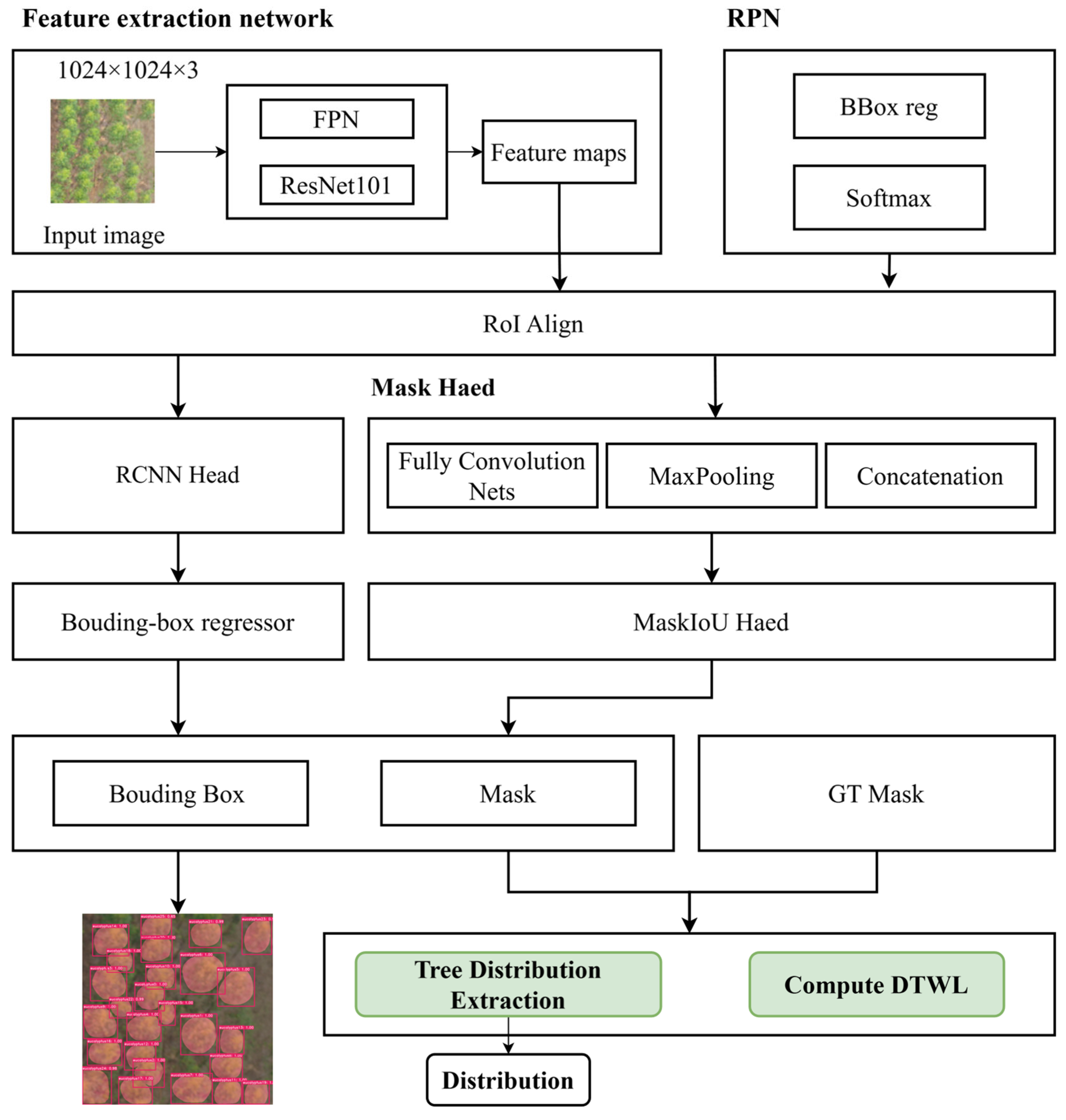
Individual tree crown images were successfully extracted using crown segmentation methods and high-resolution remote sensing data. The objective of this study was to generate a 3D mesh model for each tree based on its crown image. To accomplish this, we designed TPRN to infer key structural parameters of trees parameters from top-view crown images. Subsequently, a parameter-based tree modeling approach was applied to construct the 3D geometric model of each tree using the extracted parameters.
Most existing image-based 3D tree reconstruction methods typically rely on frontal or multi-view images of individual trees. In contrast, our approach reconstructs tree models using top-view crown images from high-resolution remote sensing data. According to forestry research, there is a strong correlation between crown size and shape and key structural parameters of trees. Crown area and crown width are among the most commonly used variables for predicting DBH and tree height. For example, Hemery et al. [
36] analyzed 11 common broadleaf tree species in the UK and established linear relationships between DBH and crown spread. Similarly, Calama et al. [
37] incorporated regional variation in developing height–DBH models for stone pine. Gonzalez et al. [
38] used field measurement data to develop models that predict DBH and usable stem volume from pine crown area. Thus, our methodology for extracting structural parameters from crown images is supported by both theoretical and empirical evidence.
4.2.1. Individual Tree Parameters
Before extracting parameters and performing parametric modeling of trees in forest scenes, it was necessary to define representative tree parameters that support model construction. Inspired by the parametric strategy of Weber et al., this study introduced a feature-parameter-based method for constructing tree models generation.
Tree parameters were classified into two main types: fixed category-level parameters and variable individual-level parameters. Category-level parameters are consistent across all trees of the same species, describing fundamental morphological and structural traits, including tree type, number of crown layers, number of lateral branches, and leaf size (see
Table 2 for examples). In contrast, individual-level parameters vary stochastically among trees, affecting overall proportions and height, thereby introducing diversity and realism to the generated models (examples listed in
Table 3).
Due to the limited structural detail contained in crown images and the relatively high dimensionality of the defined parameters, relying solely on crown images makes it challenging to accurately extract all necessary parameters. Therefore, species information was treated as a known input and combined with crown images as input to the TPRN model. This approach fixed the category-level parameters during training, reducing the complexity of the model and improving training efficiency. In the experimental dataset, the category-level parameter dictionaries for Chinese fir and eucalyptus were defined based on real-world images, while the individual-level parameters were randomly varied within a biologically reasonable range. This not only provides essential data for constructing 3D tree models but also enhances model diversity while maintaining overall structural consistency.
4.2.2. TPRN Architecture
The TPRN was designed to convert input crown images into structural feature parameters of individual trees. Consequently, its network architecture must effectively establish a mapping from images to numerical parameters. The TPRN was constructed based on a deep neural network (DNN) framework, utilizing EfficientNet-B7 [
39] as the core feature extractor. By leveraging depth wise separable convolutions and a compound scaling strategy, EfficientNet-B7 significantly improves feature representation while maintaining computational efficiency.
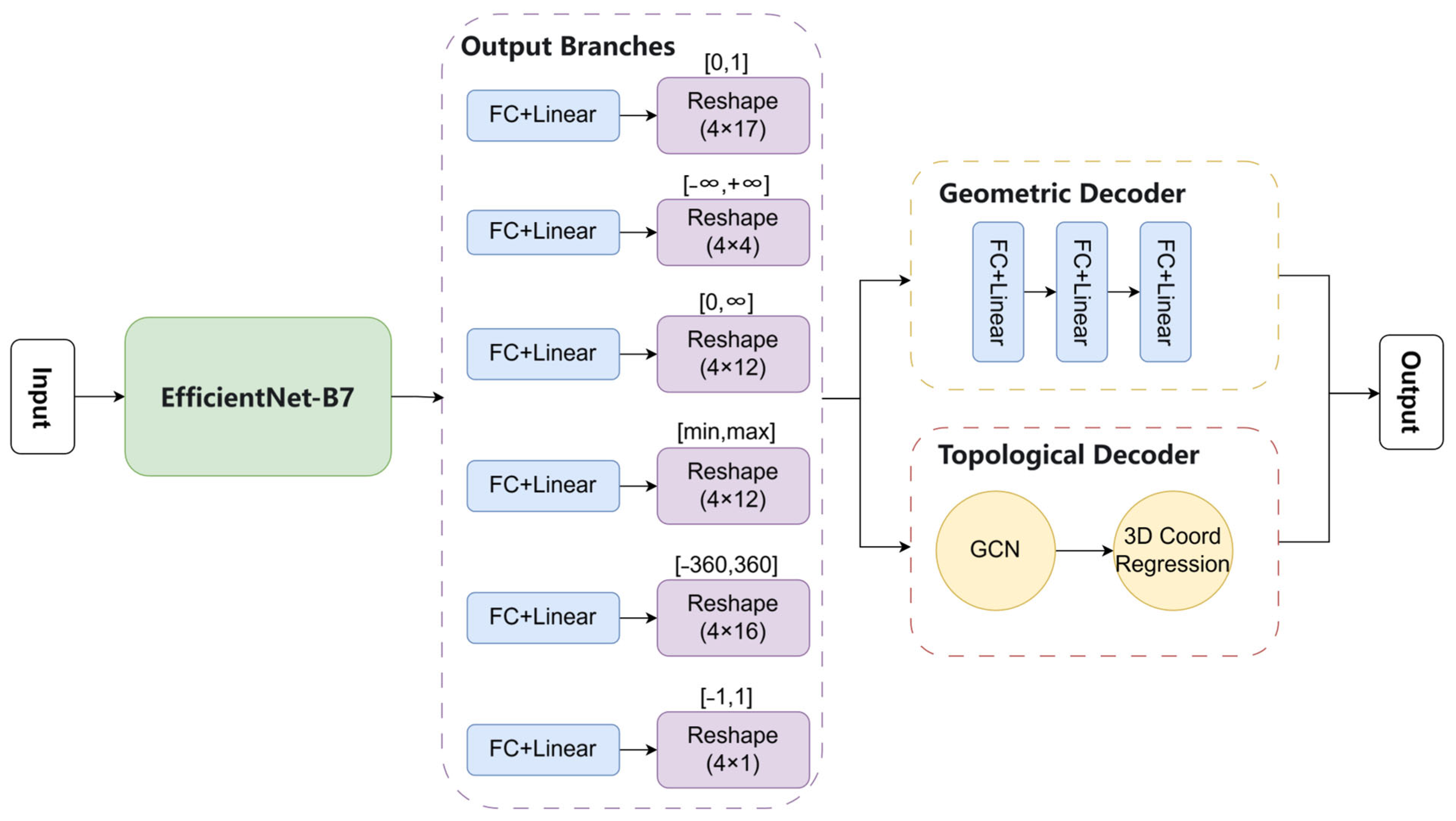
As shown in
Figure 6, the network architecture consists of multiple sub-modules, with each branch corresponding to a subset of parameters. This design accommodates varying magnitudes of feature parameters, thereby improving the model’s learning capacity and parameter fitting accuracy. To enable the high-precision reconstruction of individual trees, the network includes a dual-branch decoder after the feature extraction stage: one branch predicts key structural parameters, while the other models the tree’s topological structure.
In convolutional neural network training, lower input image resolutions often lead to improve performance [
40]. Research indicates that the accuracy improvements of the Efficient-Net baseline model tend to plateau beyond a certain resolution, suggesting that further increasing image resolution provides marginal accuracy gains while substantially raising computational cost. The input to the TPRN network consists of crown images extracted from high-resolution remote sensing imagery after segmentation, typically with a resolution not exceeding 512 × 512 pixels, aligning well with the network’s processing capabilities.
Given the strong correlation between crown area and DBH, the calculated crown area was incorporated as an additional input feature during training to enhance the model’s predictive capability. To precisely compute the actual crown area from the image, the following formula was used to determine the ratio between the number of pixels within the crown region and those within the bounding rectangle:
where
St represents the actual area of the tree crown, and
Sr represents the actual area of the bounding rectangle.
Pt and
Pr denote the number of pixels in the tree crown region and the rectangular region, respectively.
In this study, the TPRN model employ supervised learning, where each input image is paired with a corresponding set of tree feature parameters. These parameters are output in matrix form, referred to as the target matrix, which includes all category-level and individual-level feature parameters. Each row of the target matrix represents a unique feature parameter, and each column indicates the value of that parameter. The matrix has dimensions of n × m, where n is the number of parameters, and each parameter has a dimensionality of either 4 or 1. Parameters with a dimensionality of 1 are replicated across all columns.
Initially, a single-output branch was employed to directly predict the entire target matrix. However, experimental results demonstrated severe underfitting with this approach, as the model failed to effectively capture data patterns, leading to reduced parameter extraction accuracy. This limitation stemmed from the significant magnitude variations among different parameters in the target matrix. Outputting the entire matrix at once made it difficult for the network to accommodate these disparities, thereby restricting its ability to extract meaningful features.
To resolve this issue, the output structure of TPRN was refined by partitioning the original target matrix into six submatrices based on the order of magnitude of the parameter values. Each submatrix is processed by an independent output branch. The dimensionality of each submatrix is n1 × m, where n1 denotes the number of parameters within a specific value range. This partitioning strategy is guided by the heterogeneity of the tree feature parameter dictionary—different parameters exhibit significantly different data distributions. By adjusting the network to separately model these magnitudes, the representation capacity for parameters of various scales was improved.
The initial part of the network consists of a base convolutional layer to extract critical features. Subsequently, six independent output branches were deployed, each responsible for predicting parameters within a specific scale range. Each branch contains a fully connected layer with a linear activation function and a reshape layer to align the output with the target matrix dimensions.
Finally, a dual-branch decoder was introduced after the matrix outputs to extract both key structural parameters and topological features of trees. The Geometric Decoder specializes in structural parameter prediction and consists of three fully connected layers that output tree height and DBH. The Topological Decoder, on the other hand, models tree topology using a graph convolutional network (GCN) augmented with a 3D coordinate regression head to predict the spatial coordinates of branching points and their respective radii.
For feature allocation, the encoded features were uniformly distributed between the two decoder branches to ensure that the model captures both global parameter information and local structural features. The loss functions for the corresponding branches were defined as follows:
where
and
represent the predicted and ground-truth tree height, respectively;
and
represent the predicted and ground-truth DBH;
and
denote the predicted and ground-truth coordinates of the i-th branching point; and
and
represent the predicted and ground-truth radius of the i-th branch.
4.2.3. Parameter-Based 3D Reconstruction of Individual Trees
After extracting parameters using TPRN, this study proposed a feature-parameter-based tree modeling method. This method summarizes the geometric shapes and topological structures of various tree species and expresses them using a unified set of feature parameters. In combination with the growth patterns of different tree species, a general growth rule was designed to reconstruct the tree’s skeletal structure and branching architecture of the tree based on the extracted parameters. Once all branches were modeled, the total number and distribution of leaves were estimated using the Pipe Model Theory [
41]. Finally, texture details were added to improve the visual fidelity of the reconstructed tree model.
The growth direction and length of secondary branches were determined by multiple factors. According to the plant growth equation [
42], the diameter of a child branch is proportional to that of its parent branch, and a larger diameter corresponds to a longer branch length, consistent with the physiological growth principles of trees. The specific calculation formula was as follows:
where
is the length of the parent branch,
is the length of the child branch, and
represents the distance of the child branch from the base of the parent branch.
After processing the crown image through TPRN, six submatrices containing the predicted feature parameter values were generated. To utilize these predictions for tree model generation, it was first necessary to denormalize the submatrices. Specifically, each submatrix was multiplied by its corresponding normalization matrix stored during the training phase, thereby restoring the original data scale. This operation effectively transformed the predicted parameter matrix into a valid input for the tree modeling method and enables subsequent 3D reconstruction.
In terms of parameter format conversion, the six submatrices must be rearranged to match the structure of the Blender parameter dictionary. Each column of a submatrix corresponds to a specific key in the dictionary, representing an individual tree feature parameter. When a parameter is expected to be a single scalar rather than a four-element vector, only the first element of the vector is extracted as the final value to ensure data consistency and usability. After conversion, the parameter dictionary can be correctly interpreted and used to drive the generation of the 3D tree model.
According to the Pipe Model Theory, there exists a linear positive correlation between leaf biomass and the cross-sectional area of the stem at the base of the crown for a given tree species. Once all branches have been modeled, the tree model construction method calculates the total number of leaves needed based on this theory. The leaves are then generated and distributed along the branches following biologically realistic patterns.
Additionally, for different tree species, the model loads the corresponding trunk and leaf textures and applies them to the model. This ensures that both the visual appearance and structural characteristics of the reconstructed tree realistically reflect natural growth patterns.
For a given tree species, the category-level feature parameters remain consistent across all individual trees. These parameters describe the species’ typical structural characteristics. In contrast, the individual-level feature parameters are assigned randomly within predefined ranges, allowing for variation in geometric proportions, tree height, and branching structure. The individual parameters were modulated through random variation to generate representative 3D tree models. This species-driven modeling strategy not only accurately reflects key morphological traits of typical species but also demonstrates strong generalizability to a wide range of forest types and complex tree crown structures.