Evaluating the Transferability of Spectral Variables and Prediction Models for Mapping Forest Aboveground Biomass Using Transfer Learning Methods
Abstract
:1. Introduction
2. Study Area and Data
2.1. Study Area
2.2. Ground Data
2.3. Remote Sensing Data and Pre-Processing
2.4. Extraction and Selection of SV
2.4.1. Extraction of SVs
2.4.2. Selection of Optimal Variables
2.5. AGB Estimation Model and Spatial–Temporal Transfer
2.5.1. Machine Learning Models
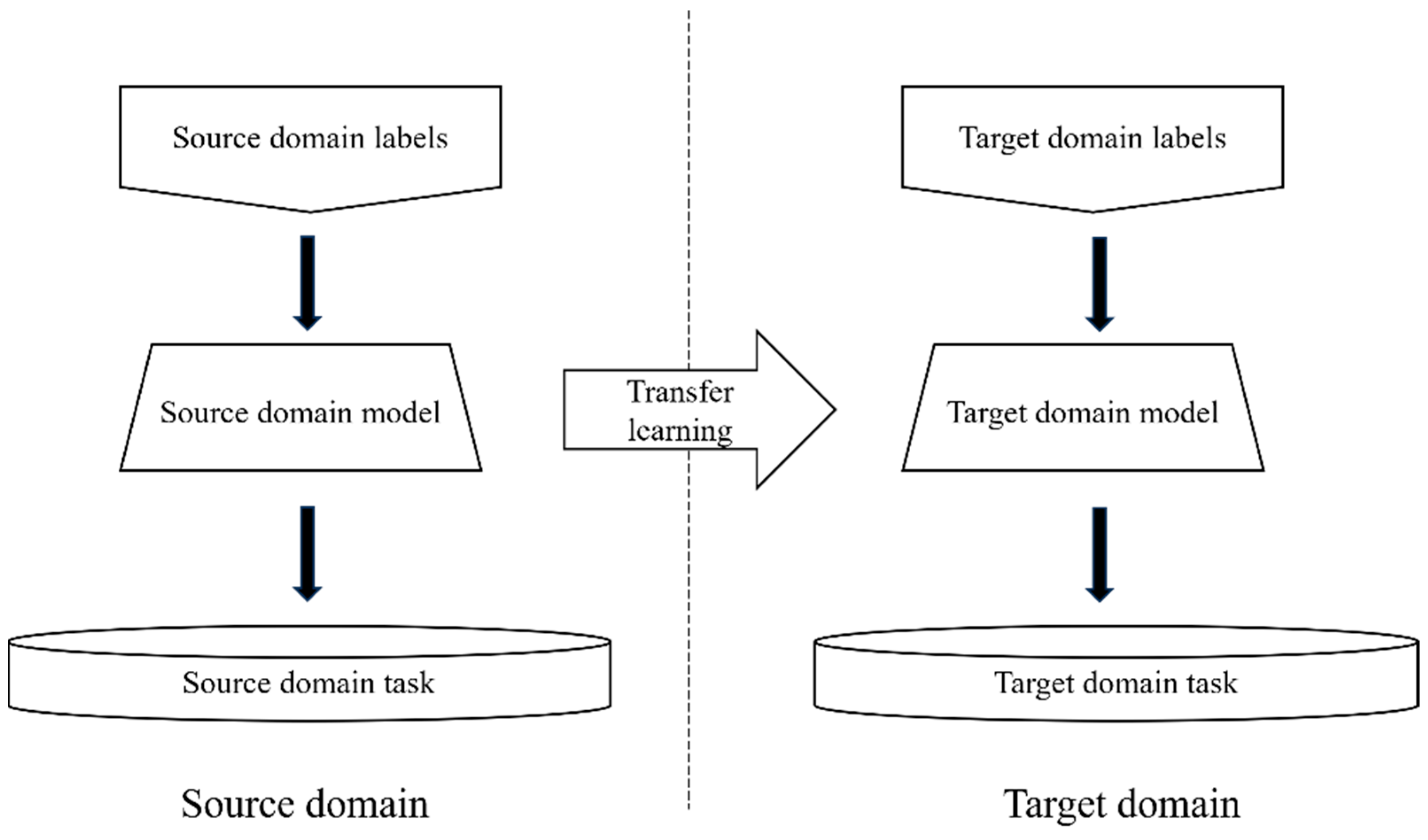
2.5.2. Transfer Learning
2.5.3. Spatial–Temporal Transfer of Mapping Forest AGB
2.6. Transfer Model Evaluation
3. Results
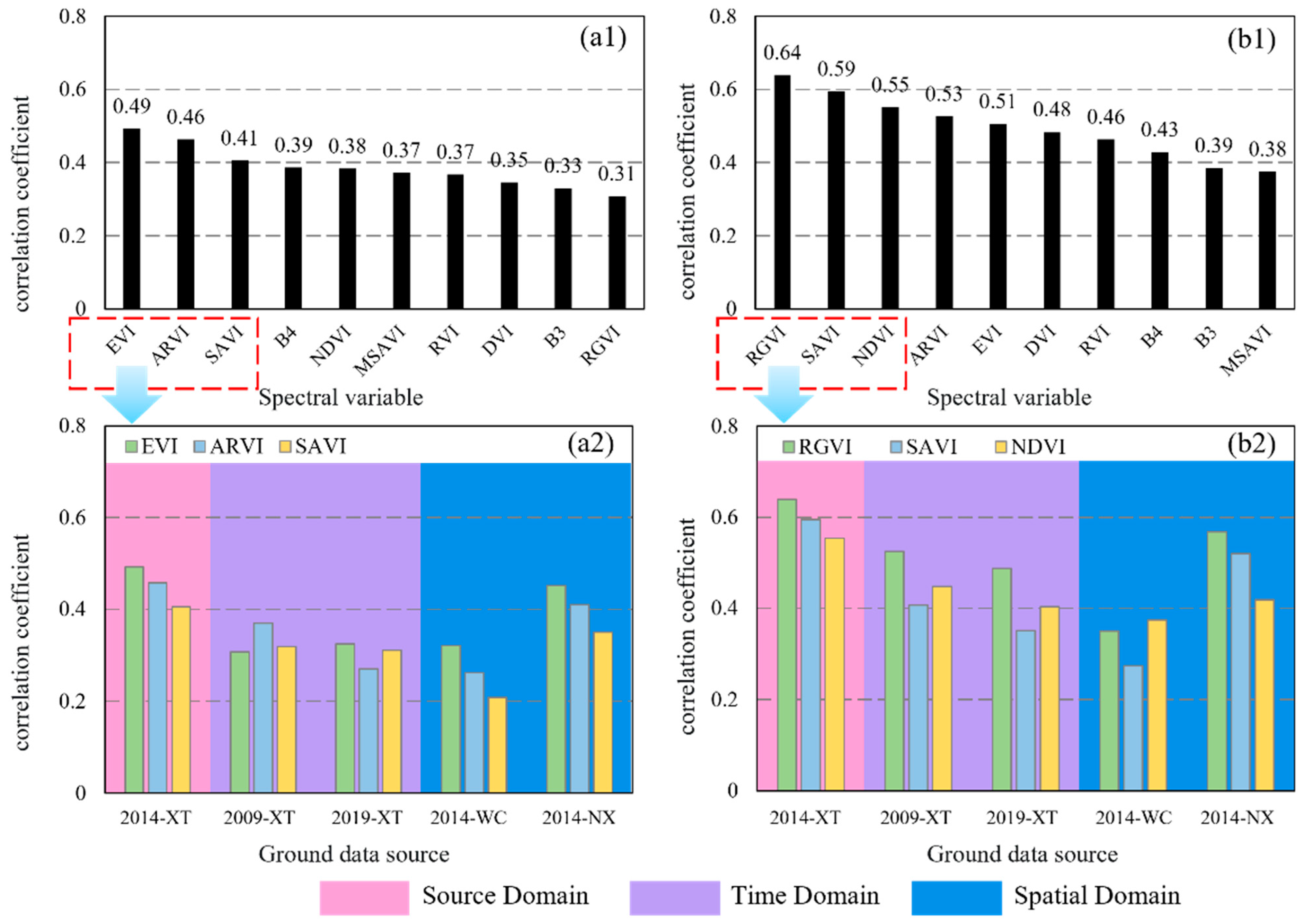
3.1. Analyzing Temporal and Spatial Variation of SVs
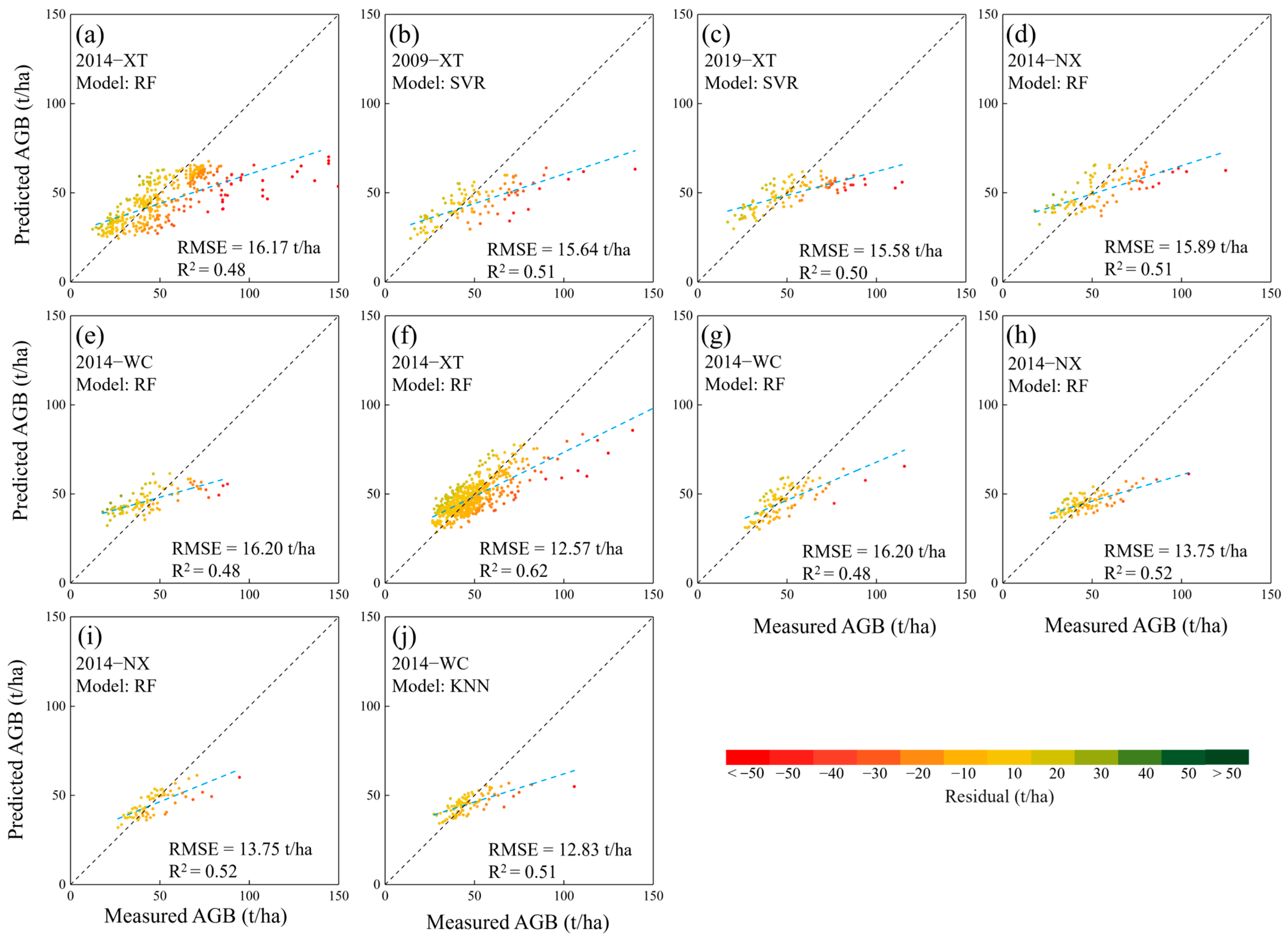
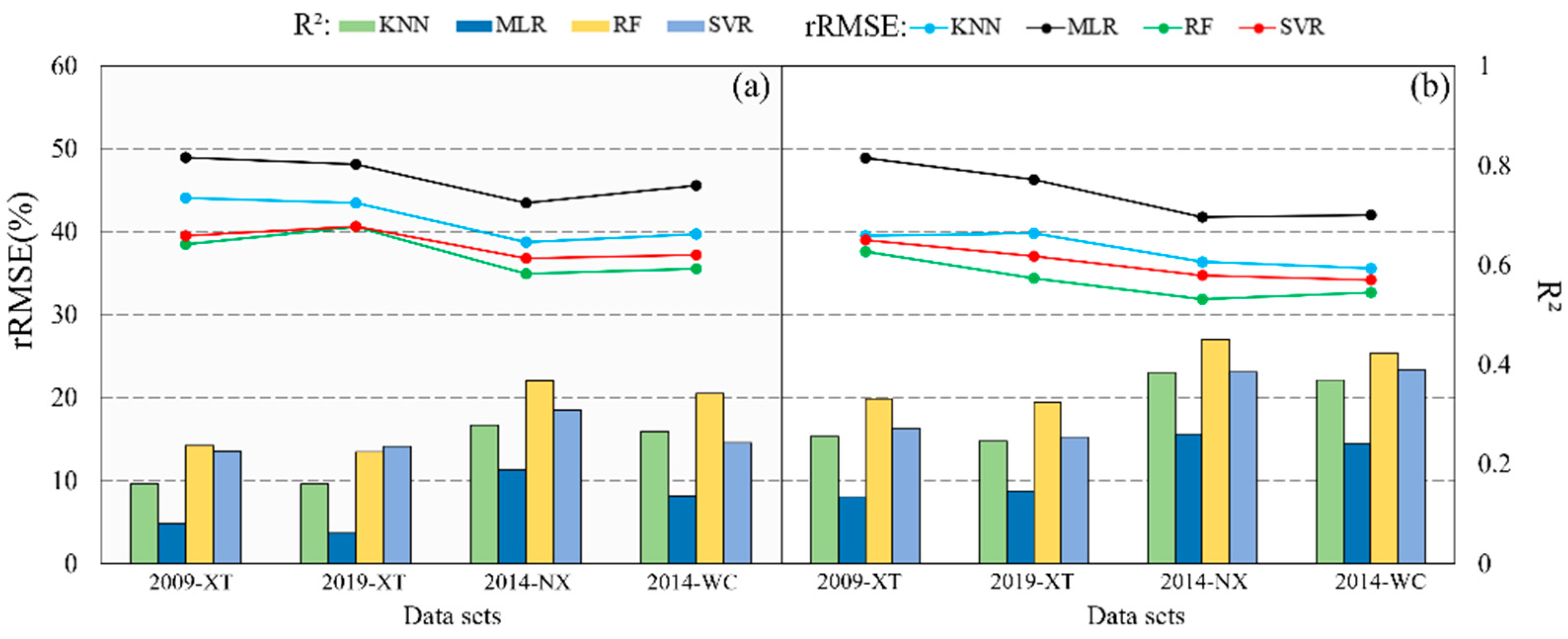
3.2. Mapping Forest AGB in Source Domains
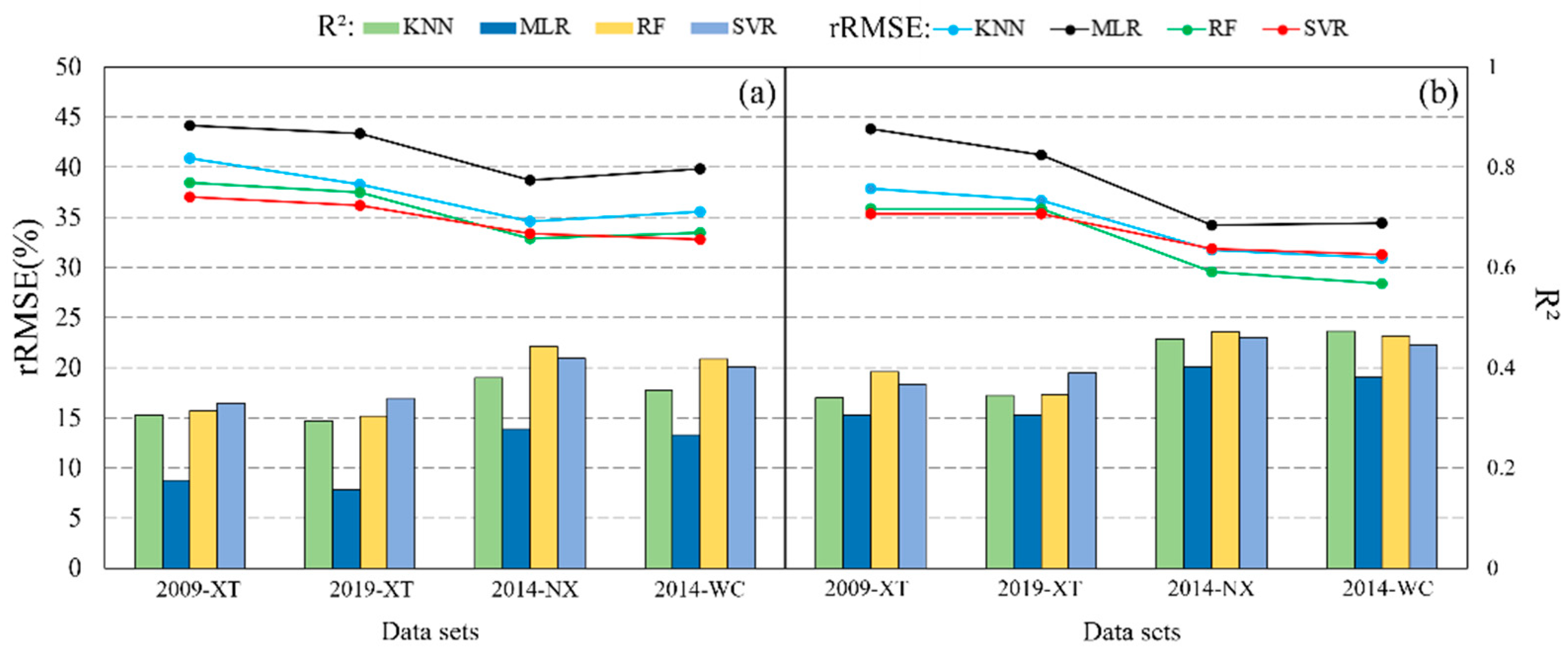
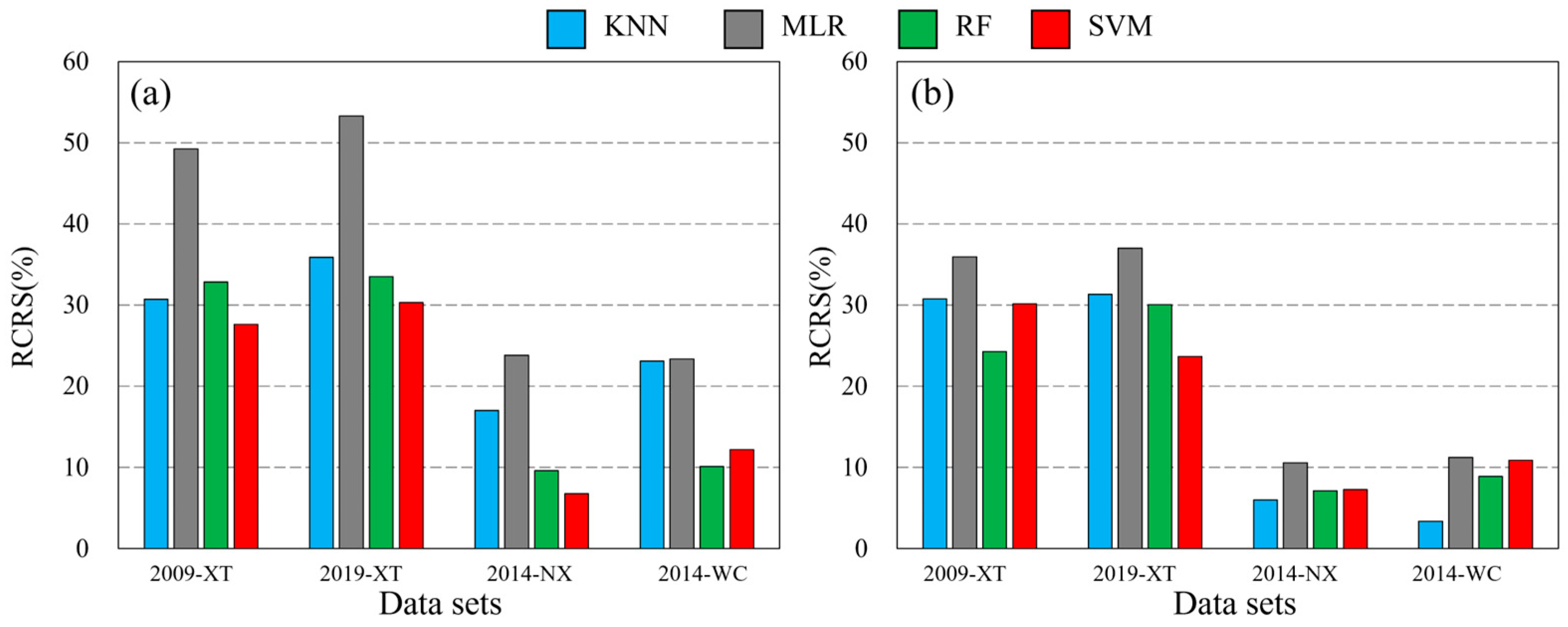
3.3. Ttransferability of Spectral Variables in Mapping Forest AGB
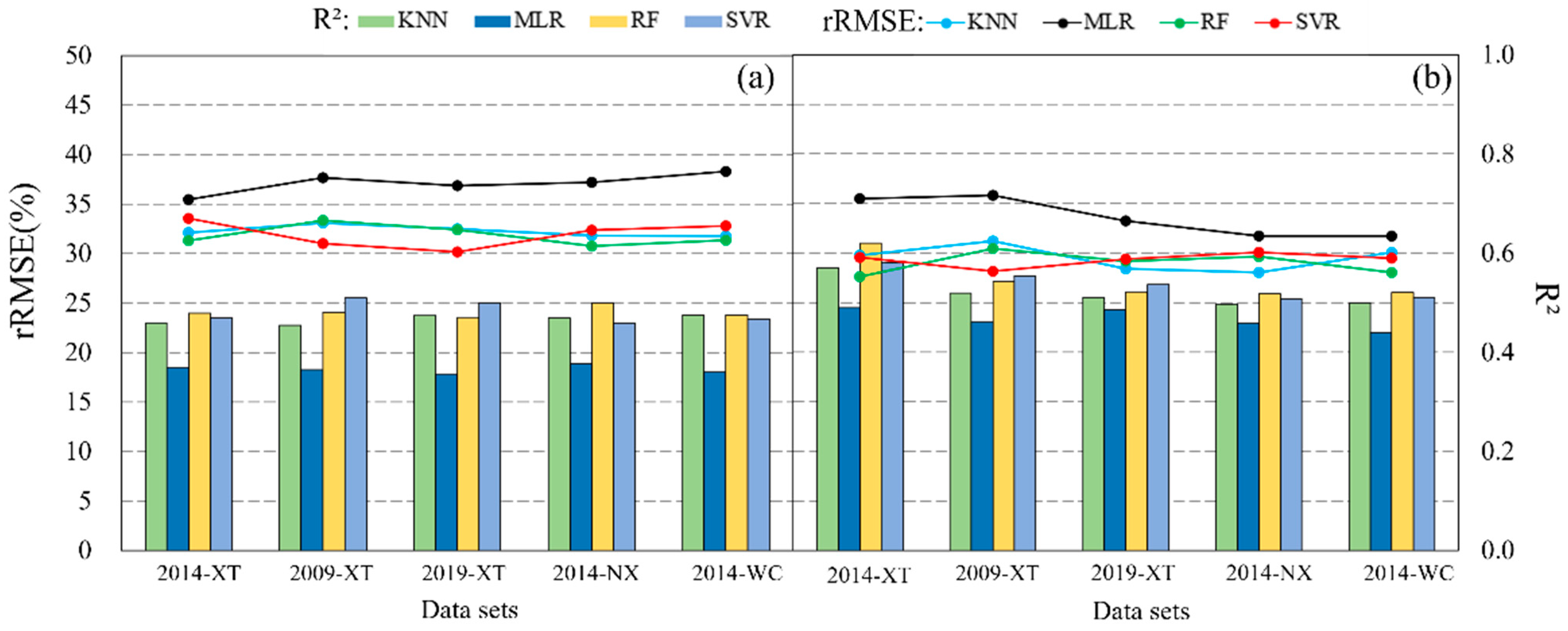
3.4. Transferability of Prediction Models in Mapping Forest AGB
4. Discussion
4.1. Factors Affecting Transferability
4.2. Potential Methods to Enhance Transferability and Generalizability
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Seidl, R.; Spies, T.A.; Peterson, D.L.; Stephens, S.L.; Hicke, J.A. Searching for resilience: Addressing the impacts of changing disturbance regimes on forest ecosystem services. J. Appl. Ecol. 2016, 53, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Schimel, D.S. Terrestrial ecosystems and the carbon cycle. Glob. Chang. Biol. 2006, 1, 77–91. [Google Scholar] [CrossRef]
- Le Toan, T.; Quegan, S.; Davidson, M.; Balzter, H.; Paillou, P.; Papathanassiou, K.; Plummer, S.; Rocca, F.; Saatchi, S.; Shugart, H.; et al. The BIOMASS mission: Mapping global forest biomass to better understand the terrestrial carbon cycle. Remote Sens. Environ. 2011, 115, 2850–2860. [Google Scholar] [CrossRef]
- Jiang, F.; Kutia, M.; Sarkissian, A.J.; Lin, H.; Long, J.; Sun, H.; Wang, G. Estimating the Growing Stem Volume of Coniferous Plantations Based on Random Forest Using an Optimized Variable Selection Method. Sensors 2020, 20, 7248. [Google Scholar] [CrossRef]
- Long, J.; Lin, H.; Wang, G.; Sun, H.; Yan, E. Estimating the Growing Stem Volume of the Planted Forest Using the General Linear Model and Time Series Quad-Polarimetric SAR Images. Sensors 2020, 20, 3957. [Google Scholar] [CrossRef]
- Jiang, F.; Kutia, M.; Ma, K.; Chen, S.; Long, J.; Sun, H. Estimating the aboveground biomass of coniferous forest in Northeast China using spectral variables, land surface temperature and soil moisture. Sci. Total Environ. 2021, 785, 147335. [Google Scholar] [CrossRef]
- Xu, X.; Lin, H.; Liu, Z.; Ye, Z.; Li, X.; Long, J. A Combined Strategy of Improved Variable Selection and Ensemble Algorithm to Map the Growing Stem Volume of Planted Coniferous Forest. Remote Sens. 2021, 13, 4631. [Google Scholar] [CrossRef]
- Li, X.; Liu, Z.; Lin, H.; Wang, G.; Sun, H.; Long, J.; Zhang, M. Estimating the Growing Stem Volume of Chinese Pine and Larch Plantations based on Fused Optical Data Using an Improved Variable Screening Method and Stacking Algorithm. Remote Sens. 2020, 12, 871. [Google Scholar] [CrossRef]
- Lu, D. The potential and challenge of remote sensing-based biomass estimation. Int. J. Remote Sens. 2006, 27, 1297–1328. [Google Scholar] [CrossRef]
- Zheng, S.; Cao, C.; Dang, Y.; Xiang, H.; Zhao, J.; Zhang, Y.; Wang, X.; Guo, H. Retrieval of forest growing stock volume by two different methods using Landsat TM images. Int. J. Remote Sens. 2014, 35, 29–43. [Google Scholar] [CrossRef]
- Yang, C.; Huang, H.; Wang, S. Estimation of tropical forest biomass using Landsat TM imagery and permanent plot data in Xishuangbanna, China. Int. J. Remote Sens. 2011, 32, 5741–5756. [Google Scholar] [CrossRef]
- Lu, D.; Chen, Q.; Wang, G.; Liu, L.; Li, G.; Moran, E. A survey of remote sensing-based aboveground biomass estimation methods in forest ecosystems. Int. J. Digit. Earth 2014, 9, 63–105. [Google Scholar] [CrossRef]
- Li, G.; Xie, Z.; Jiang, X.; Lu, D.; Chen, E. Integration of ZiYuan-3 Multispectral and Stereo Data for Modeling Aboveground Biomass of Larch Plantations in North China. Remote Sens. 2019, 11, 2328. [Google Scholar] [CrossRef]
- Li, X.; Lin, H.; Long, J.; Xu, X. Mapping the Growing Stem Volume of the Coniferous Plantations in North China Using Multispectral Data from Integrated GF-2 and Sentinel-2 Images and an Optimized Feature Variable Selection Method. Remote Sens. 2021, 13, 2740. [Google Scholar] [CrossRef]
- Liu, Z.; Ye, Z.; Xu, X.; Lin, H.; Zhang, T.; Long, J. Mapping Forest Stock Volume Based on Growth Characteristics of Crown Using Multi-Temporal Landsat 8 OLI and ZY-3 Stereo Images in Planted Eucalyptus Forest. Remote Sens. 2022, 14, 5082. [Google Scholar] [CrossRef]
- Zhu, X.; Liu, D. Improving forest aboveground biomass estimation using seasonal Landsat NDVI time-series. ISPRS J. Photogramm. Remote Sens. 2015, 102, 222–231. [Google Scholar] [CrossRef]
- Reiche, J.; Hamunyela, E.; Verbesselt, J.; Hoekman, D.; Herold, M. Improving near-real time deforestation monitoring in tropical dry forests by combining dense Sentinel-1 time series with Landsat and ALOS-2 PALSAR-2. Remote Sens. Environ. 2018, 204, 147–161. [Google Scholar] [CrossRef]
- Narine, L.L.; Popescu, S.C.; Malambo, L. Synergy of ICESat-2 and Landsat for Mapping Forest Aboveground Biomass with Deep Learning. Remote Sens. 2019, 11, 1503. [Google Scholar] [CrossRef]
- Chrysafis, I.; Mallinis, G.; Siachalou, S.; Patias, P. Assessing the relationships between growing stock volume and Sentinel-2 imagery in a Mediterranean forest ecosystem. Remote Sens. Lett. 2017, 8, 508–517. [Google Scholar] [CrossRef]
- Gasparri, N.I.; Parmuchi, M.G.; Bono, J.; Karszenbaum, H.; Montenegro, C.L. Assessing multi-temporal Landsat 7 ETM+ images for estimating above-ground biomass in subtropical dry forests of Argentina. J. Arid Environ. 2010, 74, 1262–1270. [Google Scholar] [CrossRef]
- Gu, Q.Z.J. Learning the Shared Subspace for Multi-task Clustering and Transductive Transfer Classification. In Proceedings of the International Conference on Data Mining (ICDM 2009), Miami Beach, FL, USA, 6–9 December 2009. [Google Scholar]
- Zhou, Y.; Lian, J.; Han, M. Remote Sensing Image Transfer Classification Based on Weighted Extreme Learning Machine. IEEE Geosci. Remote Sens. Lett. 2016, 13, 1405–1409. [Google Scholar] [CrossRef]
- Luo, Y.; Liu, T.; Tao, D.; Xu, C. Decomposition-Based Transfer Distance Metric Learning for Image Classification. IEEE Trans. Image Process. 2014, 23, 3789–3801. [Google Scholar] [CrossRef]
- George, D.; Shen, H.; Huerta, E.A. Classification and unsupervised clustering of LIGO data with Deep Transfer Learning. Phys. Rev. D 2018, 97, 101501. [Google Scholar] [CrossRef]
- Saba, L.; Agarwal, M.; Sanagala, S.; Gupta, S.; Sinha, G.; Johri, A.; Khanna, N.; Mavrogeni, S.; Laird, J.; Pareek, G.; et al. Brain MRI-based Wilson disease tissue classification: An optimised deep transfer learning approach. Electron. Lett. 2020, 56, 1395–1398. [Google Scholar] [CrossRef]
- Imoto, K.; Nakai, T.; Ike, T.; Haruki, K.; Sato, Y. A CNN-Based Transfer Learning Method for Defect Classification in Semiconductor Manufacturing. IEEE Trans. Semicond. Manuf. 2019, 32, 455–459. [Google Scholar] [CrossRef]
- Zhao, K.; Popescu, S.; Meng, X.; Pang, Y.; Agca, M. Characterizing forest canopy structure with lidar composite metrics and machine learning. Remote Sens. Environ. 2011, 115, 1978–1996. [Google Scholar] [CrossRef]
- Yao, J.; Raffuse, S.M.; Brauer, M.; Williamson, G.J.; Bowman, D.M.; Johnston, F.H.; Henderson, S.B. Predicting the minimum height of forest fire smoke within the atmosphere using machine learning and data from the CALIPSO satellite. Remote Sens. Environ. 2018, 206, 98–106. [Google Scholar] [CrossRef]
- Pan, S.J.; Yang, Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Ma, Y.; Luo, G.; Zeng, X.; Chen, A. Transfer learning based cross-company software defects prediction. Comput. Eng. Des. 2012, 54, 248–256. [Google Scholar] [CrossRef]
- Celestin, B.N.; Yunfei, S. The impact of learner characteristics on training transfer expectation: A survey of Thai teachers’ perception of cloud computing tools. Int. J. Train. Dev. 2018, 22, 256–273. [Google Scholar] [CrossRef]
- De Bruijne, M. Machine learning approaches in medical image analysis: From detection to diagnosis. Med. Image Anal. 2016, 33, 94–97. [Google Scholar] [CrossRef] [PubMed]
- Van Opbroek, A.; Achterberg, H.C.; Vernooij, M.W.; De Bruijne, M. Transfer Learning for Image Segmentation by Combining Image Weighting and Kernel Learning. IEEE Trans. Med. Imaging 2018, 38, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wong, L.N.Y.; Tse, K.K.C. Retraction Note: Novel Rock Image Classification: The Proposal and Implementation of RockNet. Rock Mech. Rock Eng. 2023, 56, 3859. [Google Scholar] [CrossRef]
- Dube, T.; Sibanda, M.; Shoko, C.; Mutanga, O. Stand-volume estimation from multi-source data for coppiced and high forest Eucalyptus spp. silvicultural systems in KwaZulu-Natal, South Africa. ISPRS J. Photogramm. Remote Sens. 2017, 132, 162–169. [Google Scholar] [CrossRef]
- Jiang, F.; Zhao, F.; Ma, K.; Li, D.; Sun, H. Mapping the Forest Canopy Height in Northern China by Synergizing ICESat-2 with Sentinel-2 Using a Stacking Algorithm. Remote Sens. 2021, 13, 1535. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.; Guo, Z. Estimation of tree height and aboveground biomass of coniferous forests in North China using stereo ZY-3, multispectral Sentinel-2, and DEM data. Ecol. Indic. 2021, 126, 107645. [Google Scholar] [CrossRef]
- Wu, J.; Wang, T.; Pan, K.; Li, W.; Huang, X. Assessment of forest damage caused by an ice storm using multi-temporal remote-sensing images: A case study from Guangdong Province. Int. J. Remote Sens. 2016, 37, 3125–3142. [Google Scholar] [CrossRef]
- Jucker, T.; Caspersen, J.; Chave, J.; Antin, C.; Barbier, N.; Bongers, F.; Dalponte, M.; van Ewijk, K.Y.; Forrester, D.I.; Haeni, M.; et al. Allometric equations for integrating remote sensing imagery into forest monitoring programmes. Glob. Chang. Biol. 2016, 23, 177–190. [Google Scholar] [CrossRef]
- Mauro, F.; Molina, I.; García-Abril, A.; Valbuena, R.; Ayuga-Téllez, E. Remote sensing estimates and measures of uncertainty for forest variables at different aggregation levels. Environmetrics 2016, 27, 225–238. [Google Scholar] [CrossRef]
- Gibson, R.K.; Collins, L. A remote sensing approach to mapping fire severity in south-eastern Australia using sentinel 2 and random forest. Remote Sens. Environ. 2020, 240, 111702. [Google Scholar] [CrossRef]
- Miller, Z.M.; Hupy, J.; Chandrasekaran, A.; Shao, G.; Fei, S. Application of Postprocessing Kinematic Methods with UAS Remote Sensing in Forest Ecosystems. J. For. 2021, 119, 454–466. [Google Scholar] [CrossRef]
- Elhenawy, M.; Rakotonirainy, A.; Young, K.; Haworth, N.; Grazbieta, R.; Williamson, A. Detecting Driver Distraction in the ANDS Data using Pre-trained Models and Transfer Learning. In Proceedings of the Australasian Road Safety Conference, Adelaide, SA, Australia, 25–27 September 2019. [Google Scholar]
- Petegrosso, R.; Park, S.; Hwang, T.H.; Kuang, R. Transfer learning across ontologies for phenome–genome association prediction. Bioinformatics 2016, 33, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Gao, S.; Li, C.; Wang, L.; Zhang, Q. Transfer learning with attention mechanism as predicting tool for dynamic adsorption of ammonia on MCM-41 matrix materials. Environ. Prog. Sustain. Energy 2023, 42, 14004. [Google Scholar] [CrossRef]





| Regions | Tree Species | Acquisition Date | Number of Samples | MAX (t/ha) | MIN (t/ha) | Mean (t/ha) | Coefficient of Variation (%) |
|---|---|---|---|---|---|---|---|
| XT | Chinese fir | 2009 | 98 | 115.67 | 26.54 | 47.39 | 25.24 |
| 2014 | 622 | 89.40 | 26.54 | 47.37 | 29.15 | ||
| 2019 | 98 | 86.41 | 27.18 | 46.43 | 25.26 | ||
| oak | 2009 | 107 | 139.86 | 14.27 | 49.05 | 45.01 | |
| 2014 | 411 | 149.76 | 12.36 | 51.63 | 45.51 | ||
| 2019 | 108 | 114.38 | 16.79 | 53.09 | 38.64 | ||
| WC | Chinese fir | 2014 | 65 | 105.70 | 23.58 | 48.04 | 32.11 |
| oak | 2014 | 64 | 87.84 | 20.59 | 48.43 | 33.57 | |
| NX | Chinese fir | 2014 | 93 | 74.37 | 29.53 | 47.48 | 30.46 |
| oak | 2014 | 95 | 124.67 | 17.99 | 55.09 | 36.85 |
| Area of Coverage | Image Acquisition Time | Image Category | Product Level |
|---|---|---|---|
| XT | 2009 | Landsat 5 TM | L1 |
| 2014 | Landsat 8 OLI | L1 | |
| 2019 | Landsat 8 OLI | L1 | |
| NX | 2014 | Landsat 8 OLI | L1 |
| WC | 2014 | Landsat 8 OLI | L1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Lin, H.; Long, J.; Liu, Z.; Yang, P.; Zhang, T. Evaluating the Transferability of Spectral Variables and Prediction Models for Mapping Forest Aboveground Biomass Using Transfer Learning Methods. Remote Sens. 2023, 15, 5358. https://doi.org/10.3390/rs15225358
Chen L, Lin H, Long J, Liu Z, Yang P, Zhang T. Evaluating the Transferability of Spectral Variables and Prediction Models for Mapping Forest Aboveground Biomass Using Transfer Learning Methods. Remote Sensing. 2023; 15(22):5358. https://doi.org/10.3390/rs15225358
Chicago/Turabian StyleChen, Li, Hui Lin, Jiangping Long, Zhaohua Liu, Peisong Yang, and Tingchen Zhang. 2023. "Evaluating the Transferability of Spectral Variables and Prediction Models for Mapping Forest Aboveground Biomass Using Transfer Learning Methods" Remote Sensing 15, no. 22: 5358. https://doi.org/10.3390/rs15225358
APA StyleChen, L., Lin, H., Long, J., Liu, Z., Yang, P., & Zhang, T. (2023). Evaluating the Transferability of Spectral Variables and Prediction Models for Mapping Forest Aboveground Biomass Using Transfer Learning Methods. Remote Sensing, 15(22), 5358. https://doi.org/10.3390/rs15225358






