offset that is common in methods like TreeLearn, ForAINet, and SegmentAnyTree [47,48,49] for initial proposal generation. While TreeLearn and ForAINet have significantly advanced ITS, their reliance on accurate offset prediction can be a limiting factor in dense or complex stands [45,46]. SGP module, with its novel sampling-shifting-grouping paradigm, directly generates initial candidates from point geometry. This contrasts with the aforementioned networks, which typically predict offsets for all points towards a learned instance center before clustering [44]. The SGP mechanism of refining seed points towards emergent geometric centers within a non-linearly encoded space proved more robust for delineating tree instances in our TLS datasets. As shown in Table 6, compared to the 3D offset method, the SGP module increased Prec by 1.5% and Cov by 1.3%, although Rec slightly decreased by 0.4%. Furthermore, the Affinity Aggregation (AA) module provides a sophisticated, learned refinement stage. Unlike the scoring and Non-Maximum Suppression (NMS) [50] mechanisms prevalent in PointGroup-based architectures for pruning redundant proposals, AA module learns inter-proposal affinities to merge fragmented segments actively.
The architecture of SPA-Net is a coarse-to-fine method with a clear division of labor. We do not view SGP and AA merely as two independent modules, but rather consider their combination as a unique instance branch. The effectiveness of this symbiotic design is clearly and quantitatively validated in the ablation study presented in Table 3 of the paper. Through a comparison with Table 7, we demonstrate that this combined SGP + AA paradigm exhibits stronger robustness compared to traditional methods based on offset prediction, clustering, and ScoreNet/NMS (such as ForAINet and Treelearn). The method of combining the SGP and AA modules decomposes the complex segmentation task into two more easily learnable subtasks: geometric proposal generation and contextual relationship aggregation. As a result, it demonstrates a lower risk of erroneous merging and under-segmentation when processing large trees with sparse point clouds and complex morphology.
5.3. Future Work
Our primary design and validation focus for SPA-Net was on the TLS data of the forest’s point cloud. Future research could explore the adaptation of the SGP and AA principles to other 3D sensing modalities, such as dense ALS or MLS data. Further investigation into the robustness of SPA-Net across an even broader range of forest types. Improving the model’s computational efficiency is an important next step. The KNN-transformer in the AA module is computationally expensive, so we plan to explore lightweight alternatives like linear attention to reduce this load and improve scalability without a significant loss in performance. As an initial step in assessing such generalization capabilities, we conducted experimental verification using an airborne laser scanning (ALS) dataset [61,62] acquired over a Northeast China primeval mixed forest, specifically selecting 11 natural mixed-forest subcompartments within the Shangganling Xishui Forest Farm for this purpose. Each experimental site was established as a 50 m × 50 m square standard quadrat (0.25 hectares in area), encompassing diverse stand structures and topographic characteristics.
Compared to TLS, the point density of ALS data is significantly lower, especially in the trunk and understory regions, often resulting in a severe lack of point cloud data for the tree structure, particularly at the trunk base. The top-down acquisition perspective of ALS data causes dense canopies to severely occlude the underlying trunks and branches, rendering segmentation algorithms that rely on complete tree morphology ineffective.
Offset-based methods, such as TreeLearn, typically require the prediction of an offset vector from each point to a tree’s center (e.g., the trunk base). In ALS data where trunk base points are missing, such predictions become unreliable and ill-posed, often leading to failure. In contrast, SPA-Net’s SGP module is offset-free; it aggregates seed points toward local, emergent geometric centers via centripetal shrink. This means it does not depend on a global, pre-defined target (like the trunk) but instead adapts to the locally available point cloud geometry. Building on this, the AA module further enhances adaptability to varying data quality by learning to merge fragmented instances caused by data sparsity or occlusion. Therefore, our method requires no modification for ALS data.
For natural mixed forests, as shown in Figure 10, our method maintains its performance advantage despite the increased environmental complexity.
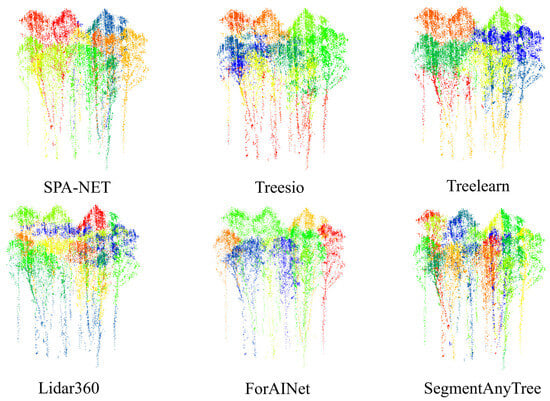
Figure 10.
Segmentation results for comparison with the existing network.
Amidst these difficulties, as shown in Table 9, our method outperforms all comparative methods, yielding a Prec of 53.1%, Rec of 92.8%, Cov of 49.7%, and an F1 of 67.4%. SegmentAnyTree performed second best, notably reaching 93.0% Rec and 49.0% Cov. The relative success of these deep learning models highlights their capacity to learn robust features suited for complex geometries, with SPA-Net benefiting from its SGP + AA architecture and SegmentAnyTree from its diverse training approach simulating ALS conditions. Conversely, methods like Treeiso struggled significantly with precision and coverage due to reliance on occluded base features critical to its algorithm. Other approaches, including Lidar360, also faced challenges with accurate delineation or data fragmentation in this complex scenario.

Table 9.
Individual tree segmentation performance on the mixed forests dataset.
When directly migrated to a new scene, SPA-Net still maintains relatively optimal performance. This strongly demonstrates the universality and potential of our method’s design. While these results highlight the generalization potential of advanced deep learning frameworks, they also reinforce that accurate individual tree segmentation from ALS of natural mixed-forest in complex, multi-layered canopies remains a significant hurdle.
Of course, we also recognize the limitations of the current research. Although SPA-Net shows generalization ability on ALS data, its performance is reduced compared to TLS data. This indicates that optimization for specific data sources is still necessary.
Future research should focus on extending SPA-Net to more diverse data scenarios, such as unmanned aerial vehicle LiDAR (UAV-LiDAR) data and more complex forest ecosystems like tropical rainforests.
By training and fine-tuning on a wider variety of datasets, we hope to build a truly cross-platform and cross-regional universal single-tree segmentation model. This would provide more powerful technical support for precision forestry management and carbon stock estimation on a global scale.
6. Conclusions
This study introduced SPA-Net, a novel deep learning architecture designed for accurate and efficient individual tree segmentation from point clouds, particularly in complex forest stands where traditional offset-based methods face limitations. The core of SPA-Net is its unique instance segmentation branch that consists of the sparse geometric proposal (SGP) module and the affinity aggregation (AA) module, which replaces conventional offset prediction and scoring/NMS mechanisms. The following conclusions are drawn from this study:
- (1)
- The SGP module, by eschewing direct 3D offset vector prediction and instead refining seed points toward emergent geometric centers within a non-linearly encoded space, proved more robust for generating high-quality initial tree instance proposals compared to traditional offset-guided clustering baselines. Ablation studies showed that SGP alone achieved high accuracy.
- (2)
- The learned affinity aggregation (AA) module significantly improved the Rec and Cov of the final segmentation by effectively merging fragmented instance proposals that can result from SGP, particularly for large or complexly structured trees. The addition of AA to SGP consistently enhanced performance across metrics.
- (3)
- The complete SPA-Net network demonstrated superior overall performance, surpassing a range of contemporary ITS methods, including recent deep learning models like TreeLearn and ForAINet, on TLS datasets, which achieve 96.0% F1 on BaiMa. It also showed promising generalization to challenging ALS data, outperforming other methods.
- (4)
- The proposed SPA-Net framework, by successfully decoupling initial proposal generation from global offset prediction and integrating a learned affinity-based refinement, offers an innovative, efficient, and more accurate approach for individual tree segmentation from TLS data.
Author Contributions
Conceptualization, Y.Z. and Z.W.; methodology, Y.Z. and Z.W.; software, Y.Z.; validation, Y.Z.; analysis, Y.Z.; data curation, Y.Z. and Z.W.; writing—original draft preparation, Y.Z.; writing—review and editing, Y.Z., Z.W., Q.Y., L.P., Q.W., X.Z. and C.H.; project administration, Q.Y. and C.H. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Jiangsu Province Carbon Reaching Peak Carbon Neutral Science and Technology Innovation Special Fund Project (Frontier Foundation), research on the evolution law of carbon sinks in typical wetland ecosystems, the potential for increasing sinks and the regulation mechanism [BK20220016] of Jiangsu Province Science and Technology Department. And the National Key Research and Development Program of China under Grant 2022YFD2201005-03 is supported by the Institute of Resource Information, Chinese Academy of Forestry, and the Jiangsu Province Graduate Research and Practice Innovation Program Project KYCX25_1457 that is supported by Jiangsu Provincial Department of Education.
Data Availability Statement
The point cloud data underlying this research are not publicly available, as they were obtained through a paid arrangement by the authors and their institutions and involve privacy considerations.
Acknowledgments
We would like to thank the Advanced Analysis and Testing Centre of Nanjing Forestry University, China and the Institute of Resource Information, Chinese Academy of Forestry for their assistance with data collection and technical support.
Conflicts of Interest
All authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
References
- Zhao, D.; Pang, Y.; Liu, L.; Li, Z. Individual Tree Classification Using Airborne LiDAR and Hyperspectral Data in a Natural Mixed Forest of Northeast China. Forests 2020, 11, 303. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, X.; Zhang, L.; Fan, X.; Ye, Q.; Fu, L. Individual tree segmentation and tree-counting using supervised clustering. Comput. Electron. Agric. 2023, 205, 107629. [Google Scholar] [CrossRef]
- Xi, Z.; Degenhardt, D. A new unified framework for supervised 3D crown segmentation (TreeisoNet) using deep neural networks across airborne, UAV-borne, and terrestrial laser scans. ISPRS J. Photogramm. Remote Sens. 2025, 15, 100083. [Google Scholar] [CrossRef]
- Cao, Y.; Ball, J.G.C.; Coomes, D.A.; Steinmeier, L.; Knapp, N.; Wilkes, P.; Disney, M.; Calders, K.; Burt, A.; Lin, Y.; et al. Benchmarking airborne laser scanning tree segmentation algorithms in broadleaf forests shows high accuracy only for canopy trees. Int. J. Appl. Earth Obs. Geoinf. 2023, 123, 103490. [Google Scholar] [CrossRef]
- Calders, K.; Adams, J.; Armston, J.; Bartholomeus, H.; Bauwens, S.; Bentley, L.P.; Chave, J.; Danson, F.M.; Demol, M.; Disney, M.; et al. Terrestrial laser scanning in forest ecology: Expanding the horizon. Remote Sens. Environ. 2020, 251, 112102. [Google Scholar] [CrossRef]
- You, H.; Liu, Y.; Lei, P.; Qin, Z.; You, Q. Segmentation of individual mangrove trees using UAV-based LiDAR data. Ecol. Inform. 2023, 77, 102200. [Google Scholar] [CrossRef]
- Zheng, J.; Yuan, S.; Li, W.; Fu, H.; Yu, L.; Huang, J. A Review of Individual Tree Crown Detection and Delineation from Optical Remote Sensing Images: Current progress and future. IEEE Geosci. Remote Sens. Mag. 2025, 13, 209–236. [Google Scholar] [CrossRef]
- Popescu, S.C.; Wynne, R.H.; Nelson, R.F. Estimating plot-level tree heights with lidar: Local filtering with a canopy-height based variable window size. Comput. Electron. Agric. 2002, 37, 71–95. [Google Scholar] [CrossRef]
- Yang, J.; Kang, Z.; Cheng, S.; Yang, Z.; Akwensi, P.H. An Individual Tree Segmentation Method Based on Watershed Algorithm and Three-Dimensional Spatial Distribution Analysis from Airborne LiDAR Point Clouds. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 1055–1067. [Google Scholar] [CrossRef]
- Hyyppa, J.; Kelle, O.; Lehikoinen, M.; Inkinen, M. A segmentation-based method to retrieve stem volume estimates from 3-D tree height models produced by laser scanners. IEEE Trans. Geosci. Remote Sens. 2001, 39, 969–975. [Google Scholar] [CrossRef]
- Li, S.; Zhao, S.; Tian, Z.; Tang, H.; Su, Z. Individual Tree Segmentation Based on Region-Growing and Density-Guided Canopy 3-D Morphology Detection Using UAV LiDAR Data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2025, 18, 8897–8909. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, X. Individual Tree Parameters Estimation for Plantation Forests Based on UAV Oblique Photography. IEEE Access 2020, 8, 96184–96198. [Google Scholar] [CrossRef]
- Ning, X.; Ma, Y.; Hou, Y.; Lv, Z.; Jin, H.; Wang, Z.; Wang, Y. Trunk-Constrained and Tree Structure Analysis Method for Individual Tree Extraction from Scanned Outdoor Scenes. Remote Sens. 2023, 15, 1567. [Google Scholar] [CrossRef]
- Wang, A.; Shi, S.; Yang, J.; Zhou, B.; Luo, Y.; Tang, X.; Du, J.; Bi, S.; Qu, F.; Gong, C.; et al. Potential of hyperspectral LiDAR in individual tree segmentation: A comparative study with multispectral LiDAR. Urban For. Urban Green. 2025, 104, 128658. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, D.; Fu, S.; Mathiopoulos, P.T.; Sui, M.; Na, J.; Peethambaran, J. Segmentation of Individual Tree Points by Combining Marker-Controlled Watershed Segmentation and Spectral Clustering Optimization. Remote Sens. 2024, 16, 610. [Google Scholar] [CrossRef]
- Fu, Y.; Niu, Y.; Wang, L.; Li, W. Individual-Tree Segmentation from UAV–LiDAR Data Using a Region-Growing Segmentation and Supervoxel-Weighted Fuzzy Clustering Approach. Remote Sens. 2024, 16, 608. [Google Scholar] [CrossRef]
- Morsdorf, F.; Meier, E.; Kötz, B.; Itten, K.I.; Dobbertin, M.; Allgöwer, B. LIDAR-based geometric reconstruction of boreal type forest stands at single tree level for forest and wildland fire management. Remote Sens. Environ. 2004, 92, 353–362. [Google Scholar] [CrossRef]
- Vo, A.-V.; Truong-Hong, L.; Laefer, D.F.; Bertolotto, M. Octree-based region growing for point cloud segmentation. ISPRS J. Photogramm. Remote Sens. 2015, 104, 88–100. [Google Scholar] [CrossRef]
- Liu, Q.; Ma, W.; Zhang, J.; Liu, Y.; Xu, D.; Wang, J. Point-cloud segmentation of individual trees in complex natural forest scenes based on a trunk-growth method. J. For. Res. 2021, 32, 2403–2414. [Google Scholar] [CrossRef]
- Wang, D. Unsupervised semantic and instance segmentation of forest point clouds. ISPRS J. Photogramm. Remote Sens. 2020, 165, 86–97. [Google Scholar] [CrossRef]
- Yang, Z.; Su, Y.; Li, W.; Cheng, K.; Guan, H.; Ren, Y.; Hu, T.; Xu, G.; Guo, Q. Segmenting Individual Trees From Terrestrial LiDAR Data Using Tree Branch Directivity. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2024, 17, 956–969. [Google Scholar] [CrossRef]
- Li, J.; Cheng, X.; Wu, Z.; Guo, W. An Over-Segmentation-Based Uphill Clustering Method for Individual Trees Extraction in Urban Street Areas from MLS Data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2021, 14, 2206–2221. [Google Scholar] [CrossRef]
- Carpenter, J.; Jung, J.; Oh, S.; Hardiman, B.; Fei, S. An Unsupervised Canopy-to-Root Pathing (UCRP) Tree Segmentation Algorithm for Automatic Forest Mapping. Remote Sens. 2022, 14, 4274. [Google Scholar] [CrossRef]
- Zhang, C.; Chengwen, S.; Aleksandra, Z.; Jiaxing, Z.; Rachel, G.; Wenxia, D.; Xiao, W. Individual tree segmentation from UAS Lidar data based on hierarchical filtering and clustering. Int. J. Digit. Earth 2024, 17, 2356124. [Google Scholar] [CrossRef]
- Hao, Y.; Widagdo, F.R.A.; Liu, X.; Liu, Y.; Dong, L.; Li, F. A Hierarchical Region-Merging Algorithm for 3-D Segmentation of Individual Trees Using UAV-LiDAR Point Clouds. IEEE Trans. Geosci. Remote Sens. 2022, 60, 1–16. [Google Scholar] [CrossRef]
- Kulicki, M.; Cabo, C.; Trzciński, T.; Będkowski, J.; Stereńczak, K. Artificial Intelligence and Terrestrial Point Clouds for Forest Monitoring. Curr. For. Rep. 2024, 11, 5. [Google Scholar] [CrossRef]
- Wang, J.; Guo, J.; Wang, R.; Zhang, Z.; Fu, L.; Ye, Q. Parameter Disentanglement for Diverse Representations. Big Data Min. Anal. 2025, 8, 606–623. [Google Scholar] [CrossRef]
- Armeni, I.; Sener, O.; Zamir, A.R.; Jiang, H.; Brilakis, I.; Fischer, M.; Savarese, S. 3D Semantic Parsing of Large-Scale Indoor Spaces. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 1534–1543. [Google Scholar]
- Zhu, W.; Zhu, C.; Zhang, Y. Research on Deep Learning Individual Tree Segmentation Method Coupling RetinaNet and Point Cloud Clustering. IEEE Access 2021, 9, 126635–126645. [Google Scholar] [CrossRef]
- Lv, L.; Li, X.; Mao, F.; Zhou, L.; Xuan, J.; Zhao, Y.; Yu, J.; Song, M.; Huang, L.; Du, H. A Deep Learning Network for Individual Tree Segmentation in UAV Images with a Coupled CSPNet and Attention Mechanism. Remote Sens. 2023, 15, 4420. [Google Scholar] [CrossRef]
- Wang, F.; Bryson, M. Tree Segmentation and Parameter Measurement from Point Clouds Using Deep and Handcrafted Features. Remote Sens. 2023, 15, 1086. [Google Scholar] [CrossRef]
- Luo, H.; Khoshelham, K.; Chen, C.; He, H. Individual tree extraction from urban mobile laser scanning point clouds using deep pointwise direction embedding. ISPRS J. Photogramm. Remote Sens. 2021, 175, 326–339. [Google Scholar] [CrossRef]
- Hu, X.D.; Hu, C.H.; Han, J.A.; Sun, H.; Wang, R. Point cloud segmentation for an individual tree combining improved point transformer and hierarchical clustering. J. Appl. Remote Sens. 2023, 17, 034505. [Google Scholar] [CrossRef]
- Chang, L.; Fan, H.; Zhu, N.; Dong, Z. A Two-Stage Approach for Individual Tree Segmentation from TLS Point Clouds. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2022, 15, 8682–8693. [Google Scholar] [CrossRef]
- Ibrahim, M.; Akhtar, N.; Anwar, S.; Mian, A. SAT3D: Slot Attention Transformer for 3D Point Cloud Semantic Segmentation. IEEE Trans. Intell. Transp. Syst. 2023, 24, 5456–5466. [Google Scholar] [CrossRef]
- Li, X.; Chen, D. A survey on deep learning-based panoptic segmentation. Digit. Signal Prog. 2022, 120, 103283. [Google Scholar] [CrossRef]
- De Carvalho, O.L.F.; de Carvalho Júnior, O.A.; Silva, C.R.E.; de Albuquerque, A.O.; Santana, N.C.; Borges, D.L.; Gomes, R.A.T.; Guimarães, R.F. Panoptic Segmentation Meets Remote Sensing. Remote Sens. 2022, 14, 965. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhang, Y.; Foroosh, H. Panoptic-PolarNet: Proposal-free LiDAR Point Cloud Panoptic Segmentation. In Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 20–25 June 2021; pp. 13189–13198. [Google Scholar] [CrossRef]
- Fong, W.K.; Mohan, R.; Hurtado, J.V.; Zhou, L.; Caesar, H.; Beijbom, O.; Valada, A. Panoptic Nuscenes: A Large-Scale Benchmark for LiDAR Panoptic Segmentation and Tracking. IEEE Robot. Autom. Lett. 2022, 7, 3795–3802. [Google Scholar] [CrossRef]
- Vu, T.; Kim, K.; Nguyen, T.; Luu, T.M.; Kim, J.; Yoo, C.D. Scalable SoftGroup for 3D Instance Segmentation on Point Clouds. IEEE Trans. Pattern Anal. Mach. Intell. 2024, 46, 1981–1995. [Google Scholar] [CrossRef]
- Schult, J.; Engelmann, F.; Hermans, A.; Litany, O.; Tang, S.; Leibe, B. Mask3D: Mask Transformer for 3D Semantic Instance Segmentation. In Proceedings of the 2023 IEEE International Conference on Robotics and Automation (ICRA), London, UK, 29 May–2 June 2023; pp. 8216–8223. [Google Scholar] [CrossRef]
- Han, L.; Zheng, T.; Xu, L.; Fang, L. OccuSeg: Occupancy-Aware 3D Instance Segmentation. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 2937–2946. [Google Scholar] [CrossRef]
- Yang, Y.; Mei, J.; Du, S.; Xiao, Y.; Wu, H.; Xu, X.; Liu, Y. DQFormer: Toward Unified LiDAR Panoptic Segmentation with Decoupled Queries for Large-Scale Outdoor Scenes. IEEE Trans. Geosci. Remote Sens. 2025, 63, 1–15. [Google Scholar] [CrossRef]
- Jiang, L.; Zhao, H.; Shi, S.; Liu, S.; Fu, C.W.; Jia, J. PointGroup: Dual-Set Point Grouping for 3D Instance Segmentation. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 4866–4875. [Google Scholar] [CrossRef]
- Xiang, B.; Peters, T.; Kontogianni, T.; Vetterli, F.; Puliti, S.; Astrup, R.; Schindler, K. Towards Accurate Instance Segmentation In Large-Scale Lidar Point Clouds. ISPRS Ann. Photogramm. Remote Sens. Spatial Inf. Sci. 2023, X-1/W1-2023, 605–612. [Google Scholar] [CrossRef]
- Xiang, B.; Yue, Y.; Peters, T.; Schindler, K. A Review of panoptic segmentation for mobile mapping point clouds. ISPRS J. Photogramm. Remote Sens. 2023, 203, 373–391. [Google Scholar] [CrossRef]
- Xiang, B.; Wielgosz, M.; Kontogianni, T.; Peters, T.; Puliti, S.; Astrup, R.; Schindler, K. Automated forest inventory: Analysis of high-density airborne LiDAR point clouds with 3D deep learning. Remote Sens. Environ. 2024, 305, 114078. [Google Scholar] [CrossRef]
- Wielgosz, M.; Puliti, S.; Xiang, B.; Schindler, K.; Astrup, R. SegmentAnyTree: A sensor and platform agnostic deep learning model for tree segmentation using laser scanning data. Remote Sens. Environ. 2024, 313, 114367. [Google Scholar] [CrossRef]
- Henrich, J.; van Delden, J.; Seidel, D.; Kneib, T.; Ecker, A.S. TreeLearn: A deep learning method for segmenting individual trees from ground-based LiDAR forest point clouds. Ecol. Inform. 2024, 84, 102888. [Google Scholar] [CrossRef]
- Zhao, W.; Yan, Y.; Yang, C.; Ye, J.; Yang, X.; Huang, K. Divide and Conquer: 3D Point Cloud Instance Segmentation with Point-Wise Binarization. In Proceedings of the 2023 IEEE/CVF International Conference on Computer Vision (ICCV), Paris, France, 1–6 October 2023; pp. 562–571. [Google Scholar] [CrossRef]
- Choy, C.; Gwak, J.; Savarese, S. 4D Spatio-Temporal ConvNets: Minkowski Convolutional Neural Networks. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 3070–3079. [Google Scholar] [CrossRef]
- Rosenfeld, A.; Pfaltz, J.L. Sequential Operations in Digital Picture Processing. JACM 1966, 13, 471–494. [Google Scholar] [CrossRef]
- Zhu, X.; Sha, C.; Niu, J. A Simple Retrieval-based Method for Code Comment Generation. In Proceedings of the 2022 IEEE International Conference on Software Analysis, Evolution and Reengineering (SANER), Honolulu, HI, USA, 15–18 March 2022; pp. 1089–1100. [Google Scholar] [CrossRef]
- Li, J.; He, X.; Wen, Y.; Gao, Y.; Cheng, X.; Zhang, D. Panoptic-PHNet: Towards Real-Time and High-Precision LiDAR Panoptic Segmentation via Clustering Pseudo Heatmap. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022; pp. 11799–11808. [Google Scholar] [CrossRef]
- Hong, F.; Kong, L.; Zhou, H.; Zhu, X.; Li, H.; Liu, Z. Unified 3D and 4D Panoptic Segmentation via Dynamic Shifting Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2024, 46, 3480–3495. [Google Scholar] [CrossRef]
- Comaniciu, D.; Meer, P. Mean shift: A robust approach toward feature space analysis. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 603–619. [Google Scholar] [CrossRef]
- Ester, M.; Kriegel, H.-P.; Sander, J.; Xu, X. A density-based algorithm for discovering clusters in large spatial databases with noise. In Proceedings of the Second International Conference on Knowledge Discovery and Data Mining, Portland, OR, USA, 2–4 August 1996; pp. 226–231. [Google Scholar]
- Wilkes, P.; Disney, M.; Armston, J.; Bartholomeus, H.; Bentley, L.; Brede, B.; Burt, A.; Calders, K.; Chavana-Bryant, C.; Clewley, D.; et al. TLS2trees: A scalable tree segmentation pipeline for TLS data. Methods Ecol. Evol. 2023, 14, 3083–3099. [Google Scholar] [CrossRef]
- Xi, Z.; Hopkinson, C. 3D Graph-Based Individual-Tree Isolation (Treeiso) from Terrestrial Laser Scanning Point Clouds. Remote Sens. 2022, 14, 6116. [Google Scholar] [CrossRef]
- Krisanski, S.; Taskhiri, M.S.; Gonzalez Aracil, S.; Herries, D.; Muneri, A.; Gurung, M.B.; Montgomery, J.; Turner, P. Forest Structural Complexity Tool—An Open Source, Fully-Automated Tool for Measuring Forest Point Clouds. Remote Sens. 2021, 13, 4677. [Google Scholar] [CrossRef]
- Ferraz, A.; Bretar, F.; Jacquemoud, S.; Gonçalves, G.; Pereira, L.; Tomé, M.; Soares, P. 3-D mapping of a multi-layered Mediterranean forest using ALS data. Remote Sens. Environ. 2012, 121, 210–223. [Google Scholar] [CrossRef]
- Windrim, L.; Bryson, M. Forest Tree Detection and Segmentation using High Resolution Airborne LiDAR. In Proceedings of the 2019 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS), Macau, China, 3–8 November 2019; pp. 3898–3904. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).