1. Introduction
Biodiversity promotes ecosystem stability and is important for maintaining ecosystem function [
1,
2]. Species recognition allows for a better understanding and documentation of the presence and distribution of different species, thereby promoting the conservation and management of biodiversity. With the development of artificial intelligence, computer scientists and taxonomists have been working to develop tools for automated species recognition [
3,
4,
5,
6,
7]. Gomez Villa et al. [
8] achieved 35.4% Top-1 accuracy (unbalanced training dataset containing empty images) and 88.9% Top-1 accuracy (balanced dataset with images containing only foreground animals) using deep convolutional neural networks on the Snapshot Serengeti dataset. Xie et al. [
9] utilized the improved YOLO V5s to recognize large mammal species in airborne thermal imagery, which improved the accuracy and reduced the recognition time for a single image. Huang et al. [
10] proposed a Part-Stacked CNN (PS-CNN) architecture by modeling subtle differences in the object portion of a given species, achieving 76.6% recognition accuracy on the CUB-200-2011 dataset. Lv et al. [
11] proposed a ViT-based multilevel feature fusion transformer that improves the recognition accuracy of the iNaturalist 2017 dataset. Li et al. [
12] achieved an average accuracy of 91.88% using a non-local mean filtering and multi-scale Retinex-based method to enhance butterfly images and introduced a cross-attention mechanism for recognition of the spatial distribution of butterfly features.
The success of the above methods cannot be achieved without large-scale datasets. However, collecting a substantial amount of image data for certain species, particularly rare and endangered ones, is challenging due to factors such as habitat constraints, living habits, and the number of species. Therefore, it is difficult to train a traditional species-recognition model. To overcome these challenges, some researchers have begun studying few-shot species-recognition methods, including meta-learning [
13], data augmentation [
14], and metric learning [
15]. Guo et al. [
16] used the Few-shot Unsupervised Image-to-image Translation (FUNIT) to expand the dataset by generating spurious fish data from the real fish dataset, thus improving the few-shot recognition accuracy. Zhai et al. [
17] introduced a sandwich-shaped attention module based on metric learning and proposed SACovaMNet for few-shot fish species recognition. Lu et al. [
18] introduced an embedding module and a metric function to improve the performance of fish recognition with limited samples. Xu et al. [
19] proposed a dual attention network to learn subtle but discriminative features from limited data. However, these methods usually perform poorly in real-world scenarios and generalize poorly.
With the rapid development of multimodal large-scale models, large vision-language models, also known as foundation models, exhibit significant performance in visual representation learning, and provide a new paradigm for downstream tasks via zero-shot or few-shot transfer learning. Contrastive Language-Image Pretraining (CLIP) [
20] utilizes large-scale image–text pairs for comparative learning and has achieved encouraging performance in a wide variety of visual classification tasks. Many studies utilized few-shot data to improve the performance of CLIP in downstream species recognition. CLIP-Adapter [
21] utilizes an additional bottleneck layer to learn new features and fuse them with the original pretrained features. Zhou et al. [
22] proposed a learning-based approach to convert CLIP’s defaults into learnable vectors and learn from the data, achieving excellent domain generalization performance on 11 datasets. Zhang et al. [
23] proposed Tip-Adapter, which constructs key-value cache models from few-shot training sets, sets up the adapter, and improves the visual representation capability of CLIP by fusing feature retrieval with pretrained features in CLIP. The above methods mainly focused on adapting the image features extracted by CLIP to the new species dataset through few-shot training. Guo et al. [
24] proposed a parameter-free attention module to improve zero-shot species recognition accuracy, but the improvement was relatively limited.
Depending solely on images makes it difficult to effectively distinguish visually similar species due to variations in the angles of photographs. To improve the recognition performance of species, utilizing the inherent attributes of species, such as morphological features, began to be investigated. Parashar et al. [
25] proposed converting scientific names to common names to improve zero-shot recognition accuracy, taking into account that species common names are used much more frequently than scientific names. Menon and Vondrick [
26] improved zero-shot species recognition performance by obtaining visual description information of species with the help of large language models. In addition, taxonomic experts have used additional information from images, such as where and when the images were captured, to assist in species recognition. Previous studies [
27,
28,
29,
30] have demonstrated that incorporating geolocation information into species-recognition models can help improve recognition performance. Liu et al. [
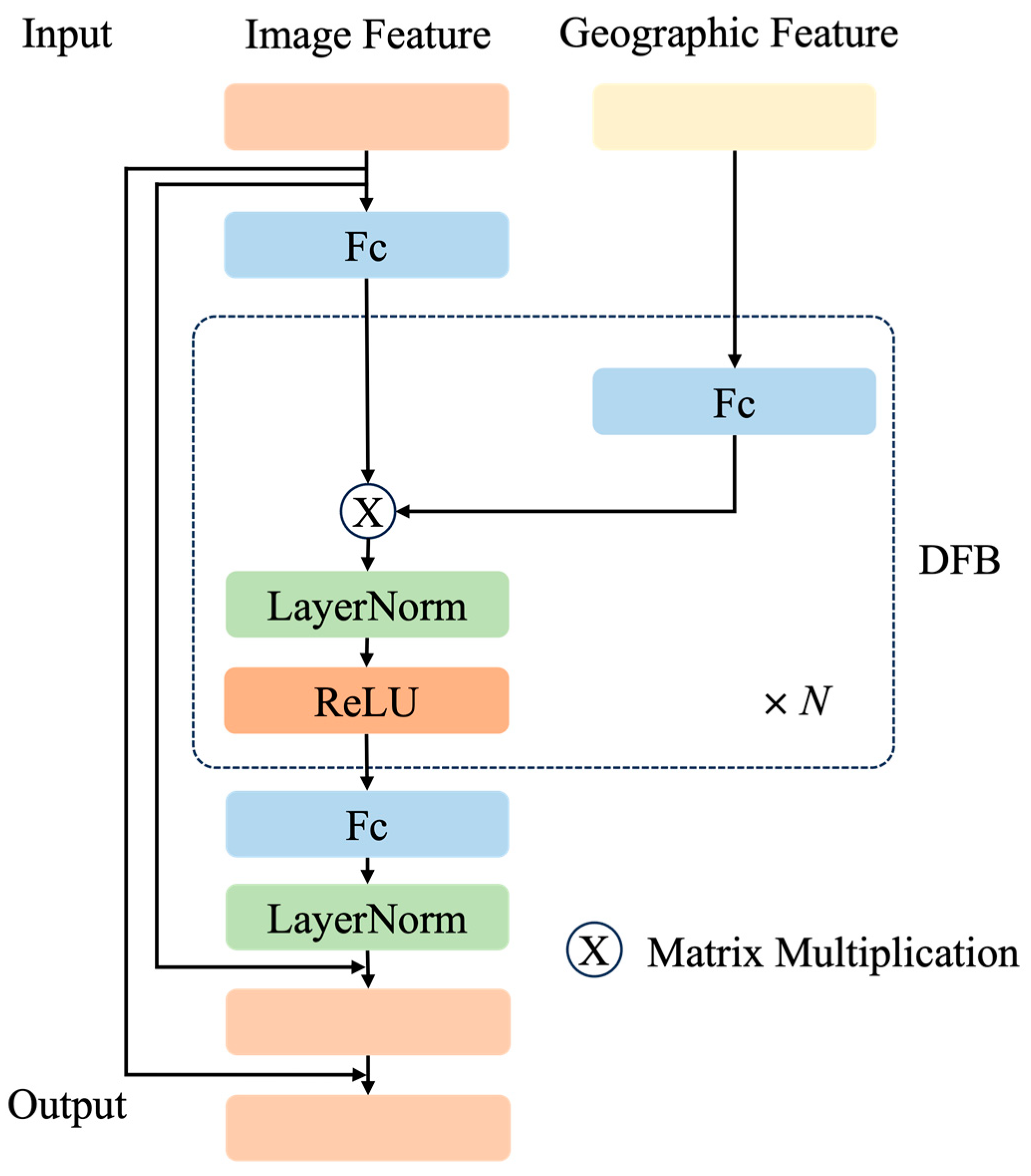
31] proposed the use of geographical distribution knowledge in textual form to improve zero-shot species recognition accuracy. However, no studies have explored the potential effects of utilizing geographic information on few-shot species recognition based on large-scale visual-language models like CLIP. To fill this gap, we proposed a few-shot species-recognition method based on species geographic information and CLIP (SG-CLIP). First, we utilized the powerful image and text feature extraction capabilities of CLIP to extract species image features and species text features through the image encoder and text encoder, respectively. Then, geographic features were driven by the geographic feature extraction module. Next, we designed an image and geographic fusion module to obtain enhanced image features. Finally, the matrix of species similarity between the enhanced image feature and the text feature was calculated to obtain recognition results. Overall, our contributions can be summarized as follows:
We proposed SG-CLIP, which integrates geographic information about species, to improve the performance of few-shot species recognition. To the best of our knowledge, this is the first work to exploit geographic information for few-shot species recognition of large vision-language models.
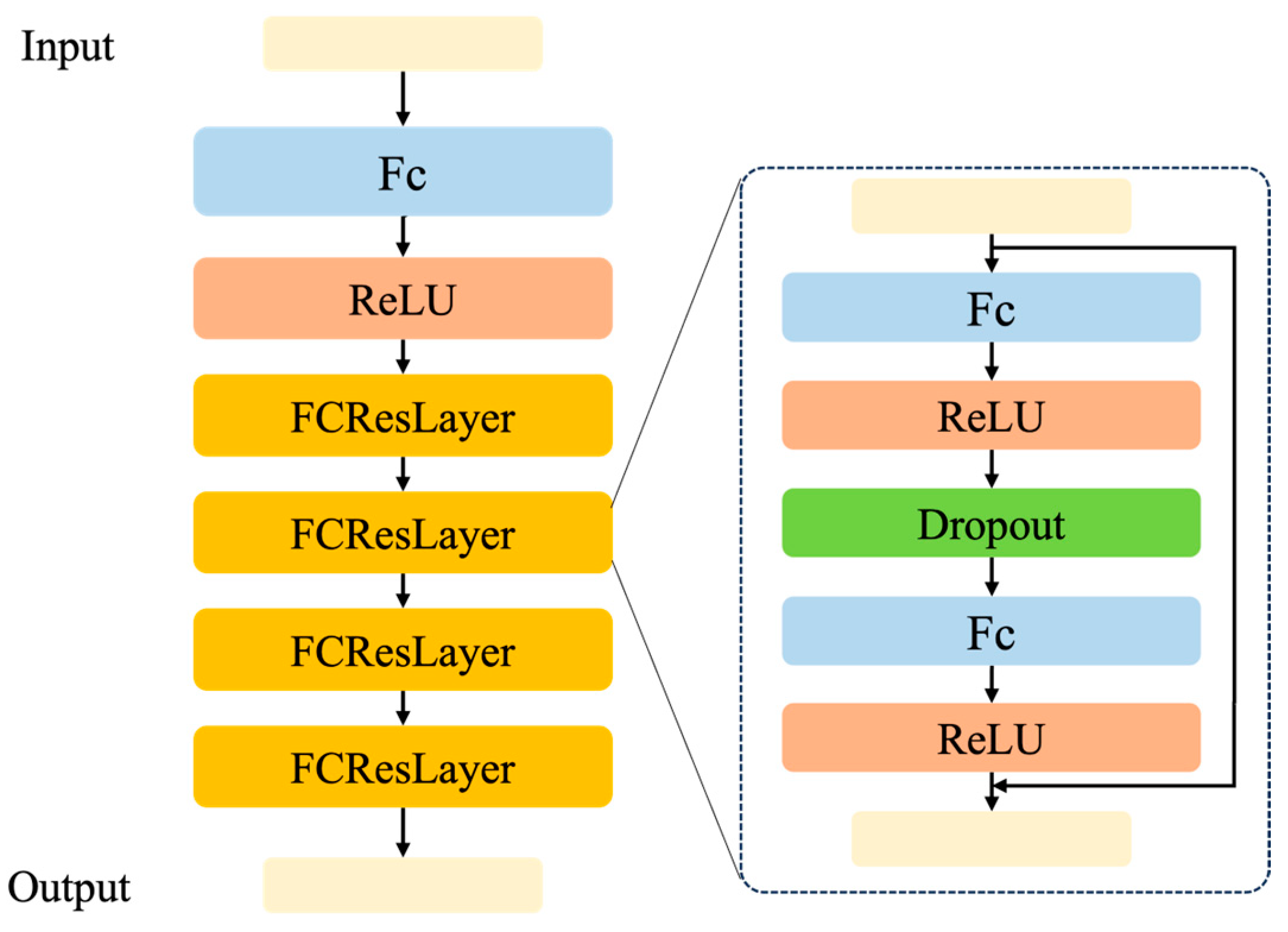
We introduced the geographic feature extraction module to better process geographic location information. Meanwhile, we designed the image and geographic feature fusion module to enhance the image representation ability.
We performed extensive experiments of SG-CLIP in the iNaturalist 2021 dataset to demonstrate its effectiveness and generalization. Under ViT-B/32 and the 16-shot training setup, compared to Linear probe CLIP, SG-CLIP improves the recognition accuracy by 15.12% on mammals, 17.51% on reptiles, and 17.65% on amphibians.
4. Results
In this section, we compare our SG-CLIP with four baseline models, Zero-shot CLIP [
20], Linear probe CLIP [
20], Tip-Adapter [
23], and Tip-Adapter-F [
23]. Zero-shot CLIP, Tip-Adapter, Tip-Adapter-F, and SG-CLIP used the same textual prompt “A photo of a [class]”. Linear probe CLIP was adapted to the target dataset by training a linear classifier. To demonstrate the effectiveness and generalization of our proposed method, we conducted a number of comparison and ablation studies on different versions of CLIP.
4.1. Performance of Different Few-Shot Learning Methods on ViT-B/32
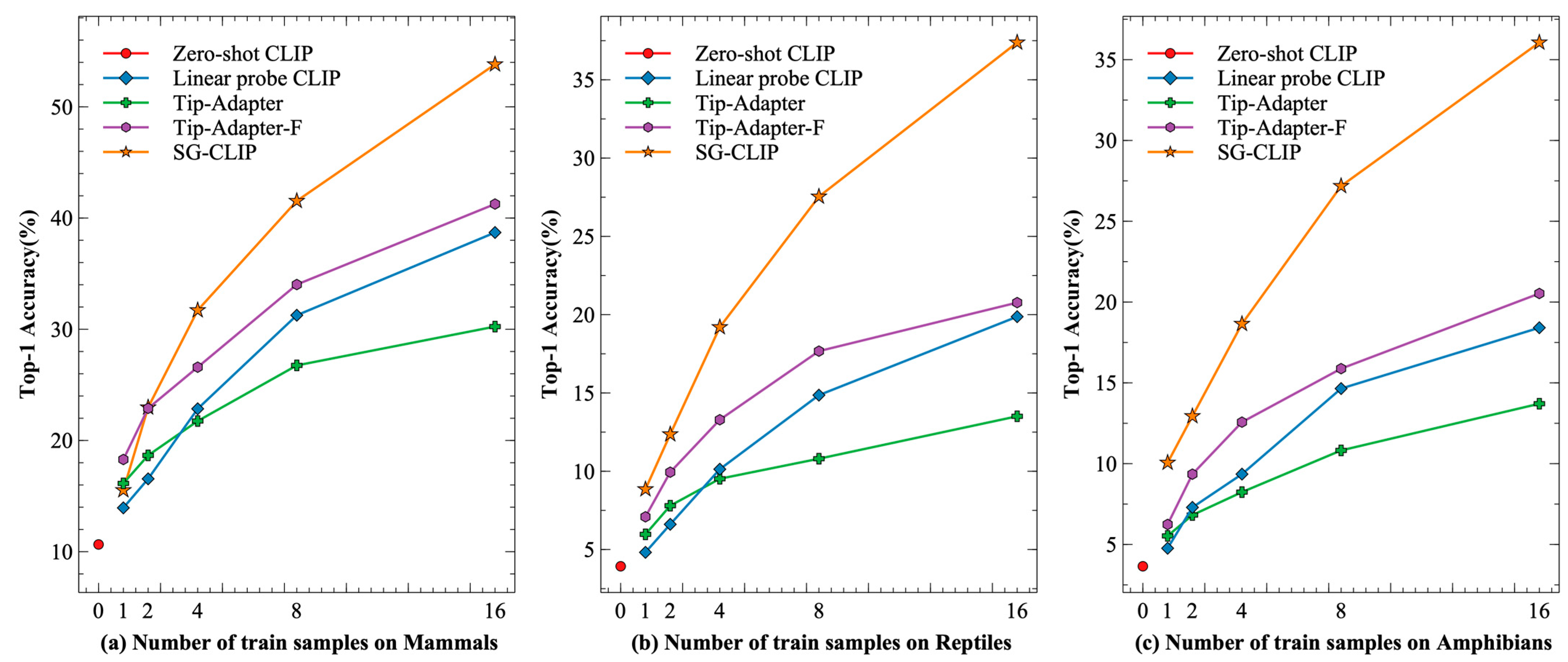
We chose the ViT-B/32 version of CLIP to compare the recognition performance of the different methods. The main results are displayed in
Table 2. The results demonstrated in
Table 2 show that SG-CLIP significantly outperforms all baseline methods in terms of recognition accuracy, proving that fusing geolocation information can improve the performance of few-shot species recognition.
Compared to Zero-shot CLIP, SG-CLIP achieved significant performance gains across all three species datasets. For the three species dataset, the performance of Zero-shot CLIP was exceeded by only the one-shot training setup. Under the 16-shot training setup, SG-CLIP boosts recognition accuracy by 5~10 times.
Compared to Linear probe CLIP, SG-CLIP achieved the best performance under all x-shot training settings, where x was 1, 2, 4, 8, and 16. As shown in
Figure 4, as the number of training samples increases, the gap between SG-CLIP and Linear probe CLIP becomes more obvious. Under the 16-shot training setup, SG-CLIP improves the recognition accuracy by 15.12% (Mammals), 17.51% (Reptiles), and 17.65% (Amphibians).
Tip-Adapter and Tip-Adapter-F outperformed SG-CLIP only on the mammal dataset with the one-shot training setup. The SG-CLIP achieved significant performance gains on all three species datasets when the training samples are set to 2 or more. Compared to Tip-Adapter, the recognition accuracy of SG-CLIP was improved by 23.58% (Mammals), 23.87 (Reptiles), and 22.35% (Amphibians) with the 16-shot training setup. Similarly, the recognition accuracy of SG-CLIP was improved by 12.56% (Mammals), 17.39 (Reptiles), and 15.53% (Amphibians) compared to Tip-Adapter-F.
As shown in
Figure 5, on the Mammals dataset, the recognition accuracy of Tip-Adapter-F was the highest, followed by Tip-Adapter, and SG-CLIP was only higher than Linear probe CLIP when the number of training samples was 1. As the number of training samples increased to more than 2, SG-CLIP gradually outperformed Tip Adapter and Tip Adapter. On the Reptiles and Amphibians datasets, SG-CLIP outperformed Tip-Adapter-F and Tip-Adapter at all settings of the number of training samples.
4.2. Performance of Few-Shot Species Recognition with Different Versions of CLIP
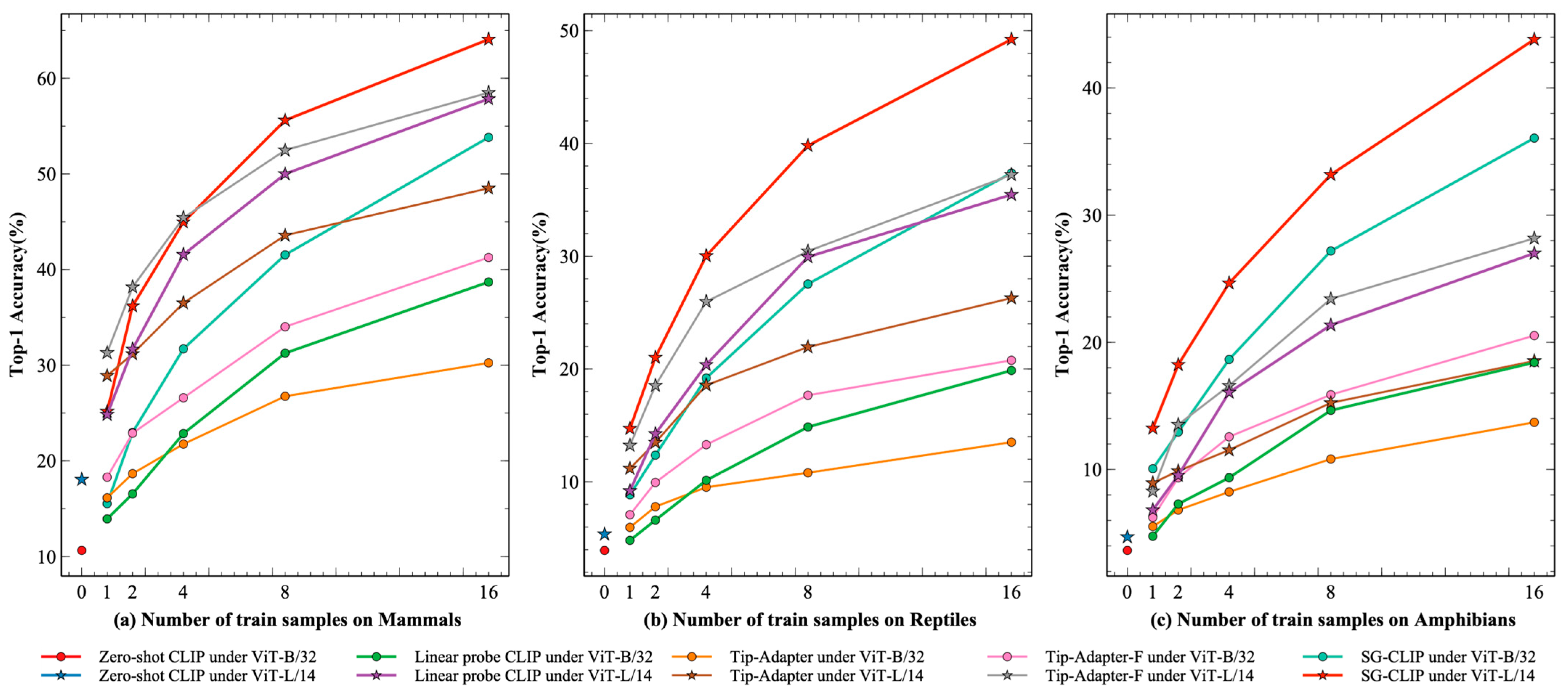
Further, we performed comparative experiments on different versions of CLIP, such as ViT-B/32 and ViT-L/14, to validate the generalizability of SG-CLIP. Compared with ViT-B/32, ViT-L/14 has more parameters and better feature extraction capability.
As can be seen from
Table 3, the recognition accuracies of Zero-shot CLIP, Linear probe CLIP, Tip-Adapter, Tip-Adapter-F, and SG-CLIP were significantly improved as the model became larger. Under the 16-shot training setup on the Mammals dataset, the recognition accuracy of SG-CLIP under ViT-L/14 improved from 53.82% to 64.07% compared to SG-CLIP under ViT-B/32, which gained a 10.25% improvement. Similarly, improvements of 11.85% and 7.22% were obtained on the Reptiles and Amphibians datasets, respectively.
From
Figure 6, it can be observed that SG-CLIP achieved the highest level of recognition accuracy when the number of training samples for each category was set to 16. Tip-Adapter-F and Linear probe CLIP followed after SG-CLIP, and their performance improved as the number of training samples increased. Finally, Tip-Adapter had the lowest recognition accuracy among all the few-shot recognition methods. On the Reptiles and Amphibians datasets, SG-CLIP achieved the best performance and substantially outperformed other methods in all training sample setups. The gap became more pronounced as the number of training samples in each class increased. On the Mammals dataset, SG-CLIP did not have the highest recognition accuracy when the training sample setting was less than 4; SG-CLIP only outperformed the other methods when it exceeded 4.
4.3. Time Efficiency Analysis
To evaluate our proposed method more comprehensively, we conducted time efficiency analysis experiments, the results of which are shown in
Table 4. The training time for SG-CLIP was far superior to that of other methods. Linear probe CLIP freezes the CLIP to extract features and adds a linear layer for classification, requiring very little training time. Tip-Adapter improves the species recognition performance by fusing the predictions of the pre-constructed key-value cache model with the predictions of Zero-shot CLIP, which is a training-free adaptation method, and thus it and Zero-shot CLIP have no additional training cost. Tip-Adapter-F further fine-tunes the keys of the cached models in Tip-Adapter as learnable parameters, thus introducing additional training time, usually within only a few minutes. SG-CLIP introduces two modules, GFEM and IGFFM, for feature extraction of geolocation information and deep fusion between images and geographic features, respectively, so it requires a lot of training time.
4.4. Ablation Studies
We conducted three ablation studies for SG-CLIP on the residual ratio α, the number of DFBs in IGFFM, and geographic information.
We first performed an ablation study by varying the residual ratio α. From
Table 5, we can see that the residual ratios were different on different datasets, where the best residual ratio on Mammals was 0.8, the best residual ratio on Reptiles was 0.8, and the best residual ratio on Amphibians was 1.0. In general, fusing image and geographic features and then adding them to the original image features better improves the performance of few-shot species recognition.
Moreover, we performed an ablation study by adjusting the number of DFBs in IGFFM. As shown in
Table 6, we observed that the number of DFBs in IGFFM is different on different datasets. Specifically, the number of DFBs in IGFFM was 2 on Mammals, 3 on Reptiles, and 4 on Amphibians. We observed that the less common the species in the CLIP training set, the more DFBs in IGFFM were required.
Finally, we conducted an ablation study on geographic information. As can be seen from
Table 7, fusing either longitude or latitude helps to improve the species recognition performance under the 16-shot training setup. Further, the recognition performance on all three species datasets when fusing longitude or latitude alone is lower than when fusing both longitude and latitude.
4.5. Visualization Analysis
SG-CLIP has been shown to improve few-shot species recognition accuracy across species datasets. To better understand the mechanism of how SG-CLIP works, we selected the top 15 species from the Mammals dataset and used t-SNE [
39] to visualize the image representations extracted by different methods as a point. As shown in
Figure 7, from ViT-B/32 to ViT-B/14, the cluster results of Zero-shot CLIP and SG-CLIP improved. SG-CLIP generated more discriminative image representations compared to Zero-shot CLIP under ViT-B/32 and ViT-L/14. This was due to the fact that SG-CLIP utilizes geolocation information for multiple interactions with image features in higher dimensions.
4.6. Case Studies
To demonstrate the effectiveness of the proposed method in practical application, we executed a case study on a subset of Mammals. First, we created Mammals-20 by selecting the top 20 species from the Mammals dataset. Then, we performed comparative experiments of different methods on Mammals-20 under ViT-L/14. From
Table 8, it can be seen that the Top-1 and Top-5 recognition accuracies of SG-CLIP were much better than the other methods under all training sample setups (1, 2, 4, 8, and 16). Specifically, SG-CLIP achieved the best Top-1 and Top-5 accuracies of 89% and 97.5%, respectively, for the 16-shot training setup. Although the Top-5 accuracy of Tip-Adapter-F was the same as the recognition accuracy of SG-CLIP, the Top-1 accuracy of 75.5% was much lower than that of SG-CLIP. Compared to the 37% Top-1 recognition accuracy of the Zero-shot CLIP on Mammals-20, the SG-CLIP with the 16-shot training setup is sufficient for real-world precision recognition applications.
5. Discussion
In this work, we explored for the first time the potential of using geographic information in foundation model-based species classification tasks, and demonstrated that fusing geolocation information from species observations can help improve performance in few-shot species recognition.
Additional information, like geographic location information and time information, can benefit species recognition. Many studies [
27,
30,
40,
41] have utilized geographic information to improve the performance of species recognition. However, the above studies on species recognition are mainly built on traditional ResNet and ViT-based visual models, while our proposed method is built on foundation models such as CLIP. In addition, the amount of data used for training is another difference; our approach requires a relatively small number of training samples.
There are many studies exploring the potential of CLIP as an emerging paradigm for zero-shot or few-shot species recognition. CLIP-Adapter [
21] constructs a new bottleneck layer to fine-tune the image or text features extracted by the pretrained CLIP to the new species dataset. Tip-Adapter [
23] utilizes the training set from the new species dataset to construct a key-value caching model and improve the performance of the CLIP for species recognition through feature retrieval. Maniparambil et al. [
36] used GPT-4 to generate species descriptions in combination with an adapter constructed by the self-attention mechanism to improve performance in zero-shot and few-shot species recognition. The above work is either improved in the image branch of CLIP or in the text branch of CLIP. Unlike the above work, we added a geographic information branch to improve species recognition performance with the help of a priori knowledge of geographic location.
First, we validated our proposed method on three species supercategories (Mammals, Reptiles, and Amphibians), demonstrating that fusing geographic information facilitates species recognition and was effective across categories. Then, we performed experiments on different versions of CLIP (ViT-B/32 and ViT-L/14), and it can be found that as the feature extraction capability of the pretrained CLIP model is enhanced, the performance of our method is enhanced accordingly. This provides strong confidence for building foundation models for taxonomic classification in the future. Next, we performed rigorous ablation experiments that demonstrate the importance of our different modules and also show that appropriate hyperparameters need to be set for different species’ supercategories. These hyperparameters are usually positively correlated with the ease of species recognition. Finally, from the visualization results in
Figure 7, it can be noticed that different species become easier to distinguish after incorporating geolocation information.
However, our work faces two limitations. First, the data used to train the CLIP model came from the Internet, where there is relatively little data on species, leading to poor performance of CLIP on the species classification task. Second, due to the difficulty of species recognition, the residual ratio and the number of DFB are variable on different species datasets, requiring a manual search for appropriate values.
In the future, we will consider allowing the proposed model to automatically learn appropriate parameters such as the residual rate and the number of DFBs. In addition, we will validate the proposed method on other foundation models.
6. Conclusions
In this paper, we proposed SG-CLIP, a few-shot species-recognition method of CLIP that leverages geographic information about species. To the best of our knowledge, we are the first to integrate geographic information for CLIP-driven few-shot species recognition. First, to harness the powerful image representation learning capabilities of foundation models such as CLIP, we used a pretrained CLIP model to extract species’ image features and corresponding textual features. Then, to better utilize geographic information, we constructed a geographic feature extraction module to transform structured geographic information into geographic features. Next, to fully exploit the potential of geographic features, we constructed a multimodal fusion module to enable deep interaction between image features and geographic features to obtain enhanced image features. Finally, we computed the similarity between the enhanced image features and the text features to obtain the predictions of the species. Through extensive experiments on different species datasets, it can be observed that utilizing geolocation information can effectively improve the performance of CLIP for species recognition, and significantly outperform other advanced methods.
For species recognition scenarios under data constraints, such as rare and endangered wildlife recognition, our model can be used as the first step for accurate recognition to improve the level of automated species recognition, accelerate species data annotation, and provide preliminary data support work for the subsequent design and iteration of models dedicated to the recognition of specific species. We hope that our model can help build better species distribution models for biodiversity research. In future work, we will focus on the impact of geographic information on species recognition at different regional scales.