Estimation of Leaf Nitrogen Content in Rice Using Vegetation Indices and Feature Variable Optimization with Information Fusion of Multiple-Sensor Images from UAV
Abstract
1. Introduction
2. Materials and Methods
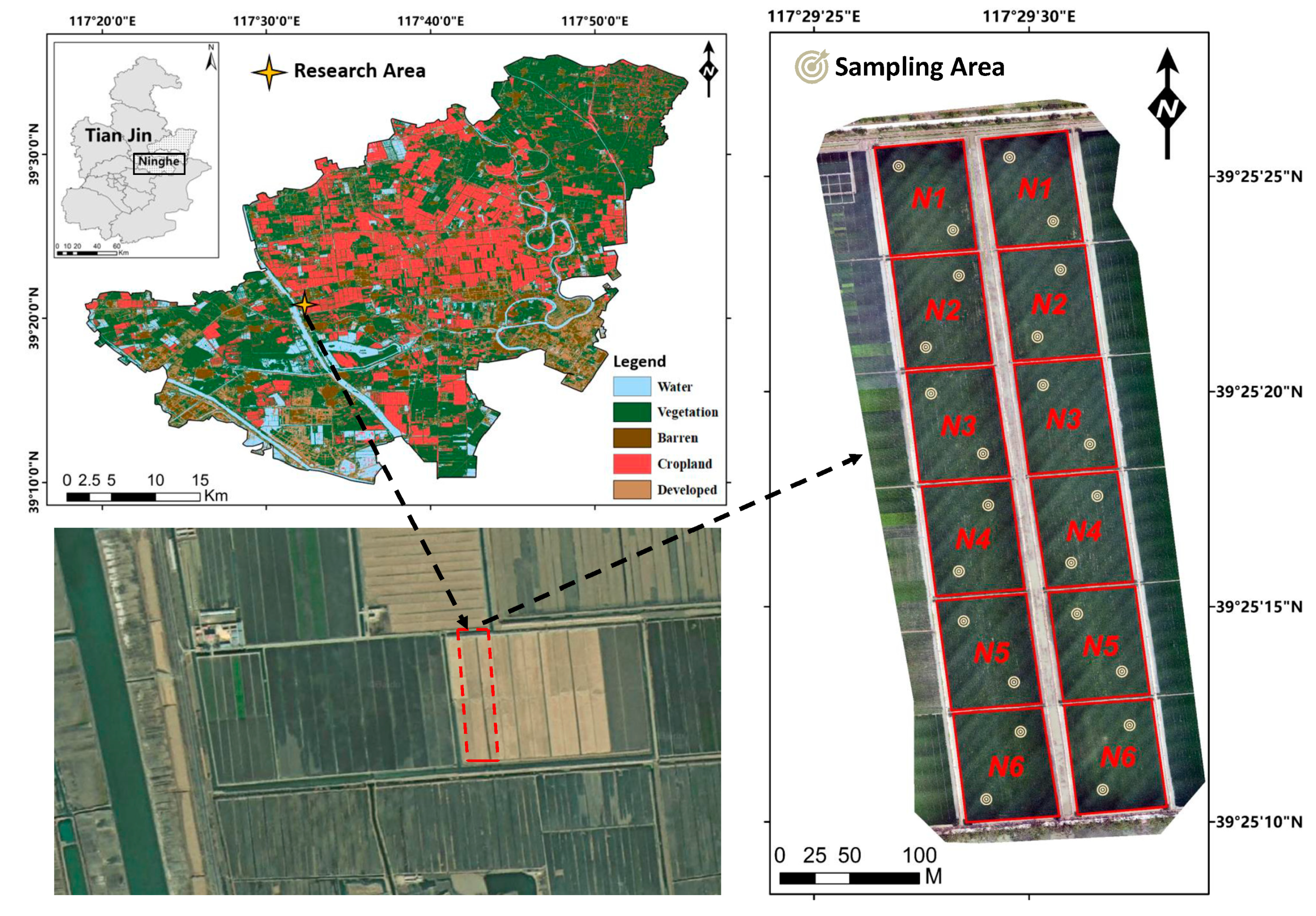
2.1. Study Area and Experimental Design
2.2. Ground Data Acquisition and LNC Determination
2.3. UAV Data Processing
2.3.1. Acquisition and Pre-Processing
2.3.2. Image Fusion
2.3.3. Removal of Background Noise
2.4. Determining Input Variables for Modeling
2.4.1. Candidate Feature Variables
2.4.2. Feature Variable Selection
- Successive Projections Algorithm (SPA)
- 2.
- Competitive Adaptative Reweighted Sampling (CARS)
2.5. Modeling Methods
2.5.1. LASSO Regression
2.5.2. RIDGE Regression
2.6. Evaluation Indicators
3. Results and Analysis
3.1. Descriptive Statistics
3.2. Correlation Analysis of Feature Variables
3.3. Extraction of Optimal Feature Variables
3.4. Modeling of LNC Using Machine Learning Algorithms
3.4.1. Results of GS Fusion
3.4.2. Results of Removing Background Noise
3.4.3. Results of the Optimal Feature Variable Prediction
3.5. Construction of the Spatial Distribution Map of LNC
4. Discussion
4.1. Nitrogen Estimation for Different Image Treatments
4.2. Nitrogen Estimation for Different Modeling Approaches
4.3. Future Research Prospects
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Inoue, Y.; Sakaiya, E.; Zhu, Y.; Takahashi, W. Diagnostic mapping of canopy nitrogen content in rice based on hyperspectral measurements. Remote Sens. Environ. 2012, 126, 210–221. [Google Scholar] [CrossRef]
- Qiu, Z.; Ma, F.; Li, Z.; Xu, X.; Ge, H.; Du, C. Estimation of nitrogen nutrition index in rice from UAV RGB images coupled with machine learning algorithms. Comput. Electron. Agric. 2021, 189, 106421. [Google Scholar] [CrossRef]
- Wu, W.; Ma, B. Integrated nutrient management (INM) for sustaining crop productivity and reducing environmental impact: A review. Sci. Total Environ. 2015, 512, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Yang, G.; Li, Z.; Song, X.; Li, Z.; Xu, X.; Wang, P.; Zhao, C. Winter Wheat Nitrogen Status Estimation Using UAV-Based RGB Imagery and Gaussian Processes Regression. Remote Sens. 2020, 12, 3778. [Google Scholar] [CrossRef]
- Shi, P.; Wang, Y.; Xu, J.; Zhao, Y.; Yang, B.; Yuan, Z.; Sun, Q. Rice nitrogen nutrition estimation with RGB images and machine learning methods. Comput. Electron. Agric. 2021, 180, 105860. [Google Scholar] [CrossRef]
- Sun, J.; Ye, M.; Peng, S.; Li, Y. Nitrogen can improve the rapid response of photosynthesis to changing irradiance in rice (Oryza sativa L.) plants. Sci. Rep. 2016, 6, srep31305. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, S.; Li, D.; Wang, C.; Jiang, H.; Zheng, Q.; Peng, Z. Estimation of Paddy Rice Nitrogen Content and Accumulation Both at Leaf and Plant Levels from UAV Hyperspectral Imagery. Remote Sens. 2021, 13, 2956. [Google Scholar] [CrossRef]
- Ge, H.; Xiang, H.; Ma, F.; Li, Z.; Qiu, Z.; Tan, Z.; Du, C. Estimating Plant Nitrogen Concentration of Rice through Fusing Vegetation Indices and Color Moments Derived from UAV-RGB Images. Remote Sens. 2021, 13, 1620. [Google Scholar] [CrossRef]
- Colorado, J.D.; Cera-Bornacelli, N.; Caldas, J.S.; Petro, E.; Rebolledo, M.C.; Cuellar, D.; Calderon, F.; Mondragon, I.F.; Jaramillo-Botero, A. Estimation of Nitrogen in Rice Crops from UAV-Captured Images. Remote Sens. 2020, 12, 3396. [Google Scholar] [CrossRef]
- Zheng, H.; Cheng, T.; Li, D.; Zhou, X.; Yao, X.; Tian, Y.; Cao, W.; Zhu, Y. Evaluation of RGB, Color-Infrared and Multispectral Images Acquired from Unmanned Aerial Systems for the Estimation of Nitrogen Accumulation in Rice. Remote Sens. 2018, 10, 824. [Google Scholar] [CrossRef]
- Loukatos, D.; Templalexis, C.; Lentzou, D.; Xanthopoulos, G.; Arvanitis, K.G. Enhancing a flexible robotic spraying platform for distant plant inspection via high-quality thermal imagery data. Comput. Electron. Agric. 2021, 190, 106462. [Google Scholar] [CrossRef]
- Kerkech, M.; Hafiane, A.; Canals, R. Deep leaning approach with colorimetric spaces and vegetation indices for vine diseases detection in UAV images. Comput. Electron. Agric. 2018, 155, 237–243. [Google Scholar] [CrossRef]
- Reza, M.N.; Na, I.; Baek, S.; Lee, I.; Lee, K. Lab Color Space based Rice Yield Prediction using Low Altitude UAV Field Image. In Proceedings of the Proceedings of the KSAM & UMRC 2017 Spring Conference, Gunwi-Gun, Republic of Korea, 7 April 2017. [Google Scholar]
- Zhou, D.; Li, M.; Li, Y.; Qi, J.; Liu, K.; Cong, X.; Tian, X. Detection of ground straw coverage under conservation tillage based on deep learning. Comput. Electron. Agric. 2020, 172, 105369. [Google Scholar] [CrossRef]
- Chen, Z.; Jia, K.; Xiao, C.; Wei, D.; Zhao, X.; Lan, J.; Wei, X.; Yao, Y.; Wang, B.; Sun, Y.; et al. Leaf Area Index Estimation Algorithm for GF-5 Hyperspectral Data Based on Different Feature Selection and Machine Learning Methods. Remote Sens. 2020, 12, 2110. [Google Scholar] [CrossRef]
- Shicheng, Q.; Youwen, T.; Qinghu, W.; Shiyuan, S.; Ping, S. Nondestructive detection of decayed blueberry based on information fusion of hyperspectral imaging (HSI) and low-Field nuclear magnetic resonance (LF-NMR). Comput. Electron. Agric. 2021, 184, 106100. [Google Scholar] [CrossRef]
- Ng, W.; Minasny, B.; Malone, B.P.; Sarathjith, M.C.; Das, B.S. Optimizing wavelength selection by using informative vectors for parsimonious infrared spectra modelling. Comput. Electron. Agric. 2019, 158, 201–210. [Google Scholar] [CrossRef]
- Jia, M.; Li, W.; Wang, K.; Zhou, C.; Cheng, T.; Tian, Y.; Zhu, Y.; Cao, W.; Yao, X. A newly developed method to extract the optimal hyperspectral feature for monitoring leaf biomass in wheat. Comput. Electron. Agric. 2019, 165, 104942. [Google Scholar] [CrossRef]
- Li, Y.; Fu, B.; Sun, X.; Fan, D.; Wang, Y.; He, H.; Gao, E.; He, W.; Yao, Y. Comparison of Different Transfer Learning Methods for Classification of Mangrove Communities Using MCCUNet and UAV Multispectral Images. Remote Sens. 2022, 14, 5533. [Google Scholar] [CrossRef]
- Guo, P.T.; Li, M.F.; Luo, W.; Cha, Z.Z. Estimation of foliar nitrogen of rubber trees using hyperspectral reflectance with feature bands. Infrared Phys. Technol. 2019, 102, 103021. [Google Scholar] [CrossRef]
- Zhang, J.; Cheng, T.; Guo, W.; Xu, X.; Ma, X. Leaf area index estimation model for UAV image hyperspectral data based on wavelength variable selection and machine learning methods. Plant Methods 2021, 17, 49. [Google Scholar] [CrossRef]
- Khaled, A.Y.; Abd Aziz, S.; Khairunniza Bejo, S.; Mat Nawi, N.; Jamaludin, D.; Ibrahim, N.U.A. A comparative study on dimensionality reduction of dielectric spectral data for the classification of basal stem rot (BSR) disease in oil palm. Comput. Electron. Agric. 2020, 170, 105288. [Google Scholar] [CrossRef]
- Samsudin, S.H.; Shafri, H.Z.M.; Hamedianfar, A.; Mansor, S. Spectral feature selection and classification of roofing materials using field spectroscopy data. J. Appl. Remote Sens. 2015, 9, 095079. [Google Scholar] [CrossRef]
- Chen, Z.; Li, S.; Ren, J.; Gong, P.; Jiang, D. Monitoring and Management of Agriculture with Remote Sensing; Springer: Dordrecht, The Netherlands, 2008. [Google Scholar]
- Rodriguez, J.C.; Duchemin, B.; Watts, C.J.; Hadria, R.; Er-Raki, S. Wheat yields estimation using remote sensing and crop modeling in Yaqui Valley in Mexico. In Proceedings of the IEEE International Geoscience & Remote Sensing Symposium, Toulouse, France, 21–25 July 2003. [Google Scholar]
- Tao, H.; Feng, H.; Xu, L.; Miao, M.; Yang, G.; Yang, X.; Fan, L. Estimation of the Yield and Plant Height of Winter Wheat Using UAV-Based Hyperspectral Images. Sensors 2020, 20, 1231. [Google Scholar] [CrossRef]
- Kefauver, S.C.; Vicente, R.; Vergara-Díaz, O.; Fernández-Gallego, J.A.; Araus, J.L. Comparative UAV and Field Phenotyping to Assess Yield and Nitrogen Use Efficiency in Hybrid and Conventional Barley. Front. Plant Sci. 2017, 8, 1733. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zhu, M.; Kang, Y.; Leung, E.L.-H.; Lei, T.; Shen, C.; Jiang, D.; Wang, Z.; Cao, D.; Hou, T. Do we need different machine learning algorithms for QSAR modeling? A comprehensive assessment of 16 machine learning algorithms on 14 QSAR data sets. Brief. Bioinform. 2020, 22, bbaa321. [Google Scholar] [CrossRef]
- Ku, N.-W.; Popescu, S.C. A comparison of multiple methods for mapping local-scale mesquite tree aboveground biomass with remotely sensed data. Biomass Bioenergy 2019, 122, 270–279. [Google Scholar] [CrossRef]
- Piepho, H.P. Ridge Regression and Extensions for Genomewide Selection in Maize. Crop Sci. 2009, 49, 1165–1176. [Google Scholar] [CrossRef]
- Ogutu, J.O.; Schulz-Streeck, T.; Piepho, H.-P. Genomic selection using regularized linear regression models: Ridge regression, lasso, elastic net and their extensions. BMC Proc. 2012, 6, S10. [Google Scholar] [CrossRef]
- Chen, S.; Guo, J.; Zhao, Y.; Li, X.; Liu, F.; Chen, Y. Evaluation and grading of climatic conditions on nutritional quality of rice: A case study of Xiaozhan rice in Tianjin. Meteorol. Appl. 2021, 28, e2021. [Google Scholar] [CrossRef]
- Nelson, D.W.; Sommers, L.E. Determination of Total Nitrogen in Plant Material1. Agron. J. 1973, 65, 109–112. [Google Scholar] [CrossRef]
- Li, D.; Song, Z.; Quan, C.; Xu, X.; Liu, C. Recent advances in image fusion technology in agriculture. Comput. Electron. Agric. 2021, 191, 106491. [Google Scholar] [CrossRef]
- Sarp, G. Spectral and spatial quality analysis of pan-sharpening algorithms: A case study in Istanbul. Eur. J. Remote Sens. 2014, 47, 19–28. [Google Scholar] [CrossRef]
- Hamuda, E.; Mc Ginley, B.; Glavin, M.; Jones, E. Automatic crop detection under field conditions using the HSV colour space and morphological operations. Comput. Electron. Agric. 2017, 133, 97–107. [Google Scholar] [CrossRef]
- Xu, X.G.; Fan, L.L.; Li, Z.H.; Meng, Y.; Feng, H.K.; Yang, H.; Xu, B. Estimating Leaf Nitrogen Content in Corn Based on Information Fusion of Multiple-Sensor Imagery from UAV. Remote Sens. 2021, 13, 340. [Google Scholar] [CrossRef]
- Guo, J.; Bai, Q.; Guo, W.; Bu, Z.; Zhang, W. Soil moisture content estimation in winter wheat planting area for multi-source sensing data using CNNR. Comput. Electron. Agric. 2022, 193, 106670. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Navarro, G.; Caballero, I.; Silva, G.; Parra, P.-C.; Vazquez, A.; Caldeira, R. Evaluation of forest fire on Madeira Island using Sentinel-2A MSI imagery. Int. J. Appl. Earth Obs. Geoinf. 2017, 58, 97–106. [Google Scholar] [CrossRef]
- Sankaran, S.; Zhou, J.; Khot, L.R.; Trapp, J.J.; Mndolwa, E.; Miklas, P.N. High-throughput field phenotyping in dry bean using small unmanned aerial vehicle based multispectral imagery. Comput. Electron. Agric. 2018, 151, 84–92. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Pattey, E.; Zarco-Tejada, P.J.; Strachan, I.B. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Hansen, P.M.; Schjoerring, J.K. Reflectance measurement of canopy biomass and nitrogen status in wheat crops using normalized difference vegetation indices and partial least squares regression. Remote Sens. Environ. 2003, 86, 542–553. [Google Scholar] [CrossRef]
- Goel, N.S.; Qin, W. Influences of canopy architecture on relationships between various vegetation indices and LAI and Fpar: A computer simulation. Remote Sens. Rev. 1994, 10, 309–347. [Google Scholar] [CrossRef]
- Roujean, J.-L.; Breon, F.-M. Estimating PAR absorbed by vegetation from bidirectional reflectance measurements. Remote Sens. Environ. 1995, 51, 375–384. [Google Scholar] [CrossRef]
- Jordan, C.F. Derivation of Leaf-Area Index from Quality of Light on the Forest Floor. Ecology 1969, 50, 663–666. [Google Scholar] [CrossRef]
- Tedesco, D.; Almeida Moreira, B.R.d.; Barbosa Júnior, M.R.; Papa, J.P.; Silva, R.P.d. Predicting on multi-target regression for the yield of sweet potato by the market class of its roots upon vegetation indices. Comput. Electron. Agric. 2021, 191, 106544. [Google Scholar] [CrossRef]
- Ihuoma, S.O.; Madramootoo, C.A. Sensitivity of spectral vegetation indices for monitoring water stress in tomato plants. Comput. Electron. Agric. 2019, 163, 104860. [Google Scholar] [CrossRef]
- Bagheri, N. Application of aerial remote sensing technology for detection of fire blight infected pear trees. Comput. Electron. Agric. 2020, 168, 105147. [Google Scholar] [CrossRef]
- Li, Z.; Li, Z.; Fairbairn, D.; Li, N.; Xu, B.; Feng, H.; Yang, G. Multi-LUTs method for canopy nitrogen density estimation in winter wheat by field and UAV hyperspectral. Comput. Electron. Agric. 2019, 162, 174–182. [Google Scholar] [CrossRef]
- Raper, T.B.; Varco, J.J. Canopy-scale wavelength and vegetative index sensitivities to cotton growth parameters and nitrogen status. Precis. Agric. 2015, 16, 62–76. [Google Scholar] [CrossRef]
- Jiang, Z.; Huete, A.R.; Didan, K.; Miura, T. Development of a two-band enhanced vegetation index without a blue band. Remote Sens. Environ. 2008, 112, 3833–3845. [Google Scholar] [CrossRef]
- Qiu, B.; Huang, Y.; Chen, C.; Tang, Z.; Zou, F. Mapping spatiotemporal dynamics of maize in China from 2005 to 2017 through designing leaf moisture based indicator from Normalized Multi-band Drought Index. Comput. Electron. Agric. 2018, 153, 82–93. [Google Scholar] [CrossRef]
- Daughtry, C.S.T.; Walthall, C.L.; Kim, M.S.; de Colstoun, E.B.; McMurtrey, J.E. Estimating Corn Leaf Chlorophyll Concentration from Leaf and Canopy Reflectance. Remote Sens. Environ. 2000, 74, 229–239. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Tremblay, N.; Zarco-Tejada, P.J.; Dextraze, L. Integrated narrow-band vegetation indices for prediction of crop chlorophyll content for application to precision agriculture. Remote Sens. Environ. 2002, 81, 416–426. [Google Scholar] [CrossRef]
- Araújo, M.C.U.; Saldanha, T.C.B.; Galvao, R.K.H.; Yoneyama, T.; Chame, H.C.; Visani, V. The successive projections algorithm for variable selection in spectroscopic multicomponent analysis. Chemom. Intell. Lab. Syst. 2001, 57, 65–73. [Google Scholar] [CrossRef]
- Jiang, X.; Zhen, J.; Miao, J.; Zhao, D.; Wang, J.; Jia, S. Assessing mangrove leaf traits under different pest and disease severity with hyperspectral imaging spectroscopy. Ecol. Indic. 2021, 129, 107901. [Google Scholar] [CrossRef]
- Xing, Z.; Du, C.; Shen, Y.; Ma, F.; Zhou, J. A method combining FTIR-ATR and Raman spectroscopy to determine soil organic matter: Improvement of prediction accuracy using competitive adaptive reweighted sampling (CARS). Comput. Electron. Agric. 2021, 191, 106549. [Google Scholar] [CrossRef]
- Sun, J.; Yang, W.; Zhang, M.; Feng, M.; Xiao, L.; Ding, G. Estimation of water content in corn leaves using hyperspectral data based on fractional order Savitzky-Golay derivation coupled with wavelength selection. Comput. Electron. Agric. 2021, 182, 105989. [Google Scholar] [CrossRef]
- Shafiee, S.; Lied, L.M.; Burud, I.; Dieseth, J.A.; Alsheikh, M.; Lillemo, M. Sequential forward selection and support vector regression in comparison to LASSO regression for spring wheat yield prediction based on UAV imagery. Comput. Electron. Agric. 2021, 183, 106036. [Google Scholar] [CrossRef]
- Endelman, J.B. Ridge Regression and Other Kernels for Genomic Selection with R Package rrBLUP. Plant Genome 2011, 4, 250–255. [Google Scholar] [CrossRef]
- Wiens, T.S.; Dale, B.C.; Boyce, M.S.; Kershaw, G.P. Three way k-fold cross-validation of resource selection functions. Ecol. Model. 2008, 212, 244–255. [Google Scholar] [CrossRef]
- Farou, B.; Rouabhia, H.; Seridi, H.; Akdag, H. Novel Approach for Detection and Removal of Moving Cast Shadows Based on RGB, HSV and YUV Color Spaces. Comput. Inform. 2017, 36, 837–856. [Google Scholar] [CrossRef] [PubMed]
- Surkutlawar, S.; Kulkarni, R.K. Shadow Suppression using RGB and HSV Color Space in Moving Object Detection. Int. J. Adv. Comput. Sci. Appl. 2013, 4, 995–1007. [Google Scholar]
- Zhang, Y.; Hartemink, A.E. A method for automated soil horizon delineation using digital images. Geoderma 2019, 343, 97–115. [Google Scholar] [CrossRef]













| Waveband | Central Wavelength (nm) | Spectral Bandwidth (nm) | Panel Reflectance |
|---|---|---|---|
| Blue | 450 ± 16 | 20 | 0.97 |
| Green | 560 ± 16 | 20 | 0.97 |
| Red | 650 ± 16 | 10 | 0.96 |
| RedEdge | 730 ± 16 | 10 | 0.95 |
| NIR | 840 ± 26 | 40 | 0.91 |
| Vegetation Index | Name | Formula | Ref |
|---|---|---|---|
| DVI | Difference Vegetation Index | Rnir − Rr | [38] |
| NDVI | Normalized Difference Vegetation Index | (Rnir − Rr)/(Rnir + Rr) | [39] |
| RDVI | Renormalized Difference Vegetation Index | (Rnir − Rr)/() | [40] |
| GNDVI | Green Normalized Difference Vegetation Index | (Rnir − Rg)/(Rnir + Rg) | [41] |
| RVI | Ratio Vegetation Index | Rnir/Rr | [42] |
| GRVI | Green-Red Vegetation Index | (Rg − Rr)/(Rg + Rr) | [43] |
| WDRVI | Wide Dynamic Range Vegetation Index | (0.12Rnir − Rr)/(0.12Rnir + Rr) | [44] |
| NLI | Nonlinear Vegetation Index | (Rnir2 − Rr)/(Rnir2 + Rr) | [45] |
| MNLI | Modified Nonlinear Vegetation Index | (1.5Rnir2 − 1.5Rg)/(Rnir2 + Rr + 0.5) | [46] |
| SAVI | Soil-Adjusted Vegetation Index | (Rnir − Rr)/1.5(Rnir + Rr + 0.5) | [47] |
| OSAVI | Optimized Soil-Adjusted Vegetation Index | (Rnir − Rr)/(Rnir + Rr + 0.16) | [48] |
| TCARI | Transformed Chlorophyll Absorption Ratio Index | 3 [(Rre − Rr) − 0.2(Rre − Rg)×(Rre/Rr)] | [49] |
| MCARI | Modified Chlorophyll Absorption Ratio Index | [(Rre − Rr) − 0.2(Rre − Rg)]×(Rre/Rr) | [50] |
| GCI | Green Chlorophyll Index | (Rnir/Rg) − 1 | [51] |
| RECI | Red Edge Chlorophyll Index | (Rnir/Rre) − 1 | [52] |
| EVI2 | Two-band Enhanced Vegetation Index | 2.5(Rnir − Rr)/(Rnir + 2.4Rr + 1) | [53] |
| NDREI | Normalized Difference Red Edge Index | (Rre − Rg)/(Rre + Rg) | [54] |
| MSRI | Modified Simple Ratio Index | (Rnir/Rr − 1)/() | [55] |
| TVI | Triangular Vegetation Index | 0.5(120(Rnir − Rre) − 200(Rr − Rre)) | [49] |
| Growth Stage | Samples | Min | Max | Mean | Standard Deviation | Coefficient of Variation (%) |
|---|---|---|---|---|---|---|
| Jointing | 24 | 3.95 | 4.65 | 4.34 | 0.21 | 4.84 |
| Booting | 24 | 3.34 | 3.89 | 3.67 | 0.16 | 4.36 |
| Filling | 24 | 2.89 | 3.52 | 3.13 | 0.17 | 5.43 |
| Growth Stage | Condition | Number of Variables | Selected Feature Variables | Method | R2 | RMSE (%) | NRMSE (%) |
|---|---|---|---|---|---|---|---|
| Jointing | Original Image | 5 | MCARI, GRVI, OSAVI, NDVI, TVI | RF | 0.43 | 14.87 | 3.43 |
| 19 (3) | MCARI, GRVI, TCARI | LASSO | 0.57 | 13.41 | 3.09 | ||
| 5 | MCARI, GRVI, OSAVI, NDVI, TVI | RIDGE | 0.52 | 14.43 | 3.32 | ||
| Fusion Image | 5 | MCARI, WDRVI, GRVI, OSAVI, MNLI | RF | 0.50 | 13.56 | 3.13 | |
| 19 (3) | MCARI, GRVI, SAVI | LASSO | 0.66 | 11.96 | 2.76 | ||
| 5 | MCARI, WDRVI, GRVI, OSAVI, MNLI | RIDGE | 0.60 | 13.48 | 3.11 | ||
| Booting | Original Image | 5 | OSAVI, NDVI, TVI, MCARI, WDRVI | RF | 0.40 | 11.72 | 3.19 |
| 19 (5) | OSAVI, TVI, MCARI, WDRVI, NLI | LASSO | 0.51 | 10.86 | 2.96 | ||
| 5 | OSAVI, NDVI, TVI, MCARI, WDRVI | RIDGE | 0.48 | 11.36 | 3.09 | ||
| Fusion Image | 5 | OSAVI, TVI, MCARI, NDVI, NLI | RF | 0.48 | 11.53 | 3.14 | |
| 19 (5) | OSAVI, TVI, NDVI, MCARI, WDRVI | LASSO | 0.57 | 11.16 | 3.04 | ||
| 5 | OSAVI, TVI, MCARI, NDVI, NLI | RIDGE | 0.55 | 11.41 | 3.11 | ||
| Filling | Original Image | 5 | SAVI, EVI2, OSAVI, TVI, MCARI | RF | 0.36 | 13.35 | 4.26 |
| 19 (4) | SAVI, WDRVI, TVI, MCARI | LASSO | 0.47 | 12.66 | 4.05 | ||
| 5 | SAVI, EVI2, OSAVI, TVI, MCARI | RIDGE | 0.44 | 13.09 | 4.18 | ||
| Fusion Image | 5 | SAVI, OSAVI, TVI, MNLI, NDVI | RF | 0.45 | 11.91 | 3.81 | |
| 19 (4) | SAVI, OSAVI, MNLI, NLI | LASSO | 0.53 | 11.13 | 3.56 | ||
| 5 | SAVI, OSAVI, TVI, MNLI, NDVI | RIDGE | 0.51 | 11.68 | 3.73 |
| Growth Stage | Condition | Number of Variables | Selected Feature Variables | Method | R2 | RMSE (%) | NRMSE (%) |
|---|---|---|---|---|---|---|---|
| Jointing | Original Image | 5 | MCARI, GRVI, OSAVI, NDVI, TVI | RF | 0.43 | 14.87 | 3.43 |
| 19 (3) | MCARI, GRVI, TCARI | LASSO | 0.57 | 13.41 | 3.09 | ||
| 5 | MCARI, GRVI, OSAVI, NDVI, TVI | RIDGE | 0.52 | 14.43 | 3.32 | ||
| Denoised Original Image | 5 | MCARI, GRVI, WDRVI, SAVI, NDVI | RF | 0.48 | 13.86 | 3.19 | |
| 19 (5) | MCARI, SAVI, NDVI, WDRVI, TVI | LASSO | 0.63 | 12.72 | 2.93 | ||
| 5 | MCARI, GRVI, WDRVI, SAVI, NDVI | RIDGE | 0.58 | 13.57 | 3.13 | ||
| Fusion Image | 5 | MCARI, WDRVI, GRVI, OSAVI, MNLI | RF | 0.50 | 13.56 | 3.13 | |
| 19 (3) | MCARI, GRVI, SAVI | LASSO | 0.66 | 11.96 | 2.76 | ||
| 5 | MCARI, WDRVI, GRVI, OSAVI, MNLI | RIDGE | 0.60 | 13.48 | 3.11 | ||
| Denoised Fusion Image | 5 | MCARI, WDRVI, GRVI, OSAVI, NLI | RF | 0.57 | 12.43 | 2.87 | |
| 19 (6) | MCARI, GRVI, SAVI, NLI, TVI, RVI | LASSO | 0.69 | 11.36 | 2.62 | ||
| 5 | MCARI, WDRVI, GRVI, OSAVI, NLI | RIDGE | 0.66 | 12.09 | 2.79 | ||
| Booting | Original Image | 5 | OSAVI, NDVI, TVI, MCARI, WDRVI | RF | 0.40 | 11.72 | 3.19 |
| 19 (5) | OSAVI, TVI, MCARI, WDRVI, NLI | LASSO | 0.51 | 10.86 | 2.96 | ||
| 5 | OSAVI, NDVI, TVI, MCARI, WDRVI | RIDGE | 0.48 | 11.36 | 3.09 | ||
| Denoised Original Image | 5 | OSAVI, NDVI, WDRVI, NLI, MNLI | RF | 0.45 | 11.59 | 3.16 | |
| 19 (3) | OSAVI, NDVI, WDRVI | LASSO | 0.55 | 10.34 | 2.82 | ||
| 5 | OSAVI, NDVI, WDRVI, NLI, MNLI | RIDGE | 0.53 | 11.24 | 3.06 | ||
| Fusion Image | 5 | OSAVI, TVI, MCARI, NDVI, NLI | RF | 0.48 | 11.53 | 3.14 | |
| 19 (5) | OSAVI, TVI, NDVI, MCARI, WDRVI | LASSO | 0.57 | 11.16 | 3.04 | ||
| 5 | OSAVI, TVI, MCARI, NDVI, NLI | RIDGE | 0.55 | 11.41 | 3.11 | ||
| Denoised Fusion Image | 5 | TVI, WDRVI, OSAVI, MCARI, NLI | RF | 0.52 | 11.17 | 3.04 | |
| 19 (4) | TVI, OSAVI, MCARI, NLI | LASSO | 0.62 | 9.79 | 2.67 | ||
| 5 | TVI, WDRVI, OSAVI, MCARI, NLI | RIDGE | 0.59 | 10.83 | 2.95 | ||
| Filling | Original Image | 5 | SAVI, EVI2, OSAVI, TVI, MCARI | RF | 0.36 | 13.35 | 4.26 |
| 19 (4) | SAVI, WDRVI, TVI, MCARI | LASSO | 0.47 | 12.66 | 4.05 | ||
| 5 | SAVI, EVI2, OSAVI, TVI, MCARI | RIDGE | 0.44 | 13.09 | 4.18 | ||
| Denoised Original Image | 5 | SAVI, TVI, OSAVI, MCARI, NLI | RF | 0.41 | 14.11 | 4.51 | |
| 19 (3) | SAVI, TVI, OSAVI | LASSO | 0.52 | 11.79 | 3.77 | ||
| 5 | SAVI, TVI, OSAVI, MCARI, NLI | RIDGE | 0.49 | 13.83 | 4.42 | ||
| Fusion Image | 5 | SAVI, OSAVI, TVI, MNLI, NDVI | RF | 0.45 | 11.91 | 3.81 | |
| 19 (4) | SAVI, OSAVI, MNLI, NLI | LASSO | 0.53 | 11.13 | 3.56 | ||
| 5 | SAVI, OSAVI, TVI, MNLI, NDVI | RIDGE | 0.51 | 11.68 | 3.73 | ||
| Denoised Fusion Image | 5 | SAVI, EVI2, GCI, OSAVI, MCARI | RF | 0.49 | 10.98 | 3.51 | |
| 19 (4) | SAVI, OSAVI, MCARI, EVI2 | LASSO | 0.58 | 9.68 | 3.09 | ||
| 5 | SAVI, EVI2, GCI, OSAVI, MCARI | RIDGE | 0.54 | 10.77 | 3.44 |
| Growth Stage | Condition | Number of Variables | Selected Feature Variables | Method | R2 | RMSE (%) | NRMSE (%) |
|---|---|---|---|---|---|---|---|
| Jointing | Denoised Original Image | 5 | MCARI, GRVI, WDRVI, SAVI, NDVI | RF | 0.48 | 13.86 | 3.19 |
| 19 (5) | MCARI, SAVI, NDVI, WDRVI, TVI | LASSO | 0.63 | 12.72 | 2.93 | ||
| 5 | MCARI, GRVI, WDRVI, SAVI, NDVI | RIDGE | 0.58 | 13.57 | 3.13 | ||
| 3 | MCARI, SAVI, WDRVI | RR-SPA | 0.68 | 12.05 | 2.78 | ||
| 5 | MCARI, GRVI, SAVI, WDRVI, NLI | RR-CARS | 0.64 | 12.22 | 2.82 | ||
| Denoised Fusion Image | 5 | MCARI, WDRVI, GRVI, OSAVI, NLI | RF | 0.57 | 12.43 | 2.87 | |
| 19 (6) | MCARI, GRVI, SAVI, NLI, TVI, RVI | LASSO | 0.69 | 11.36 | 2.62 | ||
| 5 | MCARI, WDRVI, GRVI, OSAVI, NLI | RIDGE | 0.66 | 12.09 | 2.79 | ||
| 3 | MCARI, SAVI, OSAVI | RR-SPA | 0.76 | 10.33 | 2.38 | ||
| 5 | MCARI, SAVI, GRVI, NLI, TVI | RR-CARS | 0.70 | 11.26 | 2.59 | ||
| Booting | Denoised Original Image | 5 | OSAVI, NDVI, WDRVI, NLI, MNLI | RF | 0.45 | 11.59 | 3.16 |
| 19 (3) | OSAVI, NDVI, WDRVI | LASSO | 0.55 | 10.34 | 2.82 | ||
| 5 | OSAVI, NDVI, WDRVI, NLI, MNLI | RIDGE | 0.53 | 11.24 | 3.06 | ||
| 3 | OSAVI, WDRVI, MCARI | RR-SPA | 0.62 | 9.66 | 2.63 | ||
| 7 | NDVI, NLI, TVI, RVI, MNLI, OSAVI, EVI2 | RR-CARS | 0.54 | 10.91 | 2.98 | ||
| Denoised Fusion Image | 5 | TVI, WDRVI, OSAVI, MCARI, NLI | RF | 0.52 | 11.17 | 3.04 | |
| 19 (4) | TVI, OSAVI, MCARI, NLI | LASSO | 0.62 | 9.79 | 2.67 | ||
| 5 | TVI, WDRVI, OSAVI, MCARI, NLI | RIDGE | 0.59 | 10.83 | 2.95 | ||
| 3 | TVI, WDRVI, MCARI | RR-SPA | 0.71 | 8.83 | 2.41 | ||
| 7 | NDVI, NLI, TVI, RVI, MNLI, OSAVI, GCI | RR-CARS | 0.63 | 9.74 | 2.66 | ||
| Filling | Denoised Original Image | 5 | SAVI, TVI, OSAVI, MCARI, NLI | RF | 0.41 | 14.11 | 4.51 |
| 19 (3) | SAVI, TVI, OSAVI | LASSO | 0.52 | 11.79 | 3.77 | ||
| 5 | SAVI, TVI, OSAVI, MCARI, NLI | RIDGE | 0.49 | 13.83 | 4.42 | ||
| 3 | SAVI, OSAVI, MCARI | RR-SPA | 0.58 | 11.36 | 3.63 | ||
| 6 | SAVI, WDRVI, TVI, NLI, NDVI, OSAVI | RR-CARS | 0.53 | 12.01 | 3.84 | ||
| Denoised Fusion Image | 5 | SAVI, EVI2, GCI, OSAVI, MCARI | RF | 0.49 | 10.98 | 3.51 | |
| 19 (4) | SAVI, OSAVI, MCARI, EVI2 | LASSO | 0.58 | 9.68 | 3.09 | ||
| 5 | SAVI, EVI2, GCI, OSAVI, MCARI | RIDGE | 0.54 | 10.77 | 3.44 | ||
| 3 | MCARI, SAVI, OSAVI | RR-SPA | 0.67 | 8.76 | 2.80 | ||
| 6 | EVI2, NLI, TVI, MCARI, OSAVI, RVI | RR-CARS | 0.61 | 9.30 | 2.97 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, S.; Xu, X.; Blacker, C.; Gaulton, R.; Zhu, Q.; Yang, M.; Yang, G.; Zhang, J.; Yang, Y.; Yang, M.; et al. Estimation of Leaf Nitrogen Content in Rice Using Vegetation Indices and Feature Variable Optimization with Information Fusion of Multiple-Sensor Images from UAV. Remote Sens. 2023, 15, 854. https://doi.org/10.3390/rs15030854
Xu S, Xu X, Blacker C, Gaulton R, Zhu Q, Yang M, Yang G, Zhang J, Yang Y, Yang M, et al. Estimation of Leaf Nitrogen Content in Rice Using Vegetation Indices and Feature Variable Optimization with Information Fusion of Multiple-Sensor Images from UAV. Remote Sensing. 2023; 15(3):854. https://doi.org/10.3390/rs15030854
Chicago/Turabian StyleXu, Sizhe, Xingang Xu, Clive Blacker, Rachel Gaulton, Qingzhen Zhu, Meng Yang, Guijun Yang, Jianmin Zhang, Yongan Yang, Min Yang, and et al. 2023. "Estimation of Leaf Nitrogen Content in Rice Using Vegetation Indices and Feature Variable Optimization with Information Fusion of Multiple-Sensor Images from UAV" Remote Sensing 15, no. 3: 854. https://doi.org/10.3390/rs15030854
APA StyleXu, S., Xu, X., Blacker, C., Gaulton, R., Zhu, Q., Yang, M., Yang, G., Zhang, J., Yang, Y., Yang, M., Xue, H., Yang, X., & Chen, L. (2023). Estimation of Leaf Nitrogen Content in Rice Using Vegetation Indices and Feature Variable Optimization with Information Fusion of Multiple-Sensor Images from UAV. Remote Sensing, 15(3), 854. https://doi.org/10.3390/rs15030854






