A Framework for High-Resolution Mapping of Soil Organic Matter (SOM) by the Integration of Fourier Mid-Infrared Attenuation Total Reflectance Spectroscopy (FTIR-ATR), Sentinel-2 Images, and DEM Derivatives
Abstract
1. Introduction
2. Materials and Methods
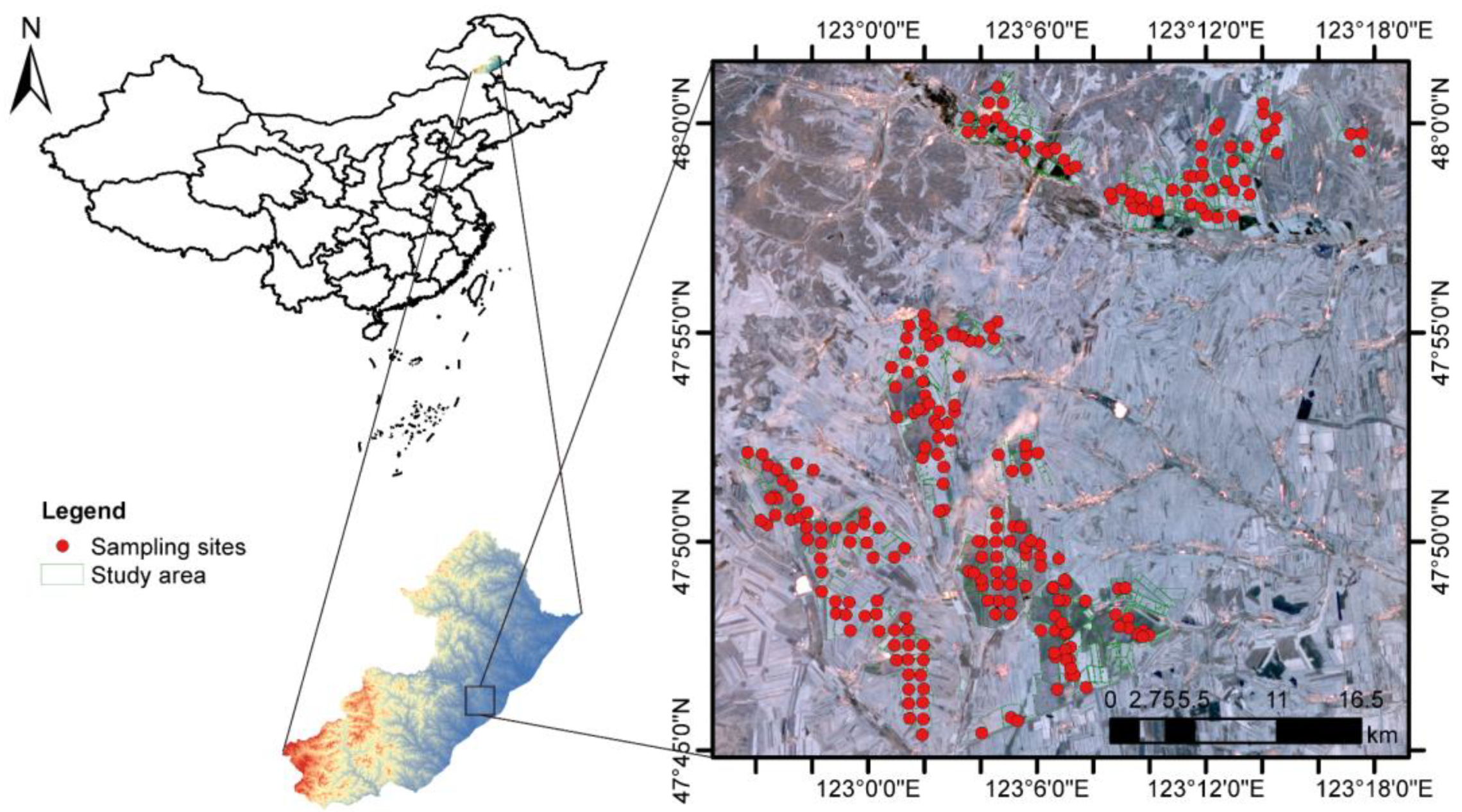
2.1. Study Area
2.2. Soil Sampling and Chemical Analysis
2.3. FTIR-ATR Spectra Acquisition and Preprocessing
2.4. Preprocessing of Sentinel-2 Images and DEM
2.5. Framework
2.6. Machine Learning Models
2.6.1. Lasso Regression
2.6.2. Partial Least Squares
2.6.3. Support Vector Regression
2.6.4. Convolutional Neural Networks
2.7. Model Evaluation
3. Results
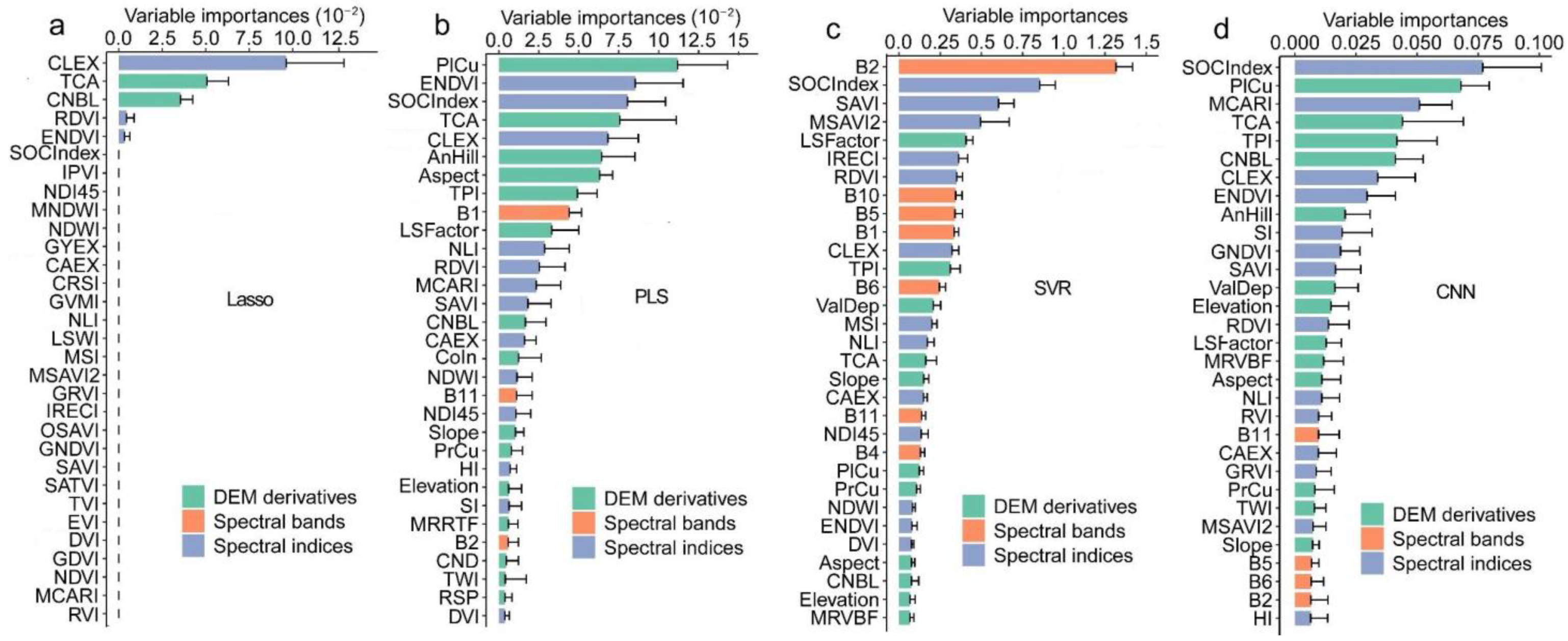
3.1. Correlation between SOM and Predictors
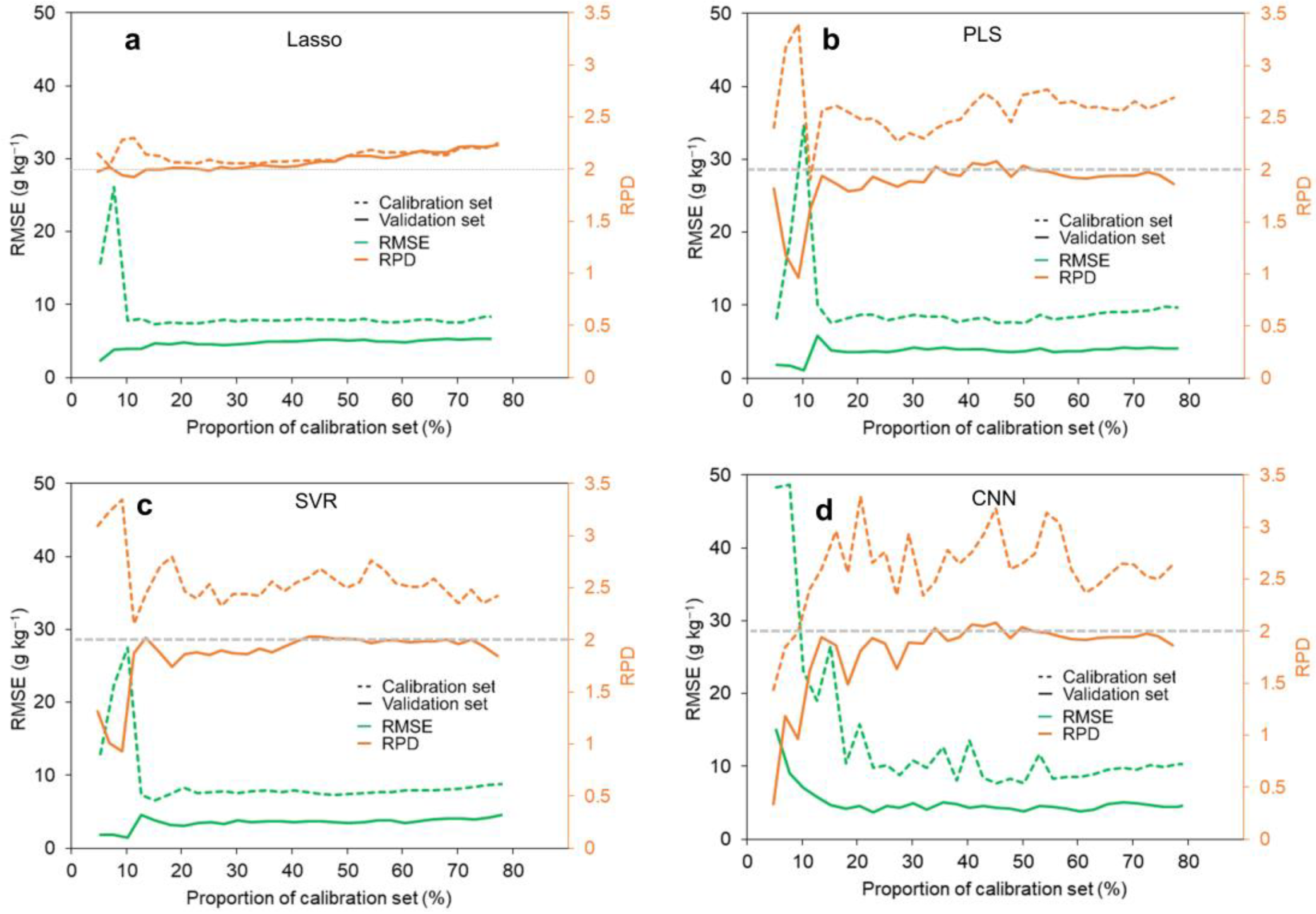
3.2. Optimal Calibration Size
3.3. SOM Prediction Using FTIR-ATR Spectra
3.4. SOM Prediction Incorporating Satellite and DEM Data
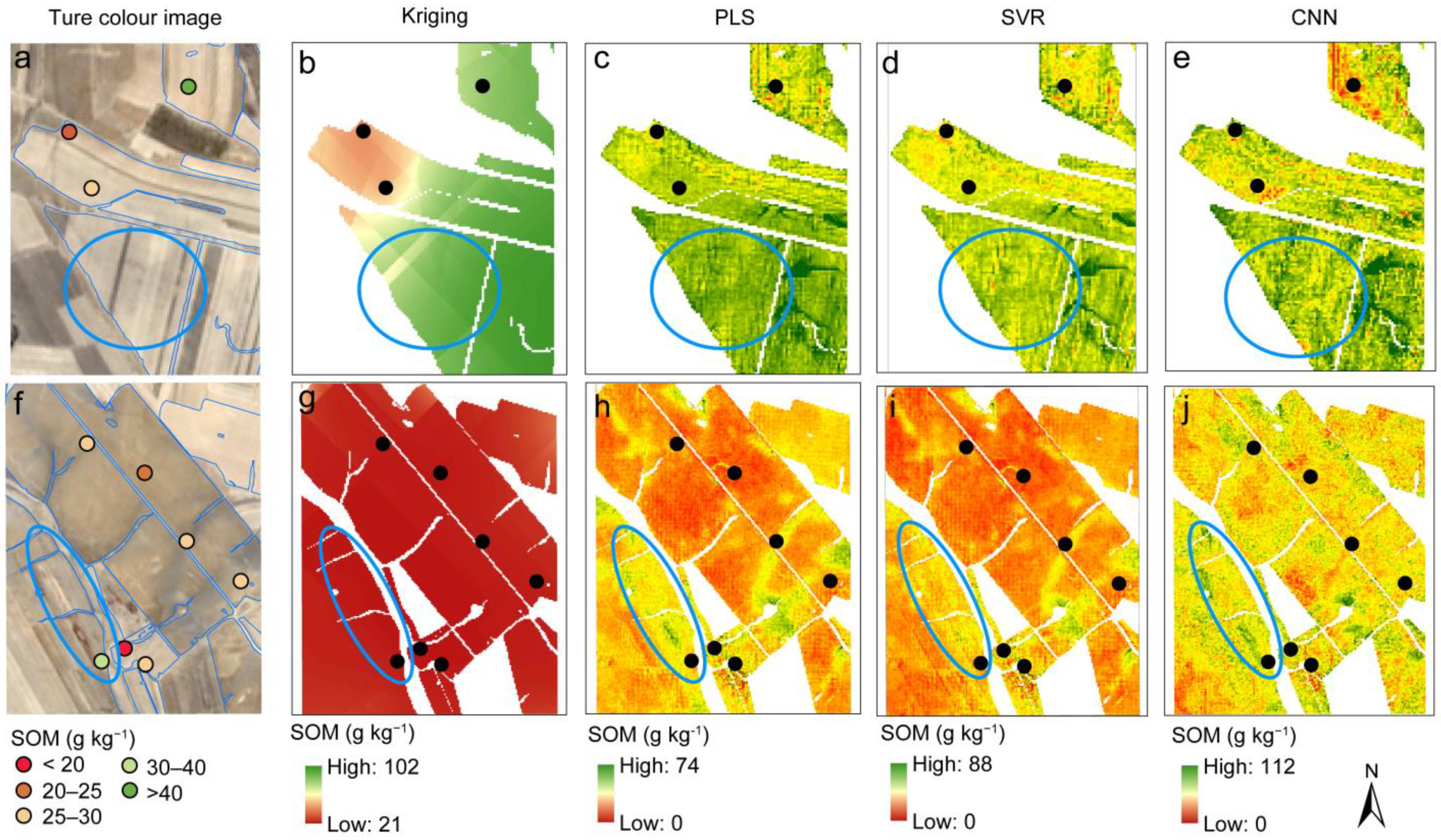
3.5. Digital Mapping of SOM
4. Discussion
4.1. Optimization of Calibration Size Promoted Analysis Efficiency
4.2. Integration of Satellite and DEM Data Enhanced Prediction Performance
4.3. Machine Learning Model Improved Mapping Accuracy and Resolution
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Guo, L.; Zhang, H.; Shi, T.; Chen, Y.; Jiang, Q.; Linderman, M. Prediction of soil organic carbon stock by laboratory spectral data and airborne hyperspectral images. Geoderma 2019, 337, 32–41. [Google Scholar] [CrossRef]
- Gomez, C.; Viscarra Rossel, R.A.; McBratney, A.B. Soil organic carbon prediction by hyperspectral remote sensing and field vis-NIR spectroscopy: An Australian case study. Geoderma 2008, 146, 403–411. [Google Scholar] [CrossRef]
- Luo, C.; Zhang, X.; Meng, X.; Zhu, H.; Ni, C.; Chen, M.; Liu, H. Regional mapping of soil organic matter content using multitemporal synthetic Landsat 8 images in Google Earth Engine. Catena 2022, 209, 105842. [Google Scholar] [CrossRef]
- Lal, R. Societal value of soil carbon. J. Soil Water Conserv. 2014, 69, 186A–192A. [Google Scholar] [CrossRef]
- Lal, R. Soil health and carbon management. Food Energy Secur. 2016, 5, 212–222. [Google Scholar] [CrossRef]
- Wang, S.; Zhao, Y.; Wang, J.; Gao, J.; Zhu, P.; Cui, X.A.; Xu, M.; Zhou, B.; Lu, C. Estimation of soil organic carbon losses and counter approaches from organic materials in black soils of northeastern China. J. Soil. Sediment. 2020, 20, 1241–1252. [Google Scholar] [CrossRef]
- Xu, X.; Du, C.; Ma, F.; Shen, Y.; Wu, K.; Liang, D.; Zhou, J. Detection of soil organic matter from laser-induced breakdown spectroscopy (LIBS) and mid-infrared spectroscopy (FTIR-ATR) coupled with multivariate techniques. Geoderma 2019, 355, 113905. [Google Scholar] [CrossRef]
- Peltre, C.; Lucas, S.; Du, C.; Thomsen, I.K.; Jensen, L.S. Assessing soil constituents and labile soil organic carbon by mid-infrared photoacoustic spectroscopy. Soil Biol. Biochem. 2014, 77, 41–50. [Google Scholar] [CrossRef]
- Huang, J.; Rinnan, Å.; Bruun, T.B.; Engedal, T.; Bruun, S. Identifying the fingerprint of permanganate oxidizable carbon as a measure of labile soil organic carbon using Fourier transform mid-infrared photoacoustic spectroscopy. Eur. J. Soil Sci. 2021, 72, 1831–1841. [Google Scholar] [CrossRef]
- McBratney, A.B.; Mendonça Santos, M.L.; Minasny, B. On digital soil mapping. Geoderma 2003, 117, 3–52. [Google Scholar] [CrossRef]
- Wadoux, A.M.J.C.; McBratney, A.B. Hypotheses, machine learning and soil mapping. Geoderma 2021, 383, 114725. [Google Scholar] [CrossRef]
- Goydaragh, S.; Kumar, L.; Wilson, B. Digital soil mapping algorithms and covariates for soil organic carbon mapping and their implications: A review. Geoderma 2019, 352, 395–413. [Google Scholar] [CrossRef]
- Wang, N.; Peng, J.; Xue, J.; Zhang, X.; Huang, J.; Biswas, A.; He, Y.; Shi, Z. A framework for determining the total salt content of soil profiles using time-series Sentinel-2 images and a random forest-temporal convolution network. Geoderma 2022, 409, 115656. [Google Scholar] [CrossRef]
- Drusch, M.; Del Bello, U.; Carlier, S.; Colin, O.; Fernandez, V.; Gascon, F.; Hoersch, B.; Isola, C.; Laberinti, P.; Martimort, P.; et al. Sentinel-2: ESA’s Optical High-Resolution Mission for GMES Operational Services. Remote Sens. Environ. 2012, 120, 25–36. [Google Scholar] [CrossRef]
- Gholizadeh, A.; Žižala, D.; Saberioon, M.; Borůvka, L. Soil organic carbon and texture retrieving and mapping using proximal, airborne and Sentinel-2 spectral imaging. Remote Sens. Environ. 2018, 218, 89–103. [Google Scholar] [CrossRef]
- Zhou, T.; Geng, Y.; Ji, C.; Xu, X.; Wang, H.; Pan, J.; Bumberger, J.; Haase, D.; Lausch, A. Prediction of soil organic carbon and the C:N ratio on a national scale using machine learning and satellite data: A comparison between Sentinel-2, Sentinel-3 and Landsat-8 images. Sci. Total Environ. 2021, 755, 142661. [Google Scholar] [CrossRef]
- Gardin, L.; Chiesi, M.; Fibbi, L.; Maselli, F. Mapping soil organic carbon in Tuscany through the statistical combination of ground observations with ancillary and remote sensing data. Geoderma 2021, 404, 115386. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Pham, T.D.; Nguyen, C.T.; Delfos, J.; Archibald, R.; Dang, K.B.; Hoang, N.B.; Guo, W.; Ngo, H.H. A novel intelligence approach based active and ensemble learning for agricultural soil organic carbon prediction using multispectral and SAR data fusion. Sci. Total Environ. 2022, 804, 150187. [Google Scholar] [CrossRef]
- Wang, B.; Waters, C.; Orgill, S.; Gray, J.; Cowie, A.; Clark, A.; Liu, D.L. High resolution mapping of soil organic carbon stocks using remote sensing variables in the semi-arid rangelands of eastern Australia. Sci. Total Environ. 2018, 630, 367–378. [Google Scholar] [CrossRef]
- Minhoni, R.T.d.A.; Scudiero, E.; Zaccaria, D.; Saad, J.C.C. Multitemporal satellite imagery analysis for soil organic carbon assessment in an agricultural farm in southeastern Brazil. Sci. Total Environ. 2021, 784, 147216. [Google Scholar] [CrossRef]
- Zhou, T.; Geng, Y.; Chen, J.; Pan, J.; Haase, D.; Lausch, A. High-resolution digital mapping of soil organic carbon and soil total nitrogen using DEM derivatives, Sentinel-1 and Sentinel-2 data based on machine learning algorithms. Sci. Total Environ. 2020, 729, 138244. [Google Scholar] [CrossRef]
- Rial, M.; Cortizas, A.M.; Rodríguez-Lado, L. Mapping soil organic carbon content using spectroscopic and environmental data: A case study in acidic soils from NW Spain. Sci. Total Environ. 2016, 539, 26–35. [Google Scholar] [CrossRef]
- Goydaragh, M.G.; Taghizadeh-Mehrjardi, R.; Jafarzadeh, A.A.; Triantafilis, J.; Lado, M. Using environmental variables and Fourier Transform Infrared Spectroscopy to predict soil organic carbon. Catena 2021, 202, 105280. [Google Scholar] [CrossRef]
- Moura-Bueno, J.M.; Dalmolin, R.S.D.; Horst-Heinen, T.Z.; Grunwald, S.; ten Caten, A. Environmental covariates improve the spectral predictions of organic carbon in subtropical soils in southern Brazil. Geoderma 2021, 393, 114981. [Google Scholar] [CrossRef]
- IUSS Working Group WRB. World Reference Base for Soil Resources; IUSS Working Group WRB: Rome, Italy, 2014. [Google Scholar]
- Walkley, A.; Black, I.A. An examination of the Degtjareff method for determining soil organic matter, and a proposed odification of the chromic acid titration method. Soil Sci. 1934, 37, 29–38. [Google Scholar] [CrossRef]
- Van Bemmelen, J.M. Über die Bestimmung des Wassers, des Humus, des Schwefels, der in den colloïdalen Silikaten gebundenen Kieselsäure, des Mangans u. s. w. im Ackerboden. Landwirthschaftlichen Vers. Station. 1890, 37, 279–290. [Google Scholar]
- Shoko, C.; Mutanga, O. Examining the strength of the newly-launched Sentinel 2 MSI sensor in detecting and discriminating subtle differences between C3 and C4 grass species. ISPRS J. Photogramm. 2017, 129, 32–40. [Google Scholar] [CrossRef]
- European Space Agency. Sentinel-2 MSI User Guide. Available online: https://sentinel.esa.int/web/sentinel/user-guides/sentinel-2-msi (accessed on 4 January 2022).
- Kennard, R.W.; Stone, L.A. Computer aided design of experiments. Technometrics 1969, 11, 137–148. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar] [CrossRef]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.s.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on heterogeneous Distributed Systems. Available online: https://www.tensorflow.org/ (accessed on 12 January 2022).
- Tibshirani, R. Regression shrinkage and selection via the Lasso. J. R. Stat. Soc. B. 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Shetty, N.; Gislum, R. Quantification of fructan concentration in grasses using NIR spectroscopy and PLSR. Field Crop. Res. 2011, 120, 31–37. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Ng, W.; Minasny, B.; Montazerolghaem, M.; Padarian, J.; Ferguson, R.; Bailey, S.; McBratney, A.B. Convolutional neural network for simultaneous prediction of several soil properties using visible/near-infrared, mid-infrared, and their combined spectra. Geoderma 2019, 352, 251–267. [Google Scholar] [CrossRef]
- Churchman, G.J.; Foster, R.C.; D’Acqui, L.P.; Janik, L.J.; Skjemstad, J.O.; Merry, R.H.; Weissmann, D.A. Effect of land-use history on the potential for carbon sequestration in an Alfisol. Soil Tillage Res. 2010, 109, 23–35. [Google Scholar] [CrossRef]
- Xing, Z.; Tian, K.; Du, C.; Li, C.; Zhou, J.; Chen, Z. Agricultural soil characterization by FTIR spectroscopy at micrometer scales: Depth profiling by photoacoustic spectroscopy. Geoderma 2019, 335, 94–103. [Google Scholar] [CrossRef]
- Viscarra Rossel, R.A.; Behrens, T. Using data mining to model and interpret soil diffuse reflectance spectra. Geoderma 2010, 158, 46–54. [Google Scholar] [CrossRef]
- Xu, X.; Du, C.; Ma, F.; Shen, Y.; Zhou, J. Forensic soil analysis using laser-induced breakdown spectroscopy (LIBS) and Fourier transform infrared total attenuated reflectance spectroscopy (FTIR-ATR): Principles and case studies. Forensic Sci. Int. 2020, 310, 110222. [Google Scholar] [CrossRef]
- Pedersen, J.A.; Simpson, M.A.; Bockheim, J.G.; Kumar, K. Characterization of soil organic carbon in drained thaw-lake basins of Arctic Alaska using NMR and FTIR photoacoustic spectroscopy. Org. Geochem. 2011, 42, 947–954. [Google Scholar] [CrossRef]
- Ellerbrock, R.H.; Gerke, H.H. Characterizing organic matter of soil aggregate coatings and biopores by Fourier transform infrared spectroscopy. Eur. J. Soil Sci. 2004, 55, 219–228. [Google Scholar] [CrossRef]
- Soriano-Disla, J.M.; Janik, L.J.; Viscarra Rossel, R.A.; Macdonald, L.M.; McLaughlin, M.J. The performance of visible, near-, and mid-infrared reflectance spectroscopy for prediction of soil physical, chemical, and biological properties. Appl. Spectrosc. Rev. 2014, 49, 139–186. [Google Scholar] [CrossRef]
- Janik, L.J.; Skjemstad, J.O.; Shepherd, K.D.; Spouncer, L.R. The prediction of soil carbon fractions using mid-infrared-partial least square analysis. Soil Res. 2007, 45, 73–81. [Google Scholar] [CrossRef]
- Du, C.; Goyne, K.W.; Miles, R.J.; Zhou, J. A 1915–2011 microscale record of soil organic matter under wheat cultivation using FTIR-PAS depth-profiling. Agron. Sustain. Dev. 2014, 34, 803–811. [Google Scholar] [CrossRef]
- Du, C.; Linker, R.; Shaviv, A. Characterization of soils using photoacoustic mid-infrared spectroscopy. Appl. Spectrosc. 2007, 61, 1063–1067. [Google Scholar] [CrossRef]
- Calderón, F.; Haddix, M.; Conant, R.; Magrini-Bair, K.; Paul, E. Diffuse-reflectance fourier-transform mid-infrared spectroscopy as a method of characterizing changes in soil organic matter. Soil Sci. Soc. Am. J. 2013, 77, 1591–1600. [Google Scholar] [CrossRef]
- Leifeld, J. Application of diffuse reflectance FT-IR spectroscopy and partial least-squares regression to predict NMR properties of soil organic matter. Eur. J. Soil Sci. 2006, 57, 846–857. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; ur Rehman, D.I. Fourier transform infrared (FTIR) dpectroscopy of biological tissues. Appl. Spectrosc. Rev. 2008, 43, 134–179. [Google Scholar] [CrossRef]
- Madejová, J. FTIR techniques in clay mineral studies. Vib. Spectrosc. 2003, 31, 1–10. [Google Scholar] [CrossRef]
- Nayak, P.S.; Singh, B.K. Instrumental characterization of clay by XRF, XRD and FTIR. Bull. Mater. Sci. 2007, 30, 235–238. [Google Scholar] [CrossRef]
- Ma, F.; Du, C.; Zhou, J.; Shen, Y. Optimized self-adaptive model for assessment of soil organic matter using Fourier transform mid-infrared photoacoustic spectroscopy. Chemometr. Intell. Lab. Syst. 2017, 171, 9–15. [Google Scholar] [CrossRef]
- Kuang, B.; Mouazen, A.M. Influence of the number of samples on prediction error of visible and near infrared spectroscopy of selected soil properties at the farm scale. Eur. J. Soil Sci. 2012, 63, 421–429. [Google Scholar] [CrossRef]
- Lucà, F.; Conforti, M.; Castrignanò, A.; Matteucci, G.; Buttafuoco, G. Effect of calibration set size on prediction at local scale of soil carbon by Vis-NIR spectroscopy. Geoderma 2017, 288, 175–183. [Google Scholar] [CrossRef]
- Ramirez-Lopez, L.; Wadoux, A.M.J.C.; Franceschini, M.H.D.; Terra, F.S.; Marques, K.P.P.; Sayão, V.M.; Demattê, J.A.M. Robust soil mapping at the farm scale with vis–NIR spectroscopy. Eur. J. Soil Sci. 2019, 70, 378–393. [Google Scholar] [CrossRef]
- Chen, S.; Xu, H.; Xu, D.; Ji, W.; Li, S.; Yang, M.; Hu, B.; Zhou, Y.; Wang, N.; Arrouays, D.; et al. Evaluating validation strategies on the performance of soil property prediction from regional to continental spectral data. Geoderma 2021, 400, 115159. [Google Scholar] [CrossRef]
- Du, C.; Zhou, J.; Wang, H.; Chen, X.; Zhu, A.; Zhang, J. Determination of soil properties using Fourier transform mid-infrared photoacoustic spectroscopy. Vib. Spectrosc. 2009, 49, 32–37. [Google Scholar] [CrossRef]
- Xing, Z.; Du, C.; Tian, K.; Ma, F.; Shen, Y.; Zhou, J. Application of FTIR-PAS and Raman spectroscopies for the determination of organic matter in farmland soils. Talanta 2016, 158, 262–269. [Google Scholar] [CrossRef]
- Ni, W.; Nørgaard, L.; Mørup, M. Non-linear calibration models for near infrared spectroscopy. Anal. Chim. Acta 2014, 813, 1–14. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Castaldi, F.; Palombo, A.; Santini, F.; Pascucci, S.; Pignatti, S.; Casa, R. Evaluation of the potential of the current and forthcoming multispectral and hyperspectral imagers to estimate soil texture and organic carbon. Remote Sens. Environ. 2016, 179, 54–65. [Google Scholar] [CrossRef]
- Bangroo, S.A.; Najar, G.R.; Achin, E.; Truong, P.N. Application of predictor variables in spatial quantification of soil organic carbon and total nitrogen using regression kriging in the North Kashmir forest Himalayas. Catena 2020, 193, 104632. [Google Scholar] [CrossRef]
- Beguin, J.; Fuglstad, G.A.; Mansuy, N.; Paré, D. Predicting soil properties in the Canadian boreal forest with limited data: Comparison of spatial and non-spatial statistical approaches. Geoderma 2017, 306, 195–205. [Google Scholar] [CrossRef]
- Castaldi, F.; Hueni, A.; Chabrillat, S.; Ward, K.; Buttafuoco, G.; Bomans, B.; Vreys, K.; Brell, M.; van Wesemael, B. Evaluating the capability of the Sentinel 2 data for soil organic carbon prediction in croplands. ISPRS J. Photogramm. 2019, 147, 267–282. [Google Scholar] [CrossRef]
- Padarian, J.; Minasny, B.; McBratney, A.B. Using deep learning to predict soil properties from regional spectral data. Geoderma Reg. 2019, 16, e00198. [Google Scholar] [CrossRef]
- Song, X.D.; Brus, D.J.; Liu, F.; Li, D.C.; Zhao, Y.G.; Yang, J.L.; Zhang, G.L. Mapping soil organic carbon content by geographically weighted regression: A case study in the Heihe River Basin, China. Geoderma 2016, 261, 11–22. [Google Scholar] [CrossRef]
- Li, X.; Gao, B.; Pan, Y.; Bai, Z.; Gao, Y.; Dong, S.; Li, S. Multi-objective optimization sampling based on Pareto optimality for soil mapping. Geoderma 2022, 425, 116069. [Google Scholar] [CrossRef]
- Wadoux, A.M.J.C.; Brus, D.J.; Heuvelink, G.B.M. Sampling design optimization for soil mapping with random forest. Geoderma 2019, 355, 113913. [Google Scholar] [CrossRef]
- Conforti, M.; Buttafuoco, G. Insights into the effects of study area size and soil sampling density in the prediction of soil organic carbon by vis-NIR diffuse reflectance spectroscopy in two forest areas. Land 2023, 12, 44. [Google Scholar] [CrossRef]
- Escadafal, R. Remote sensing of arid soil surface color with Landsat thematic mapper. Adv. Space Res. 1989, 9, 159–163. [Google Scholar] [CrossRef]
- Srisomkiew, S.; Kawahigashi, M.; Limtong, P.; Yuttum, O. Digital soil assessment of soil fertility for Thai jasmine rice in the Thung Kula Ronghai region, Thailand. Geoderma 2022, 409, 115597. [Google Scholar] [CrossRef]
- Metternicht, G.I.; Zinck, J.A. Remote sensing of soil salinity: Potentials and constraints. Remote Sens. Environ. 2003, 85, 1–20. [Google Scholar] [CrossRef]
- Yu, H.; Liu, M.; Du, B.; Wang, Z.; Hu, L.; Zhang, B. Mapping soil salinity/sodicity by using Landsat OLI imagery and PLSR algorithm over semiarid west Jilin Province, China. Sensors 2018, 18, 1048. [Google Scholar] [CrossRef]
- Wang, F.; Shi, Z.; Biswas, A.; Yang, S.; Ding, J. Multi-algorithm comparison for predicting soil salinity. Geoderma 2020, 365, 114211. [Google Scholar] [CrossRef]
- Wang, F.; Yang, S.; Wei, Y.; Shi, Q.; Ding, J. Characterizing soil salinity at multiple depth using electromagnetic induction and remote sensing data with random forests: A case study in Tarim River Basin of southern Xinjiang, China. Sci. Total Environ. 2021, 754, 142030. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Al-Shafie, W.M.; Mhaimeed, A.S.; Ziadat, F.; Nangia, V.; Payne, W.B. Soil salinity mapping by multiscale remote sensing in Mesopotamia, Iraq. IEEE J. Sel. Top. Appl. Earth Observ. Remote Sens. 2014, 7, 4442–4452. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Nellis, M.D.; Briggs, J.M. Transformed vegetation index for measuring spatial variation in drought impactedbiomass on Konza Prairie, Kansas. Trans. Kans. Acad. Sci. 1992, 95, 93–99. [Google Scholar] [CrossRef]
- Marsett, R.C.; Qi, J.; Heilman, P.; Biedenbender, S.H.; Carolyn Watson, M.; Amer, S.; Weltz, M.; Goodrich, D.; Marsett, R. Remote sensing for grassland management in the arid southwest. Rangeland Ecol. Manag. 2006, 59, 530–540. [Google Scholar] [CrossRef]
- Huete, A.R. A soil-adjusted vegetation index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Qi, J.; Kerr, Y.; Chehbouni, A. External factor consideration in vegetation index development. In Proceedings of the 6th International Symposium on Physical Measurements and Signatures in Remote Sensing, Val d’Isere, France, 17–21 January 1994. [Google Scholar]
- Rondeaux, G.; Steven, M.; Baret, F. Optimization of soil-adjusted vegetation indices. Remote Sens. Environ. 1996, 55, 95–107. [Google Scholar] [CrossRef]
- Crippen, R.E. Calculating the vegetation index faster. Remote Sens. Environ. 1990, 34, 71–73. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Goffart, D.; Curnel, Y.; Planchon, V.; Goffart, J.-P.; Defourny, P. Field-scale assessment of Belgian winter cover crops biomass based on Sentinel-2 data. Eur. J. Agron. 2021, 126, 126278. [Google Scholar] [CrossRef]
- Xiao, X.; Zhang, Q.; Braswell, B.; Urbanski, S.; Boles, S.; Wofsy, S.; Moore, B.; Ojima, D. Modeling gross primary production of temperate deciduous broadleaf forest using satellite images and climate data. Remote Sens. Environ. 2004, 91, 256–270. [Google Scholar] [CrossRef]
- Gao, B. NDWI—A normalized difference water index for remote sensing of vegetation liquid water from space. Remote Sens. Environ. 1996, 58, 257–266. [Google Scholar] [CrossRef]
- Ceccato, P.; Gobron, N.; Flasse, S.; Pinty, B.; Tarantola, S. Designing a spectral index to estimate vegetation water content from remote sensing data: Part 1: Theoretical approach. Remote Sens. Environ. 2002, 82, 188–197. [Google Scholar] [CrossRef]
- Scudiero, E.; Skaggs, T.H.; Corwin, D.L. Regional scale soil salinity evaluation using Landsat 7, western San Joaquin Valley, California, USA. Geoderma Reg. 2014, 2–3, 82–90. [Google Scholar] [CrossRef]
- Thaler, E.A.; Larsen, I.J.; Yu, Q. A new index for remote sensing of soil organic carbon based solely on visible wavelengths. Soil Sci. Soc. Am. J. 2019, 83, 1443–1450. [Google Scholar] [CrossRef]
- Taghizadeh-Mehrjardi, R.; Minasny, B.; Sarmadian, F.; Malone, B.P. Digital mapping of soil salinity in Ardakan region, central Iran. Geoderma 2014, 213, 15–28. [Google Scholar] [CrossRef]
- Xu, H. Modification of normalised difference water index (NDWI) to enhance open water features in remotely sensed imagery. Int. J. Remote Sens. 2006, 27, 3025–3033. [Google Scholar] [CrossRef]
- Delegido, J.; Verrelst, J.; Alonso, L.; Moreno, J. Evaluation of Sentinel-2 red-edge bands for empirical estimation of green LAI and chlorophyll content. Sensors 2011, 11, 7063–7081. [Google Scholar] [CrossRef]
- Frampton, W.J.; Dash, J.; Watmough, G.; Milton, E.J. Evaluating the capabilities of Sentinel-2 for quantitative estimation of biophysical variables in vegetation. ISPRS J. Photogramm. 2013, 82, 83–92. [Google Scholar] [CrossRef]
- Daughtry, C.S.T.; Walthall, C.L.; Kim, M.S.; de Colstoun, E.B.; McMurtrey, J.E. Estimating corn leaf chlorophyll concentration from leaf and canopy reflectance. Remote Sens. Environ. 2000, 74, 229–239. [Google Scholar] [CrossRef]




| Band | Central Wavelengths (nm) | Bandwidth (nm) | Spatial Resolution (m) | SNR 1 (at Lref) | Description |
|---|---|---|---|---|---|
| B1 | 443 | 20 | 60 | 129 | Coastal aerosol |
| B2 | 490 | 65 | 10 | 154 | Blue |
| B3 | 560 | 35 | 10 | 168 | Green |
| B4 | 665 | 30 | 10 | 142 | Red |
| B5 | 705 | 15 | 20 | 117 | Vegetation red edge |
| B6 | 740 | 15 | 20 | 59 | Vegetation red edge |
| B7 | 783 | 20 | 20 | 105 | Vegetation red edge |
| B8 | 842 | 115 | 10 | 172 | NIR |
| B8A | 865 | 20 | 20 | 72 | Narrow NIR |
| B10 | 1375 | 30 | 60 | 50 | SWIR-Cirrus |
| B11 | 1610 | 90 | 20 | 10 | SWIR1 |
| B12 | 2190 | 180 | 20 | 10 | SWIR2 |
| Attributes | Definition |
|---|---|
| Elevation | Elevation |
| Slope | Slope |
| Aspect | Aspect |
| AnHill | Analytical Hillshading |
| LSFactor | LS Factor |
| TCA | Total catchment area |
| TWI | Topographic wetness index |
| CoIn | Convergence index |
| RSP | Relative slope position |
| PrCu | Profile curvature |
| PlCu | Plan curvature |
| CND | Chanel network distance |
| ValDep | Valley depth |
| TPI | Topographic position index |
| CNBL | Chanel network base level |
| MRRTF | Multi-resolution of ridge top flatness index |
| MRVBF | Multi-resolution of valley bottom flatness index |
| Wavenumber (cm−1) | Vibration | Functional Group or Component |
|---|---|---|
| 620 | νO–H | Clay minerals [40] |
| 3600–3000 | νO–H, N–H | Water, alcohols, and phenols; carboxyl and hydroxyl groups, amides [41,42] |
| 3000–2800 | νC–H | Aliphatic methyl and methylene groups [42,43] |
| 2200–2000 | Overtone νCOH | Carbohydrates [44] |
| 1720–1600 | νC=O, νC=C | Carboxylic acids; amides; Aromatics [45,46] |
| 1570–1540 | νN–H, νC–N in plane | Amide II [47,48] |
| 1515 | νC=C | Aromatics [47] |
| 1500–1300 | νCO32− | Carbonates [39] |
| 1445–1350 | νC–H | Methyls [44,39] |
| 1160 | νC–O | Polysaccharides, nucleic acids, proteins [47] |
| 1050 | δC–O | Carbohydrates [39,49] |
| 990 | νSi–O | Clay minerals [50] |
| 915 | δAl–OH | Kaolinite and smectite [39] |
| 875 | C–O out of plane | Carbonates |
| 770 | NH2 out of plane | Primary amine [51] |
| Model | Spectra | Satellite and DEM Data | Fused | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RMSE | R2 | RPD | Rv2/Rc2 | RMSE | R2 | RPD | Rv2/Rc2 | RMSE | R2 | RPD | Rv2/Rc2 | |
| Lasso | 7.6 | 0.651 | 1.921 | 0.770 | 13.0 | 0.121 | 1.123 | 0.951 | 7.1 | 0.700 | 2.056 | 1.132 |
| PLS | 7.1 | 0.701 | 2.056 | 0.950 | 12.0 | 0.215 | 1.217 | 0.914 | 6.9 | 0.713 | 2.116 | 1.021 |
| SVR | 7.4 | 0.643 | 1.973 | 0.809 | 11.9 | 0.299 | 1.232 | 0.931 | 7.2 | 0.688 | 2.028 | 1.010 |
| CNN | 7.7 | 0.635 | 1.896 | 0.781 | 12.1 | 0.204 | 1.205 | 0.896 | 7.5 | 0.632 | 1.947 | 0.996 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Du, C.; Ma, F.; Qiu, Z.; Zhou, J. A Framework for High-Resolution Mapping of Soil Organic Matter (SOM) by the Integration of Fourier Mid-Infrared Attenuation Total Reflectance Spectroscopy (FTIR-ATR), Sentinel-2 Images, and DEM Derivatives. Remote Sens. 2023, 15, 1072. https://doi.org/10.3390/rs15041072
Xu X, Du C, Ma F, Qiu Z, Zhou J. A Framework for High-Resolution Mapping of Soil Organic Matter (SOM) by the Integration of Fourier Mid-Infrared Attenuation Total Reflectance Spectroscopy (FTIR-ATR), Sentinel-2 Images, and DEM Derivatives. Remote Sensing. 2023; 15(4):1072. https://doi.org/10.3390/rs15041072
Chicago/Turabian StyleXu, Xuebin, Changwen Du, Fei Ma, Zhengchao Qiu, and Jianmin Zhou. 2023. "A Framework for High-Resolution Mapping of Soil Organic Matter (SOM) by the Integration of Fourier Mid-Infrared Attenuation Total Reflectance Spectroscopy (FTIR-ATR), Sentinel-2 Images, and DEM Derivatives" Remote Sensing 15, no. 4: 1072. https://doi.org/10.3390/rs15041072
APA StyleXu, X., Du, C., Ma, F., Qiu, Z., & Zhou, J. (2023). A Framework for High-Resolution Mapping of Soil Organic Matter (SOM) by the Integration of Fourier Mid-Infrared Attenuation Total Reflectance Spectroscopy (FTIR-ATR), Sentinel-2 Images, and DEM Derivatives. Remote Sensing, 15(4), 1072. https://doi.org/10.3390/rs15041072







