Abstract
Accurate identification of individual tree species (ITS) is crucial to forest management. However, current ITS identification methods are mainly based on traditional image features or deep learning. Traditional image features are more interpretative, but the generalization and robustness of such methods are inferior. In contrast, deep learning based approaches are more generalizable, but the extracted features are not interpreted; moreover, the methods can hardly be applied to limited sample sets. In this study, to further improve ITS identification, typical spectral and texture image features were weighted to assist deep learning models for ITS identification. To validate the hybrid models, two experiments were conducted; one on the dense forests of the Huangshan Mountains, Anhui Province and one on the Gaofeng forest farm, Guangxi Province, China. The experimental results demonstrated that with the addition of image features, different deep learning ITS identification models, such as DenseNet, AlexNet, U-Net, and LeNet, with different limited sample sizes (480, 420, 360), were all enhanced in both study areas. For example, the accuracy of DenseNet model with a sample size of 480 were improved to 87.67% from 85.41% in Huangshan. This hybrid model can effectively improve ITS identification accuracy, especially for UAV aerial imagery or limited sample sets, providing the possibility to classify ITS accurately in sample-poor areas.
1. Introduction
Forests are the basis for the survival and development of humans and many organisms [1], and forestry surveys are valuable for monitoring and maintaining sustainable forest resources as well as for biodiversity research [2]. Identifying forest species using remote sensing imagery is an important component of forestry surveys and is key to the utilization and conservation of forest resources [3]. Such technology can be used to assess the current status and distribution of forest resources in a timely manner and carry out dynamic observations and forest research to enable the sustainable development of forestry [4].
Many studies have used remote sensing imagery for tree species identification. For instance, Nijland et al. [5] used Landsat ETM+ and airborne laser scanning (ALS)-derived long-term image sequences of topography and vegetation structure to identify 4 forest cover types in northwestern Alberta, Canada, with a minimum root mean square error (RMSE) of 10.9%. Fedrigo et al. [6] identified 3 forest stand types in southeastern Australia using the random forest (RF) methodology and light detection and ranging (LiDAR) data with an overall accuracy (OA) of 84%. Grabska et al. [7] used the RF method and Sentinel-2 time-series imagery to identify 9 tree species in mixed forests of the Polish Carpathian Mountains with an OA greater than 90%. Oreti et al. [8] used the K-nearest neighbors (KNN) method and multispectral imagery acquired by Airborne Digital Sensor 40 to identify mixed forests of the Sila Plateau with an OA up to 85%. Wan et al. [9] used a curve matching based method and fused data from UAV imagery, Sentinel-2 time-series imagery and LiDAR data to identify 11 tree species in the Gaofeng forest farm in Guangxi Province, China, with an OA greater than 90%.
With the development of remote sensing, the focus of tree species identification is changing from stands to individual trees; this approach is more accurate and more conducive to the fine management of forest resources. Since individual trees represent the smallest forest unit, individual tree species (ITS) identification is important for forestry research and forest resource management [10]. ITS identification using remote sensing imagery mainly consists of two steps: individual tree crown (ITC) delineation and ITS identification using image classification algorithms.
ITC delineation is the automatic delineation of the location, size, and shape of each ITC in remote sensing imagery [11]. Accurate ITC delineation is critical for extracting spectral data, texture, shape, internal structure, and other information regarding tree crowns with high accuracy, reducing the interference of noncanopy image elements and improving the ITS identification accuracy [12]. Gougeon et al. [13] proposed a valley-following method for ITC delineation using intertree shading. Culvenor [14] proposed a new ITC delineation algorithm that employs local radiometric maxima and minima in images to determine treetops and tree boundaries. Jing et al. [15] used multiscale low-pass filtering to extract the treetops of multiscale tree crowns by referring to the geometrical shape and multiscale properties of tree crowns, and then delineated crowns of different scales from aerial imagery and LiDAR data using the marker-controlled watershed segmentation algorithm. Hamraz et al. [16] vertically stratified the forest LiDAR point cloud and delineated ITCs from multiple canopy layers. Qiu et al. [17] improved the ITC delineation algorithm proposed by Jing et al. [15] to determine treetops with respect to image brightness and spectra and delineated multiscale ITCs in dense forests. The improved algorithm offered an ITC delineation accuracy of up to 76% in the rainforest and 63% in the deciduous forest.
Current ITS identification methods can be divided into two main categories. The first category is based on traditional image features; that is, after delineating ITCs in remote sensing imagery, the spectral and textural features of the ITCs are extracted and applied to ITS identification [18,19,20,21]. The spectral features derived from images are the albedo of surface objects. Because the internal structure and physiological and biochemical parameters of the same type of objects are normally similar, objects of the same type also exhibit similar spectral characteristics in images. In contrast, objects of different types exhibit different spectral characteristics in imagery due to their different structures, compositions, and physical and chemical properties [22]. Using this principle, objects can be distinguished from images based on their spectral characteristics. The texture features of images reflect the properties of the image grayscales and the spatial topological relationships. They are a kind of locally structured feature of images, and are expressed in images as the grayscale or color variation in image elements within a certain neighborhood of image element points [23]. Objects have consistent internal texture features, and the internal texture features of different objects differ; as a result, it is possible to distinguish different objects using the texture features in images. Traditional image features are widely used in ITS classification. For instance, Maschler et al. [24] used the spectral and textural features of hyperspectral data to identify 13 ITS with an OA of 91.7% in the UNESCO Biosphere Reserve Wienerwald, which is located in the colline and submontane altitudinal belt. Mishra et al. [25] employed unmanned aerial system imagery and eCognition software for segmentation and designed spectral, textural, and other features to identify ITS located in the Himalayan plateau region using the ensemble decision tree algorithm with an OA up to 73%. Sothe et al. [26] utilized texture, spectral data, and other features to identify 16 tree species in 2 mountainous subtropical forest fragments in southern Brazil with an OA greater than 74%. Xu et al. [27] used multispectral image point clouds to extract spectral and textural features, along with additional features, to identify 8 species of trees in Xiangguqing, Yunnan Province, China, located on the plateau using RF with an OA of 66.34%.
The second category of ITS identification methods uses deep learning models to identify ITS after delineating ITCs in images. With the development of deep learning, convolutional neural networks (CNNs) have frequently been applied to image classification. AlexNet, a classic CNN, won the 2012 ImageNet Large-Scale Visual Recognition Challenge (ILSVRC) [28]. ResNet is based on the improved AlexNet and successfully avoids the problems of forward parameter explosion and backward parameter vanishing [29]. In 2017, the DenseNet model [30] was improved and optimized based on ResNet; the changes significantly improved the generalization performance of ResNet and alleviated the problem of gradient disappearance in the deep network. The DenseNet model has been tested on several image sets and has achieved excellent classification results. In view of the superior performance of deep learning classification, researchers have begun to use deep learning for ITS identification [31,32,33,34,35]. Nezami et al. [36] used 3D-CNN, hyperspectral, and red-green-blue (RGB) imagery to identify 3 ITS in the Vesijako research forest area, with an OA of 98.3%. Yan et al. [37] used AlexNet, GoogLeNet, and ResNet models to identify six ITS from WorldView-3 imagery of the Beijing Olympic Forest Park. The OA of GoogLeNet reached 82.7%. Ouyang et al. [38] used a total of five deep learning models, including AlexNet and DenseNet, to identify ITS from WorldView-3 imagery of the Huangshan Scenic Area in the mountain area. DenseNet achieved an OA of 94.14%. Zhang et al. [39] used an improved masked R-CNN and UAV aerial imagery to identify 8 ITS in the Jingyue Eco-Forest in Changping District, Beijing, located on the plain with an OA of 90.13%.
Both types of ITS identification methods have strengths and weaknesses. In traditional image feature-based ITS identification methods, traditional image features are mostly selected based on expert knowledge and experience, with high interpretability. These methods require a smaller sample size and achieve stable and reliable identification accuracy [40,41,42]. However, these methods require experts with extensive expertise and complex exploration processes; furthermore, each method was designed for a specific application with limited generalizability and robustness. [43,44]. In contrast, deep learning methods automatically extract features and rely mainly on data-driven results, which can obtain deep, dataset-specific feature representations based on the learning of a large number of samples, with a more efficient and accurate representation of the dataset. Therefore, the extracted abstract features are more robust and have better generalizability, but the features are more easily influenced by the sample set and are not interpretable. Moreover, the model is computationally demanding and requires a large sample set for training [45].
To address these problems, some researchers have explored combining traditional imagery identification with deep learning and proposed hybrid models to obtain more accurate identification. For example, Jin et al. [46] extracted the features of original images using AlexNet, used the ReliefF algorithm to rank the extracted features, and then used trial-and-error to select the n most important features to reduce the feature dimensionality. They used the SVM classifier to provide identification results based on the n selected features. The results showed that the proposed model exhibited a significant improvement in identification accuracy and identification time efficiency compared to the performance of existing models. Saini et al. [47] used a ResNet-50 pretrained network to extract low-level deep features and then used k-means clustering followed by SVM for identification. The new model obtained better identification results with higher F1-scores and greater area under the curve (AUC) metrics compared to an existing model. Bakour et al. [48] first extracted local and global features from images to form a feature vector, and then input the feature vector into the deep learning model for identification. The identification accuracy was greater than 98%. Dey et al. [49] fused the manually extracted features with the features extracted by deep learning before using RF for identification; the identification accuracy was 97.94%.
The current hybrid models have the following drawbacks: (1) traditional image features are often extracted manually, with less automation; (2) most hybrid models utilize only deep learning features or traditional image features, rarely considering the combination of the two; and (3) identification features are mostly used for traditional machine learning identification, resulting in less automation and incomplete identification feature extraction.
To solve the above problems and obtain more reliable identification results, we used the deep learning model as the basic network, with a traditional image feature-assisted basic network to identify ITS (the traditional image feature-assisted module is referred to as the explore module). The hybrid model was constructed to complete the identification of ITS in the study area to explore the feasibility of traditional image feature-assisted identification and obtain an ITS identification model with higher identification accuracy and greater interpretability. In addition, in order to explore the applicability of the explore module, the accuracy improvement results were compared before and after introducing the explore module for different sample sets or deep learning models.
2. Data
2.1. Study Area
Two primary study areas were selected for the experiment, and the study areas are shown in Figure 1. The first study area is located in the Huangshan Mountains area, Anhui Province, with a geographical range of 118°9′16″E to 118°11′24″E latitude and 30°7′8″N to 30°10′37″N longitude, and is found in the subtropical monsoon climate zone. There are more natural forests and mixed forests in the area and fewer artificial forests and pure forests. The main tree species include Phyllostachys heterocycla, Cunninghamia lanceolata, Pinus hwangshanensis, other evergreen arbors, and other deciduous arbors. The second study area is located in the central area of the Gaofeng forest farm, Guangxi Province, China, with a geographical range of 108°21′36″E to 108°23′38″E latitude and 22°57′43″N to 22°58′59″N longitude, and a tropical northern edge climate. This study area is rich in forest cover types, primarily secondary plantation forests, and the main tree species include Eucalyptus urophylla S.T.Blake, Cunninghamia lanceolata, Pinus massoniana Lamb, Eucalyptus grandis x urophylla, and Illicium verum Hook. f.
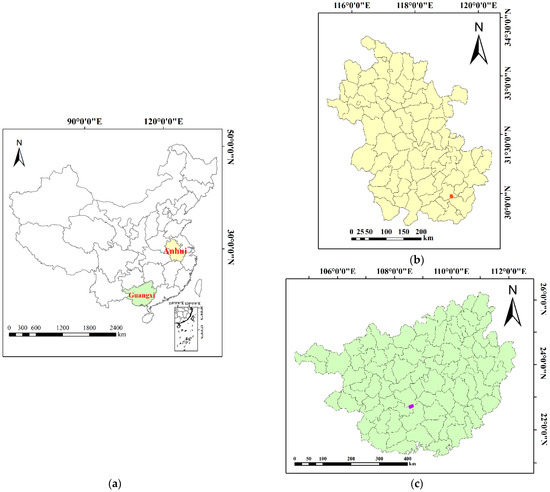
Figure 1.
Map of the study areas. (a) Location of the study areas: the yellow area is Anhui Province where the Huangshan Mountains area is located, and the green area is Guangxi Province where the Gaofeng forest farm is located. (b) Anhui Province where the Huangshan Mountains area is located; the red area is the Huangshan study area. (c) Guangxi Province where the Gaofeng forest farm is located; the purple area is the Gaofeng study area.
2.2. Experimental Data
A WorldView-3 image of the Huangshan study area obtained on 10 March 2019, was also selected. This image has 1 panchromatic band with a spatial resolution of 0.31 m and 8 multispectral bands with a spatial resolution of 1.24 m [50].
An unmanned aerial vehicle (UAV) aerial image of the Gaofeng study area obtained on 9 October 2016, with a spatial resolution of 0.2 m in the red, green, and blue bands, was selected for study.
The study area and the image used are shown in Figure 2 and Figure 3; Figure 2 shows the red, green, and blue bands as the 5th, 3rd, and 2nd bands in WorldView-3, respectively, and Figure 3 shows the red, green, and blue bands.
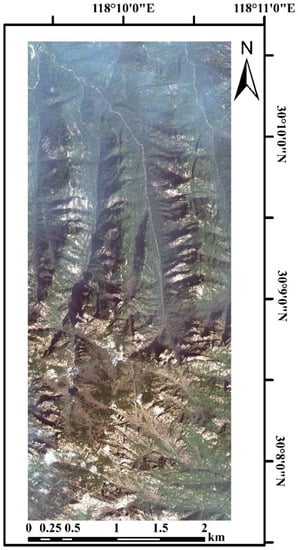
Figure 2.
WorldView-3 image of the Huangshan study area.
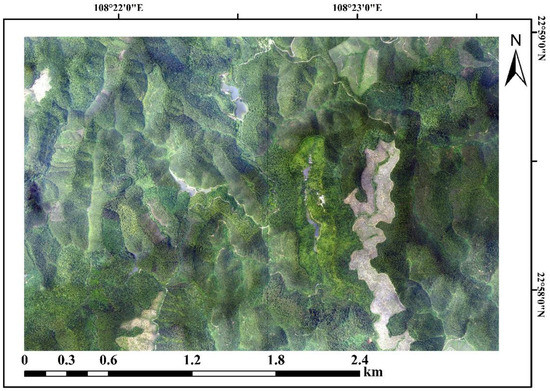
Figure 3.
UAV aerial image of the Gaofeng study area.
2.3. Data Preprocessing
The UAV aerial images of the Gaofeng study area were preprocessed for subsequent identification studies by radiometric calibration, orthorectification, atmospheric correction, and image cropping.
After preprocessing with radiometric calibration, orthorectification, atmospheric correction, and image cropping, the WorldView-3 image of the Huangshan study area was fused using the haze-and-ratio-based (HR) [51] method proposed by Jing et al. Then, an 8-band multispectral fusion image with a spatial resolution of 0.31 m was obtained.
2.4. Sample Set Construction and Enhancement
In this study, the main steps for constructing the remote sensing image sample sets of ITS were as follows:
(1) After assessing the study areas in advance, the field survey study areas were determined, and samples of the main tree species category were collected in each study area. Based on the results of the field collection, the main tree species types in the samples from the Huangshan study area were Phyllostachys heterocycla, Cunninghamia lanceolata, Pinus hwangshanensis, other evergreen arbors, and deciduous arbors; the main tree species types in the samples from the Gaofeng study area were Eucalyptus urophylla S.T.Blake, Cunninghamia lanceolata, Pinus massoniana Lamb, Eucalyptus grandis x urophylla, and Illicium verum Hook. f.
(2) Crown slices from imagery (CSI) [52], a method designed specifically for ITC delineation and proposed by Jing, was used to carry out multiscale segmentation and ITC delineation of remote sensing images of the study areas in Gaofeng and Huangshan.
(3) By combining the geographical location and tree species of the collected sample points with manual visual interpretation, more accurate ITC delineation results were selected, and the tree species of ITC were labeled.
(4) According to the minimum bounding rectangular of the labeled ITCs, ITS images that are beneficial for model training were outputted.
(5) The ITS remote sensing images were categorized based on the tree species categories to construct ITS remote sensing image sample sets containing the main tree species in the study areas.
(6) The images in original ITS sample set were rotated by 90°, 180°, and 270°, flipped horizontally, and turned upside down, individually, to expand the number of samples by 6 times [53].
The main steps are shown in Figure 4.
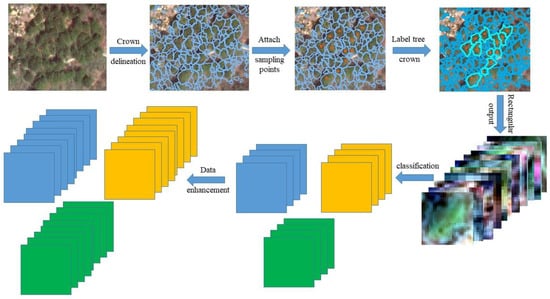
Figure 4.
The steps for building the ITS sample set.
The enhanced ITS sample set was proportionally divided into training, validation, and test sample sets. The training and validation sets were used for training deep learning models dynamically, while the test set was used to test the final classification accuracy of the model. The ratio of Huangshan training, validation, and test sample set was 3:1:1, as shown in Table 1. Due to the small number of samples obtained in the Gaofeng study area, to better train the deep learning model, the Gaofeng sample set was divided into a training sample set, validation sample set, and test sample set at a ratio of 6:2:1, as shown in Table 2.

Table 1.
ITS samples from the Huangshan study area.

Table 2.
ITS sample set of the Gaofeng study area.
3. Model Structure
3.1. Basic Network
In the field of deep learning, the CNN is the most famous and commonly employed algorithm [54,55,56,57]. A CNN is mainly composed of a convolutional layer, a pooling layer, a fully connected layer, and an output layer. The convolutional layer is the most significant component in CNN architecture and it consists of a collection of convolutional filters. In the convolutional layer, the input image is convolved with these filters to generate the output feature map. The main task of the pooling layer is the sub-sampling of the feature maps by following the convolutional operations. Subsequently, the fully connected layer is utilized as the CNN classifier. At last, the final classification is achieved from the output layer [58].
The DenseNet model, which performed well in classification, was used in this experiment.
The DenseNet model is divided into two major modules: the dense module and the compression layer. The dense module achieves feature reuse by densely connecting the features on the channels to further alleviate the gradient disappearance problem in the deep network, while the compression layer reduces the parameter computation by decreasing the size of the feature map. In addition, a bottleneck layer can be introduced in the dense module to reduce the parameter computation.
We modified the DenseNet model to meet the identification needs of a small ITS sample set. The input layer was changed to 32 × 32 × 8, which is suitable for multispectral imagery (32 × 32 × 3 for UAV aerial image identification). The number of convolution filters in the dense module was adjusted from 12 to 6 for each layer to prevent overfitting. The repetition parameter of the dense module with bottleneck layer processing was set to 5, and the compression layer coefficient was set to 0.5.
The improved DenseNet model contains an input layer, a convolutional layer, 5 dense modules, 4 compression layers, a fully connected layer, and an output layer. The second layer of the DenseNet model is the convolutional layer, which contains 12 convolutional filters with 3 × 3 convolutional kernels, and the ReLU function was chosen as the activation function. Dense modules (1) to (5) are then alternately superimposed with compression layers (1) to (4), and the compression layer is composed of a convolutional layer with 1 × 1 convolutional kernels and a 2 × 2 average pooling layer. The global average pooling layer replaced the original fully connected layer with a global average pooling of 99 output neuron nodes, and the final output layer is a softmax function as the classifier. The model uses the categorical_crossentropy function as the loss function and Adam as the iterative optimizer.
For the dense module, in addition to using a dense connection to establish the relationship between all the previous layers and the later layers, a bottleneck layer is used to reduce the dimensionality before the convolutional filter of size 3 × 3 within the module. Figure 5 and Figure 6 show schematic diagrams of the improved DenseNet model and the structure of the Dense module, respectively.
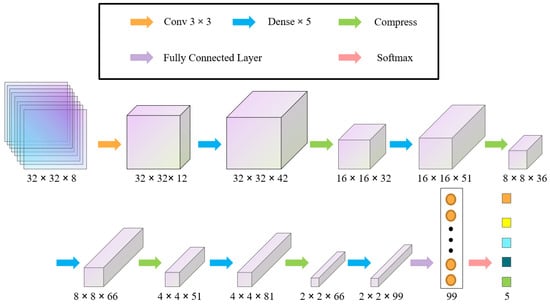
Figure 5.
The improved DenseNet model.

Figure 6.
Structure of the dense module.
3.2. Explore Module
Using traditional image features to assist deep learning networks for decision identification, the constructed explore module was added to the underlying deep learning model to assist in decision making to obtain better identification results. The specific implementation of the explore module is divided into two parts: traditional image feature selection and explore module construction.
3.2.1. Feature Selection
The main traditional image identification features used in this study are spectral and texture features.
The commonly used spectral features are generally the mean and variance in each band, as well as the brightness and maximum difference.
A common algorithm for extracting texture features of an object is to compute the gray-level cooccurrence matrix (GLCM) of the object [59]. The GLCM of an image reflects the comprehensive information of the image grayscale regarding the direction, adjacent spacing, and change magnitude, and is the basis for analyzing the local features and alignment rules of the image.
In this study, the spectral features used and the texture features extracted from GLCM [60] are shown in Table 3.

Table 3.
The main traditional image identification features.
3.2.2. Explore Module Construction
After inputting the image of sample into the hybrid model, it first extracts the traditional image features (mean value, standard deviation, brightness, maximum difference, ASM, CON, and other features). Subsequently, the extracted features are fully concatenated, and then classification is implemented by softmax to obtain the identification results of traditional image features, which are then multiplied by the weight (W) to assist with identification by the deep learning network.
where n is the number of samples.
This weight is inversely proportional to the number of samples. Since the deep learning model needs to learn plenty of parameters from the samples, when the sample number is small, the deep learning model cannot train enough parameters, leading to the occurrence of overfitting. In contrast, using traditional image features for classification requires fewer samples and can maintain stable identification accuracy with limited samples. By introducing the explore module into the deep learning model, the weights value is larger with a limited sample set, the explore module participates in a larger proportion of decision making, and the classification results of traditional image features takes a larger weight. Thus it adjusts the classification results of deep learning and avoids the occurrence of deep learning overfitting.
When the sample number is large, the parameters of the deep learning model can be fully trained and the classification accuracy is high. At this time, the explore module engages in a smaller proportion of decision making, the classification results of traditional image features only account for a smaller portion, and more classification results of deep learning are adopted to ensure high-precision classification results.
With the introduction of the explore module, the classification results of deep learning can be dynamically adjusted, making the classification results more stable and reliable with higher accuracy, which is more suitable for individual tree species classification where collecting samples is difficult.
3.3. Whole Model Construction
The identification results obtained after identification by the basic deep learning network are multiplied by (1 − W) and then added to the auxiliary identification results from the explore module to obtain the final identification results. The entire architecture of the model is shown in Figure 7.
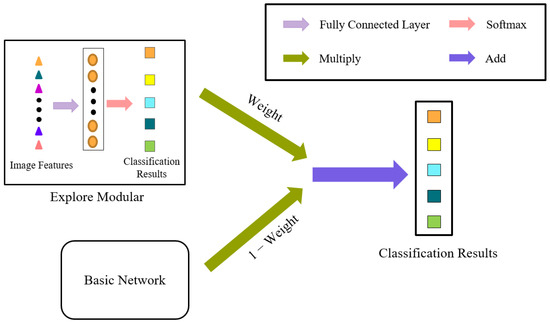
Figure 7.
The ExploreNet model.
The explore module represents the identification results of traditional image features. Specifically, the image spectral features and texture features are first extracted from the imported image of the sample, and softmax identification is applied after the features are fully concatenated, in order to obtain the identification results of the traditional image features.
The basic network represents the identification results of basic deep learning networks. In this study, DenseNet was used as the basic network, and the identification results of deep learning for ITS were obtained after deep learning training.
The identification results of the traditional image features (explore module) were multiplied by W and added with the identification results of deep learning multiplied by (1 − W) to obtain the final identification results.
When using traditional image features to assist deep learning for identification, when the number of samples is small, the weights are large, and the traditional image features are given greater weight in the decision-making process to avoid overfitting or underfitting by deep learning due to the small sample size. When the number of samples is large, the weight of each is small, and the traditional image features are given less weight in the decision-making process so that the identification advantages of deep learning can be fully realized.
3.4. Experimental Settings
In this study, the deep learning environment was built using the pattern of front-end Keras 2.2.4 and back-end TensorFlow 1.14.0, with programs written in Python 3.6.8. The operating system was Windows 10, the graphics card was NVIDIA GTX1060, and the parallel computing of the graphics card was realized by CUDA10.0.
In the study, 60 batches were trained at a time for a total of 500 iterations. During the iterations, the tolerated number of times that the performance of the network did not improve (performance improvement was defined as an accuracy improvement greater than 0.001) was set to 5, and the learning rate was reduced by 0.005 after 5 times, with a lower limit of the learning rate of 0.5 × 10−6. Test results were obtained on the validation set at the end of each epoch, and the network stopped training if the test error was found to increase on the validation set or if the accuracy did not improve after 10 times. The model with the highest identification accuracy on the validation set was saved.
3.5. Accuracy Assessment
To assess the effectiveness of the model, an accuracy assessment was performed. After training the model using training samples and validation samples, the classification results of the model were tested using real test samples, and the confusion matrix was calculated with the four metrics of producer’s accuracy (PA), user’s accuracy (UA), OA, and Macro-F1 obtained based on the confusion matrix to evaluate the accuracy of the model. The PA refers to the agreement between the result of tree species identification and actual tree species; the UA shows the usability of the result from the perspective of using the maps; the (OA) and Macro-F1 measure the overall type consistency between the identification result and the real tree species distribution. The indicators PA, UA, OA can be calculated using the following formulas [61,62], and the indicator Macro-F1 is defined in Refs. [63,64]:
where r denotes the number of tree species, n denotes the total number of samples in the study area, denotes the number of samples that have been classified correctly as tree species type i, denotes the number of samples in the validation sample sets, and denotes the number of samples in the reference data corresponding to the sample set.
4. Results
4.1. Training and Validation Accuracies
The divided training sample set was used to train the DenseNet and ExploreDenseNet (ED) models, and the validation sample set was used during the training process to verify the accuracy of the trained models. The training and validation accuracies of different models in different study areas are shown in Table 4.

Table 4.
The accuracies of model training and validation.
By comparing the training results of the models, we found that the training and validation accuracies of the DenseNet model with the addition of the explore module were higher than those of the original DenseNet model in the two study areas, indicating that the introduction of the explore module can effectively extract the identification information and improve the identification accuracy of the models.
4.2. Identification Accuracy Evaluation
By comparing the identification accuracies of the two models using different data sources in different study areas (as shown in Table 5 and Table 6), we found that the OA of the DenseNet model increased after introducing the explore module. In the Huangshan study area, the OA increased from 93.53% to 94.14%, and the user’s accuracy and producer’s accuracy of the five tree species improved after introducing the explore module. For example, the UA of other evergreen trees increased from 83.75% to 88.00%; the PA of Cunninghamia lanceolata improved from 78.57% to 82.14%, and the PA of other deciduous trees improved from 97.58% to 99.19%.

Table 5.
The identification accuracies of different models for the Huangshan study area.

Table 6.
The identification accuracies of different models for the Gaofeng study area.
The OA improved from 91.54% to 93.38% in the Gaofeng study area. The producer’s accuracy of all five species improved to different degrees after introducing the explore module. The producer’s accuracy of Eucalyptus grandis x urophylla improved from 89.58% to 93.40%, and the producer’s accuracy of Illicium verum Hook. f. improved from 90.80% to 94.83%, with a lower sample miss-score error. The user precision of Eucalyptus urophylla S.T.Blake, Cunninghamia lanceolata, and Eucalyptus grandis x urophylla also improved to different degrees. The user precision of Cunninghamia lanceolata increased from 88.76% to 93.39%, and that of Eucalyptus grandis x urophylla increased from 97.36% to 100.00%, with a smaller sample misidentification error rate.
The improvement of the overall identification accuracy in the Huangshan and Gaofeng study areas proved that the introduction of the explore module helped to obtain more accurate identification results, and the interpretation of the identification results was also enhanced due to the inclusion of traditional image features.
The improvement of the identification accuracy of the DenseNet model in UAV aerial imagery after adding the explore module was higher than that of multispectral identification results, indicating that introducing the explore module to the deep learning network can improve the accuracy in areas with less information to a greater degree, improving the identification accuracy effectively without adding additional data and enhancing the application value.
4.3. ITS Classification Map of Study Areas
The DenseNet and ExploreDenseNet models were used to classify the ITS in the Huangshan and Gaofeng study areas, and ITS classification maps were obtained, as shown in Figure 8 and Figure 9, respectively. In Figure 8, “other” represents areas without trees.
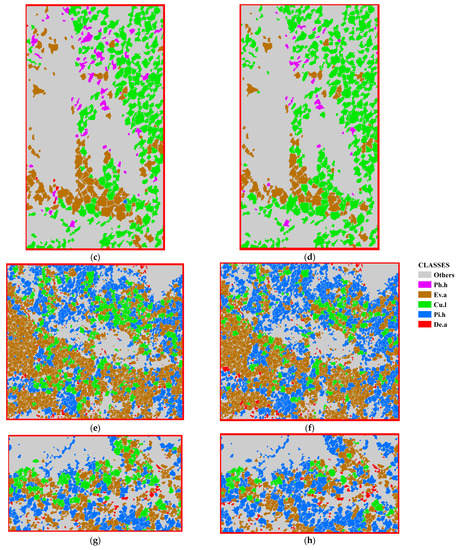
Figure 8.
Classification map of different models in the Huangshan study area, (a–c) in the maps are demonstration areas. (a) ITS classification map of the Huangshan study area using the DenseNet model; (b) ITS classification map of the Huangshan study area using the ExploreDenseNet model; (c) ITS classification map of area a using the DenseNet model; (d) ITS classification map of area a using the ExploreDenseNet model; (e) ITS classification map of area b using the DenseNet model; (f) ITS classification map of area b using the ExploreDenseNet model; (g) ITS classification map of area c using the DenseNet model; (h) ITS classification map of area c using the ExploreDenseNet model.
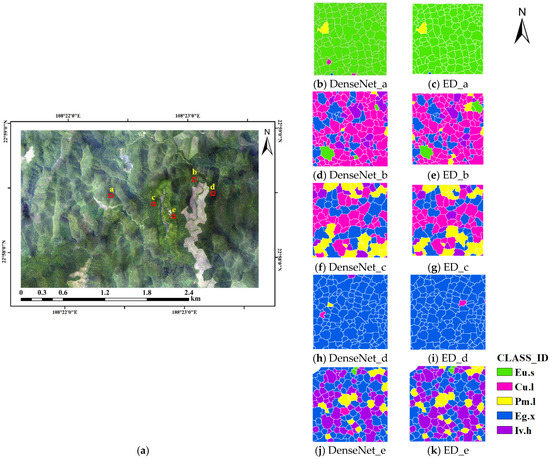
Figure 9.
Classification map of the Gaofeng study area. (a) The imagery of the study area; a–e in the imagery are pure forests of Eucalyptus urophylla S.T.Blake, Cunninghamia lanceolata, Pinus massoniana Lamb, Eucalyptus grandis x urophylla, and Illicium verum Hook. f., respectively; (b–k) the classification maps of DenseNet and ED of study areas a–e, respectively.
4.3.1. ITS Classification Map of the Huangshan Study Area
Due to the vertical variation in climate in the Huangshan area, the tree species distribution in the study area should be clearly vertically distributed with altitude. The classification result of the DenseNet model (Figure 8a) has obvious errors in the vertical distribution of tree species. Compared with the classification result of the DenseNet model, the classification result of the ED model (Figure 8b) is more consistent with the realistic vertical distribution of tree species. The comparison between Figure 8d,c shows that the classification results of the ED model have significantly reduced the erroneous distribution of Phyllostachys heterocycla in region a. The comparisons between Figure 8f,e and between Figure 8h,g show that the classification results of the ED model significantly reduce the misdistribution of Cunninghamia lanceolata in area b and area c, which is more consistent with the vertical distribution of different tree species in Huangshan with altitude.
In addition, Figure 8 shows that in the Huangshan study area, from north to south, there are mixed forests of Phyllostachys heterocycla and Cunninghamia lanceolata, mixed forests of evergreen arbors and Cunninghamia lanceolata, mixed forests of evergreen arbors and Pinus hwangshanensis, and mixed forests of Pinus hwangshanensis and deciduous arbors. Mixed forests of evergreen trees and Pinus hwangshanensis were distributed in the southeastern part of the study area.
4.3.2. ITS Classification Map of the Gaofeng Study Area
It can be seen in Figure 9 that for Eucalyptus urophylla S.T.Blake (Figure 9b,c) and Eucalyptus grandis x urophylla (Figure 9h,i), the ED model has higher classification accuracy and less misclassified canopy compared to the DenseNet model, so is capable of identifying eucalyptus species more easily. For Cunninghamia lanceolata (Figure 9d,e), there is misclassification with the other 4 tree species, but it can be seen in Figure 9d,e that the ED model can identify more Cunninghamia lanceolata canopies than the DenseNet model, with 113 Cunninghamia lanceolata canopies identified by the DenseNet model and 119 Cunninghamia lanceolata canopies identified by the ED model. For Pinus massoniana Lamb (Figure 9f,g), which is often misclassified as Cunninghamia lanceolata, Eucalyptus grandis x urophylla, or Illicium verum Hook. f., both the DenseNet and ED models can identify 31 Pinus massoniana Lamb canopies. For Illicium verum Hook. f. (Figure 9j,k), the ED model can identify more Illicium verum Hook. f. canopies than the DenseNet model, with 66 canopies, while the DenseNet model can identify only 61 canopies.
4.4. Applicability of the Explore Module for Limited Sample Sets
To evaluate the applicability of the explore module with a limited sample set (we define the limited sample set as the sample set with training samples less than or equal to 80 to simulate sample-poor areas), we selected sample subsets with only 80, 70, and 60 training samples (the number of training samples after sample enhancement is 480, 420, and 360, respectively) for ITS classification in the Huangshan and Gaofeng regions. The same test samples as in Section 2.4 were still selected to obtain the classification results in the two study areas using the limited sample set. It is very difficult to classify ITS using these limited training samples, and the classification results of different limited sample sets are shown in Table 7 and Table 8.

Table 7.
The classification accuracies in the Huangshan study area using limited sample sets.

Table 8.
The classification accuracies in the Gaofeng study area using limited sample sets.
4.4.1. ITS Classification Results in the Huangshan Study Area Using Limited Sample Sets
The classification results show that after introducing the explore module, the overall classification accuracy of the DenseNet model significantly improved with the limited sample set in the Huangshan area, from 85.41% to 87.67% for the training sample of 480, from 86.17% to 89.62% for the training sample of 420, and the overall classification accuracy for the training sample of 360 from 85.71% to 87.52%.
Among different tree species, the classification accuracy of Pinus hwangshanensis improved more significantly. For example, when the training sample was 480, the producer’s accuracy of other evergreen arbors improved from 78.65% to 82.02%, and the user’s accuracy improved from 53.85% to 57.94%; the user’s accuracy of Cunninghamia lanceolata improved from 61.54% to 82.76%, and the producer’s accuracy of Pinus hwangshanensis improved from 87.53% to 90.02%. When the training sample was 420, the producer’s accuracy of Pinus hwangshanensis increased from 90.02% to 93.02%; the producer’s accuracy of other deciduous arbors increased from 75.00% to 88.71%. When the training sample was 360, the producer’s accuracy of Pinus hwangshanensis improved from 89.78% to 94.76%.
4.4.2. ITS Classification Results in the Gaofeng Study Area Using Limited Sample Sets
In the Gaofeng area, the overall classification accuracy improved after introducing the explore module to the DenseNet model when the training sample was 420, from 85.85% to 89.62%, and the overall classification accuracy did not improve significantly when the training sample was 480 and 360. This may be related to the low number of UAV image bands and classification information.
Among different tree species, with the introduction of the explore module, the classification accuracy of Pinus massoniana Lamb improved, from 80.06% to 80.65% for the producer’s accuracy and from 94.39% to 96.79% for the user’s accuracy using the training sample of 480. With a training sample of 420, the producer’s accuracy improved from 77.38% to 78.57%, and the user’s accuracy improved from 93.53% to 98.51% for Pinus massoniana Lamb. When the training sample was 360, the producer’s accuracy of Pinus massoniana Lamb improved from 72.32% to 74.11%. The producer’s accuracy of giant-tailed eucalyptus increased from 86.46% to 90.28%, 87.85% to 89.24%, and 80.56% to 89.24% with different training samples, respectively. The user’s accuracy of Cunninghamia lanceolata ranged from 76.40% to 87.36%, and 77.59% to 81.85% for training samples of 420 and 360, respectively. The producer’s accuracy of Illicium verum Hook. f. increased from 77.01% to 89.66% when the training sample was 420.
By comparing the classification results of the two study areas, it was found that at limited sample sets, the UAV images in the Gaofeng area with fewer bands and fewer traditional classification features could be introduced; thus, the accuracy improvement effect of the DenseNet model was not as obvious compared with the multispectral image in the Huangshan area. In addition, the classification accuracy of Pinus hwangshanensis and other deciduous trees in the Huangshan region has a better improvement after the introduction of the explore module.
4.5. Applicability of the Explore Module for Different Deep Learning Models
To compare the applicability of the explore module for different deep learning models, AlexNet, U-Net [64], and LeNet [65] were selected as the base networks for the experiments (we scaled down the three models appropriately for better comparison). Table 9 and Table 10 indicate the ITS classification of the three models before and after the introduction of the explore module in the Huangshan and Gaofeng study areas, respectively.

Table 9.
The classification accuracies of the three models in the Huangshan study area.

Table 10.
The classification accuracies of the three models in the Gaofeng study area.
4.5.1. ITS Classification Results of Three Models in the Huangshan Study Area
In the Huangshan study area, with the introduction of the explore module, the overall classification accuracy of the U-Net model improved from 83.01% to 85.11%, and that of the other 2 models also improved to different degrees. In the 3 models, the classification accuracy of Pinus hwangshanensis improved with the introduction of the explore module; for example, the producer’s accuracy of Pinus hwangshanensis improved from 90.77% to 95.26% in the U-Net model. This may be because traditional image features such as the texture of pine trees are more easily recognized compared to other tree species.
4.5.2. ITS Classification Results of the Three Models in the Gaofeng Study Area
In the Gaofeng area, the overall classification accuracy of the three models increased to a significant level after the introduction of the explore module. For example, the classification accuracy of AlexNet increased from 87.19% to 90.37%, the classification accuracy of U-Net increased from 54.27% to 64.15%, and the classification accuracy of LeNet increased from 80.74% to 83.50%. Similar to the classification results after introducing the explore module when using limited sample sets in the Gaofeng area, the classification accuracy of Pinus massoniana Lamb in all three models increased after introducing the explore module. For example, the producer’s accuracy of Pinus massoniana Lamb in AlexNet increased from 86.01% to 89.29%, and the user’s accuracy increased from 88.65% to 92.59%; in U-Net, the producer precision of Pinus massoniana Lamb improved from 54.46% to 61.90%, and the user precision improved from 55.45% to 68.42%; and the user precision of Pinus massoniana Lamb improved from 80.00% to 85.76% in LeNet. With the exception of Pinus massoniana Lamb, the classification accuracy of other tree species also improved to different degrees.
By comparing the OA of the three models in the two areas, it was found that the classification accuracy of the models all improved after the introduction of the explore module. The degree of improvement of the three models was higher in the Gaofeng region than in the Huangshan region, and this finding was similar to the ITS classification results using the DenseNet model in both study areas. Combining the ITS classification results of the DenseNet model using different limited sample sets, it was found that the introduction of the explore module helps to classify ITS. Moreover, when sufficient traditional image classification features were provided, the improvement after introducing the explore module based on the UAV aerial image sample set was greater than that based on the multispectral image, probably due to the higher spatial resolution and decreased spectral information of UAV aerial imagery, which enabled the traditional image features to play a greater role. For UAV aerial images with fewer spectral bands, the introduction of the explore module can still lead to a greater identification performance improvement, proving that the introduction of the explore module can be valuable in areas with less information and can effectively improve the identification accuracy without adding additional data. However, if too few traditional classification features are provided, the improvement in model accuracy is more pronounced for imagery that can provide more traditional classification features.
5. Discussion
5.1. The Influencing Factors of Tree Species Classification
In the Huangshan area, because of the many tree species included in other evergreen arbors, there is low classification accuracy, which is improved with the introduction of the explore module to ensure the user’s reliability when using the classification results. The accuracy of Cunninghamia lanceolata is overly low, which may be because the ITCs of Cunninghamia lanceolata are typically narrow and cover less pixel area than the wide canopies of broad-leaved trees [66]. Thus, this may lead to the overestimation of broadleaf species and underestimation of Cunninghamia lanceolata, which has a smaller canopy [67].
The low users accuracy of Eucalyptus urophylla S.T.Blake and Illicium verum Hook. f. in the Gaofeng area is presumably related to the difficulty of accurate ITC delineation for broadleaf trees. This is because deciduous forests have more complex crown structures and unclear inter-canopy boundaries, making it more difficult to perform accurate ITS delineation, which will affect the broadleaf tree classification. In the future, we need to further improve the ITC delineation algorithm to enhance the classification accuracy of broadleaf trees.
In addition, since different tree species are distributed vertically with altitude in the Huangshan area, the tree species differ greatly in different altitude areas, so the difficulty of classification may be lower than that in Gaofeng area with relatively consistent topography.
5.2. The Weight Setting
In Section 3.2.2, it was mentioned that 500/n can be chosen as the weight to control the proportion of the explore module, that is, the traditional image features involved in decision making. In fact, this weight can be adjusted; for example, in the Huangshan area, it was noted that the weight selected at 500/n was more suitable, and the weight selected at 100/n in the Gaofeng area was more accurate in the same classification model. This is because the number of UAV image bands in the Gaofeng area is less than that in the Huangshan area with multiple spectra, so the weight of the Gaofeng area should be set smaller than that of the Huangshan area in the same batch. In testing the limited sample set, the weight selection can have more possibilities, but for easier comparison, the weight in the Huangshan area was still selected as 500/n, and the weight in the Gaofeng area was still set as 100/n. The setting of the weight is closely related to the number of bands of imagery, the number of samples, the input batch, the image resolution, and other factors that determine the amount of classification information. In future similar experiments, the weight can be chosen flexibly according to the amount of classification information in the experimentally available imagery.
5.3. Introduction of Traditional Image Classification Methods
In this study, a softmax classifier was used to classify traditional image features to better fit deep learning networks. However, there are many other traditional image classification methods, such as SVM classification [68] and KNN [69]. Different classification methods have different benefits, and combining them with deep learning networks may result in different properties that need to be explored more deeply in the future. In addition, there are various methods to combine traditional image features with deep learning; for example, traditional image features can be used as input vectors for deep learning networks for training and classification to reduce the interference of redundant information, or the identification results of traditional image features can be used to assist deep learning networks for classification. Different combinations generate different results, which is a topic that should be actively explored in the future.
6. Conclusions
In this study, we constructed individual tree species sample sets from Huangshan multispectral and Gaofeng UAV imagery, respectively. Some traditional image features were used to build the explore module to assist the deep learning network in making individual tree species identification decisions.
We found that the identification accuracy of the four deep learning models for different data sources were improved to varying degrees by adding the explore module, especially for UAV aerial samples. In addition, introducing the explore module can effectively address the deficiency of deep learning in the limited sample sets and improve ITS identification accuracy. These improvements prove that introducing the explore module allows for classifying ITS more accurately in sample-poor areas.
However, only traditional image features assisted by classical CNNs for identification decisions were researched in this study. In future experiments, the changes brought by different combinations of conventional image features and deep learning networks will be explored more deeply.
Author Contributions
C.C. designed and completed the experiment; L.J., H.L., Y.T. and F.C. provided comments on the method; L.J., H.L., Y.T. and F.C. revised the manuscript and provided feedback on the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Innovative Research Program of the International Research Center of Big Data for Sustainable Development Goals (CBAS2022IRP03); the National Natural Science Foundation of China (41972308); the Jiangxi Provincial Technology Innovation Guidance Program (National Science and Technology Award Reserve Project Cultivation Program) (20212AEI91006); and the Second Tibetan Plateau Scientific Expedition and Research (2019QZKK0806).
Data Availability Statement
Data sharing is not applicable to this article.
Acknowledgments
We thank Guang Ouyang, Haoming Wan, and Xianfei Guo, who graduated from this research group, for their contributions to this research.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dixon, R.K.; Solomon, A.M.; Brown, S.; Houghton, R.A.; Trexier, M.C.; Wisniewski, J. Carbon Pools and Flux of Global Forest Ecosystems. Science 1994, 263, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Anitha, K.; Joseph, S.; Chandran, R.J.; Ramasamy, E.V.; Prasad, S.N. Tree species diversity and community com-position in a human-dominated tropical forest of Western Ghats biodiversity hotspot. India. Ecol. Complex. 2010, 7, 217–224. [Google Scholar] [CrossRef]
- Yin, D.M.; Wang, L. How to assess the accuracy of the individual tree-based forest inventory derived from remotely sensed data: A review. Int. J. Remote Sens. 2016, 37, 4521–4553. [Google Scholar] [CrossRef]
- Madonsela, S.; Cho, M.A.; Mathieu, R.; Mutanga, O.; Ramoelo, A.; Kaszta, Z.; Van de Kerchove, R.; Wolff, E. Multi-phenology WorldView-2 imagery improves remote sensing of savannah tree species. Int. J. Appl. Earth Obs. Geoinf. 2017, 58, 65–73. [Google Scholar] [CrossRef]
- Nijland, W.; Coops, N.C.; Macdonald, S.E.; Nielsen, S.E.; Bater, C.W.; White, B.; Ogilvie, J.; Stadt, J. Remote sensing proxies of productivity and moisture predict forest stand type and recovery rate following experimental harvest. For. Ecol. Manag. 2015, 357, 239–247. [Google Scholar] [CrossRef]
- Fedrigo, M.; Newnham, G.J.; Coops, N.C.; Culvenor, D.S.; Bolton, D.K.; Nitschke, C.R. Predicting temperate for-est stand types using only structural profiles from discrete return airborne lidar. ISPRS J. Photogramm. Remote Sens. 2018, 136, 106–119. [Google Scholar] [CrossRef]
- Grabska, E.; Hostert, P.; Pflugmacher, D.; Ostapowicz, K. Forest Stand Species Mapping Using the Sentinel-2 Time Series. Remote Sens. 2019, 11, 1197. [Google Scholar] [CrossRef]
- Oreti, L.; Giuliarelli, D.; Tomao, A.; Barbati, A. Object Oriented Classification for Mapping Mixed and Pure Forest Stands Using Very-High Resolution Imagery. Remote Sens. 2021, 13, 2508. [Google Scholar] [CrossRef]
- Wan, H.M.; Tang, Y.W.; Jing, L.H.; Li, H.; Qiu, F.; Wu, W.J. Tree species classification of forest stands using multi-source remote sensing data. Remote Sens. 2021, 13, 144. [Google Scholar] [CrossRef]
- Holmgren, J.; Persson, Å. Identifying species of individual trees using airborne laser scanner. Remote Sens. Environ. 2004, 90, 415–423. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Tree species classification in the Southern Alps based on the fusion of very high geometrical resolution multispectral/hyperspectral images and LiDAR data. Remote Sens. Environ. 2012, 123, 258–270. [Google Scholar] [CrossRef]
- Immitzer, M.; Atzberger, C.; Koukal, T. Tree Species Classification with Random Forest Using Very High Spatial Resolution 8-Band WorldView-2 Satellite Data. Remote Sens. 2012, 4, 2661–2693. [Google Scholar] [CrossRef]
- Gougeon, F.A. A crown-following approach to the automatic delineation of individual tree crowns in high spatial resolution aerial images. Can. J. Remote. Sens. 1995, 21, 274–284. [Google Scholar] [CrossRef]
- Culvenor, D.S. TIDA: An algorithm for the delineation of tree crowns in high spatial resolution remotely sensed imagery. Comput. Geosci. 2002, 28, 33–44. [Google Scholar] [CrossRef]
- Jing, L.H.; Hu, B.X.; Li, J.L.; Noland, T. Automated delineation of individual tree crowns from Lidar data by multi-scale analysis and segmentation. Photogramm. Eng. Remote Sen. 2012, 78, 1275–1284. [Google Scholar] [CrossRef]
- Hamraz, H.; Contreras, M.A.; Zhang, J. Vertical stratification of forest canopy for segmentation of understory trees within small-footprint airborne LiDAR point clouds. ISPRS J. Photogramm. Remote. Sens. 2017, 130, 385–392. [Google Scholar] [CrossRef]
- Qiu, L.; Jing, L.H.; Hu, B.X.; Li, H.; Tang, Y.W. A new individual tree crown delineation method for high resolution multispectral imagery. Remote Sens. 2020, 12, 585. [Google Scholar] [CrossRef]
- Cho, M.A.; Malahlela, O.; Ramoelo, A. Assessing the utility WorldView-2 imagery for tree species mapping in South African subtropical humid forest and the conservation implications: Dukuduku forest patch as case study. Int. J. Appl. Earth Obs. Geoinf. 2015, 38, 349–357. [Google Scholar] [CrossRef]
- Lee, J.; Cai, X.H.; Lellmann, J.; Dalponte, M.; Malhi, Y.; Butt, N.; Morecroft, M.; Schonlieb, C.B.; Coomes, D.A. Individual tree species classification from airborne multisensor imagery using robust PCA. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 9, 2554–2567. [Google Scholar] [CrossRef]
- Sedliak, M.; Sačkov, I.; Kulla, L. Classification of tree species composition using a combination of multispectral imagery and airborne laser scanning data. For. J. 2017, 63, 1–9. [Google Scholar] [CrossRef]
- Chenari, A.; Erfanifard, Y.; Dehghani, M.; Pourghasemi, H.R. Woodland mapping at single-tree levels using object-oriented classification of unmanned aerial vehicle (UAV) images. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2017, 42–44, 43–49. [Google Scholar] [CrossRef]
- Lu, D.; Weng, Q. A survey of image classification methods and techniques for improving classification performance. Int. J. Remote Sens. 2007, 28, 823–870. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Zhou, C.; Yin, L.; Feng, X. Urban forest monitoring based on multiple features at the single tree scale by UAV. Urban. For. Urban. Green. 2021, 58, 126958. [Google Scholar] [CrossRef]
- Maschler, J.; Atzberger, C.; Immitzer, M. Individual Tree Crown Segmentation and Classification of 13 Tree Species Using Airborne Hyperspectral Data. Remote Sens. 2018, 10, 1218. [Google Scholar] [CrossRef]
- Mishra, N.B.; Mainali, K.P.; Shrestha, B.B.; Radenz, J.; Karki, D. Species-Level Vegetation Mapping in a Himalayan Treeline Ecotone Using Unmanned Aerial System (UAS) Imagery. ISPRS Int. J. Geo-Inf. 2018, 7, 445. [Google Scholar] [CrossRef]
- Sothe, C.; De Almeida, C.M.; Schimalski, M.B.; La Rosa, L.E.C.; Castro, J.D.B.; Feitosa, R.Q.; Dalponte, M.; Lima, C.L.; Liesenberg, V.; Miyoshi, G.T.; et al. Comparative performance of convolutional neural network, weighted and conventional support vector machine and random forest for classifying tree species using hyperspectral and photogrammetric data. GIScience Remote Sens. 2020, 57, 369–394. [Google Scholar] [CrossRef]
- Xu, Z.; Shen, X.; Cao, L.; Coops, N.C.; Goodbody, T.R.H.; Zhong, T.; Zhao, W.D.; Sun, Q.L.; Ba, S.; Zhang, Z.N.; et al. Tree species classification using UAS-based digital aerial photogrammetry point clouds and multispectral imageries in subtropical natural forests. Int. J. Appl. Earth Obs. Geoinf. 2020, 92, 102173. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- He, K.M.; Zhang, X.Y.; Ren, S.Q.; Sun, J. Deep residual learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Fujimoto, A.; Haga, C.; Matsui, T.; Machimura, T.; Hayashi, K.; Sugita, S.; Takagi, H. An end to end process development for UAV-SfM based forest monitoring: Individual tree detection, species classification and carbon dynamics simulation. Forests 2019, 10, 680. [Google Scholar] [CrossRef]
- Egli, S.; Höpke, M. CNN-Based Tree Species Classification Using High Resolution RGB Image Data from Automated UAV Observations. Remote Sens. 2020, 12, 3892. [Google Scholar] [CrossRef]
- Cao, K.; Zhang, X. An Improved Res-UNet Model for Tree Species Classification Using Airborne High-Resolution Images. Remote Sens. 2020, 12, 1128. [Google Scholar] [CrossRef]
- Osco, L.P.; Arruda, M.D.S.D.; Gonçalves, D.N.; Dias, A.; Batistoti, J.; de Souza, M.; Gomes, F.D.G.; Ramos, A.P.M.; Jorge, L.A.D.C.; Liesenberg, V.; et al. A CNN approach to simultaneously count plants and detect plantation-rows from UAV imagery. ISPRS J. Photogramm. Remote Sens. 2021, 174, 1–17. [Google Scholar] [CrossRef]
- Miraki, M.; Sohrabi, H.; Fatehi, P.; Kneubuehler, M. Individual tree crown delineation from high-resolution UAV images in broadleaf forest. Ecol. Inform. 2021, 61, 101207. [Google Scholar] [CrossRef]
- Nezami, S.; Khoramshahi, E.; Nevalainen, O.; Polonen, I.; Honkavaara, E. Tree species classification of drone hyperspectral and RGB imagery with deep learning convolutional neural networks. Remote Sens. 2020, 12, 1070. [Google Scholar] [CrossRef]
- Yan, S.; Jing, L.; Wang, H. A New Individual Tree Species Recognition Method Based on a Convolutional Neural Network and High-Spatial Resolution Remote Sensing Imagery. Remote Sens. 2021, 13, 479. [Google Scholar] [CrossRef]
- Ouyang, G.; Jing, L.H.; Yan, S.J.; Li, H.; Tang, Y.W.; Tan, B.X. Classification of individual tree species in high-resolution remote sensing imagery based on convolution neural network. Laser Optoelectron. Prog. 2021, 58, 349–362. [Google Scholar]
- Zhang, C.; Zhou, J.W.; Wang, H.W.; Tan, T.Y.; Cui, M.C.; Huang, Z.L.; Wang, P.; Zhang, L. Multi-species individual tree segmentation and identification based on improved mask R-CNN and UAV imagery in mixed forests. Remote Sens. 2022, 14, 874. [Google Scholar] [CrossRef]
- Mallinis, G.; Koutsias, N.; Tsakiri-Strati, M.; Karteris, M. Object-based classification using Quickbird imagery for delineating forest vegetation polygons in a Mediterranean test site. ISPRS J. Photogramm. Remote Sens. 2008, 63, 237–250. [Google Scholar] [CrossRef]
- Zhang, C.Y.; Qiu, F. Mapping Individual tree species in an urban forest using airborne Lidar data and hyperspectral imagery. Photogramm. Eng. Remote Sens. 2012, 78, 1079–1087. [Google Scholar] [CrossRef]
- Pant, P.; Heikkinen, V.; Hovi, A.; Korpela, I.; Hauta-Kasari, M.; Tokola, T. Evaluation of simulated bands in air-borne optical sensors for tree species identification. Remote Sens. Environ. 2013, 138, 27–37. [Google Scholar] [CrossRef]
- Naidoo, L.; Cho, M.A.; Mathieu, R.; Asner, G. Classification of savanna tree species, in the Greater Kruger National Park region, by integrating hyperspectral and LiDAR data in a Random Forest data mining environment. ISPRS J. Photogramm. Remote Sens. 2012, 69, 167–179. [Google Scholar] [CrossRef]
- Dalponte, M.; Orka, H.O.; Ene, L.T.; Gobakken, T.; Naesset, E. Tree crown delineation and tree species classification in boreal forests using hyperspectral and ALS data. Remote Sens. Environ. 2014, 140, 306–317. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Jin, W.; Dong, S.; Dong, C.; Ye, X. Hybrid ensemble model for differential diagnosis between COVID-19 and common viral pneumonia by chest X-ray radiograph. Comput. Biol. Med. 2021, 131, 104252. [Google Scholar] [CrossRef]
- Saini, M.; Susan, S. Bag-of-Visual-Words codebook generation using deep features for effective classification of imbalanced multi-class image datasets. Multimed. Tools Appl. 2021, 80, 20821–20847. [Google Scholar] [CrossRef]
- Bakour, K.; Ünver, H.M. DeepVisDroid: Android malware detection by hybridizing image-based features with deep learning techniques. Neural Comput. Appl. 2021, 33, 11499–11516. [Google Scholar] [CrossRef]
- Dey, N.; Zhang, Y.-D.; Rajinikanth, V.; Pugalenthi, R.; Raja, N.S.M. Customized VGG19 Architecture for Pneumonia Detection in Chest X-Rays. Pattern Recognit. Lett. 2021, 143, 67–74. [Google Scholar] [CrossRef]
- Varin, M.; Chalghaf, B.; Joanisse, G. Object-Based Approach Using Very High Spatial Resolution 16-Band WorldView-3 and LiDAR Data for Tree Species Classification in a Broadleaf Forest in Quebec, Canada. Remote Sens. 2020, 12, 3092. [Google Scholar] [CrossRef]
- Jing, L.; Cheng, Q. Two improvement schemes of PAN modulation fusion methods for spectral distortion minimization. Int. J. Remote Sens. 2009, 30, 2119–2131. [Google Scholar] [CrossRef]
- Jing, L.; Hu, B.; Li, J.; Noland, T.; Guo, H. Automated tree crown delineation from imagery based on morphological techniques. In Proceedings of the 35th International Symposium on Remote Sensing of Environment (ISRSE35), Beijing, China, 22–26 April 2013. [Google Scholar]
- Shorten, C.; Khoshgoftaar., T. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Zhou, D.-X. Theory of deep convolutional neural networks: Downsampling. Neural Netw. 2020, 124, 319–327. [Google Scholar] [CrossRef]
- Al-Azzawi, A.; Ouadou, A.; Max, H.; Duan, Y.; Tanner, J.J.; Cheng, J. DeepCryoPicker: Fully automated deep neural network for single protein particle picking in cryo-EM. BMC Bioinform. 2020, 21, 509. [Google Scholar] [CrossRef]
- Wang, T.; Lu, C.; Yang, M.; Hong, F.; Liu, C. A hybrid method for heartbeat classification via convolutional neural networks, multilayer perceptrons and focal loss. PeerJ Comput. Sci. 2020, 6, 324. [Google Scholar] [CrossRef]
- Li, G.; Zhang, M.; Li, J.; Lv, F.; Tong, G. Efficient densely connected convolutional neural networks. Pattern Recognit. 2020, 109, 107610. [Google Scholar] [CrossRef]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, A.; Al-Amidie, M.; Farhan, L. Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions. J. Big Data 2021, 8, 53. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I.H. Textural Features for Image Classification. IEEE Trans. Syst. Man. Cybern. 1973, 6, 610–621. [Google Scholar] [CrossRef]
- Haralick, R.M. Statistical and structural approaches to texture. Proc. IEEE 1979, 67, 786–804. [Google Scholar] [CrossRef]
- Fung, T.; Ellsworth, L. For Change Detection Using Various Accuracy. Photogramm. Eng. Remote Sens. 1988, 54, 1449–1454. [Google Scholar]
- Shi, W.; Zhao, X.; Zhao, J.; Zhao, S.; Guo, Y.; Liu, N.; Sun, N.; Du, X.; Sun, M. Reliability and consistency assessment of land cover products atmacro and local scales in typical cities. Int. J. Digit. Earth 2023, 16, 486–508. [Google Scholar] [CrossRef]
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar] [CrossRef]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Caudullo, G.; Tinner, W.; De Rigo, D. Picea abies in Europe: Distribution, habitat, usage and threats. Eur. Atlas For. Tree Species 2016, 1, 114–116. [Google Scholar]
- Modzelewska, A.; Fassnacht, F.E.; Stereńczak, K. Tree species identification within an extensive forest area with diverse management regimes using airborne hyperspectral data. Int. J. Appl. Earth Obs. Geoinf. 2020, 84, 101960. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Franco-Lopez, H.; Ek, A.R.; Bauer, M.E. Estimation and mapping of forest stand density, volume, and cover type using the k-nearest neighbors method. Remote Sens. Environ. 2001, 77, 251–274. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).