Determining the Capability of the Tree-Based Pipeline Optimization Tool (TPOT) in Mapping Parthenium Weed Using Multi-Date Sentinel-2 Image Data
Abstract
:1. Introduction
2. Materials and Methods
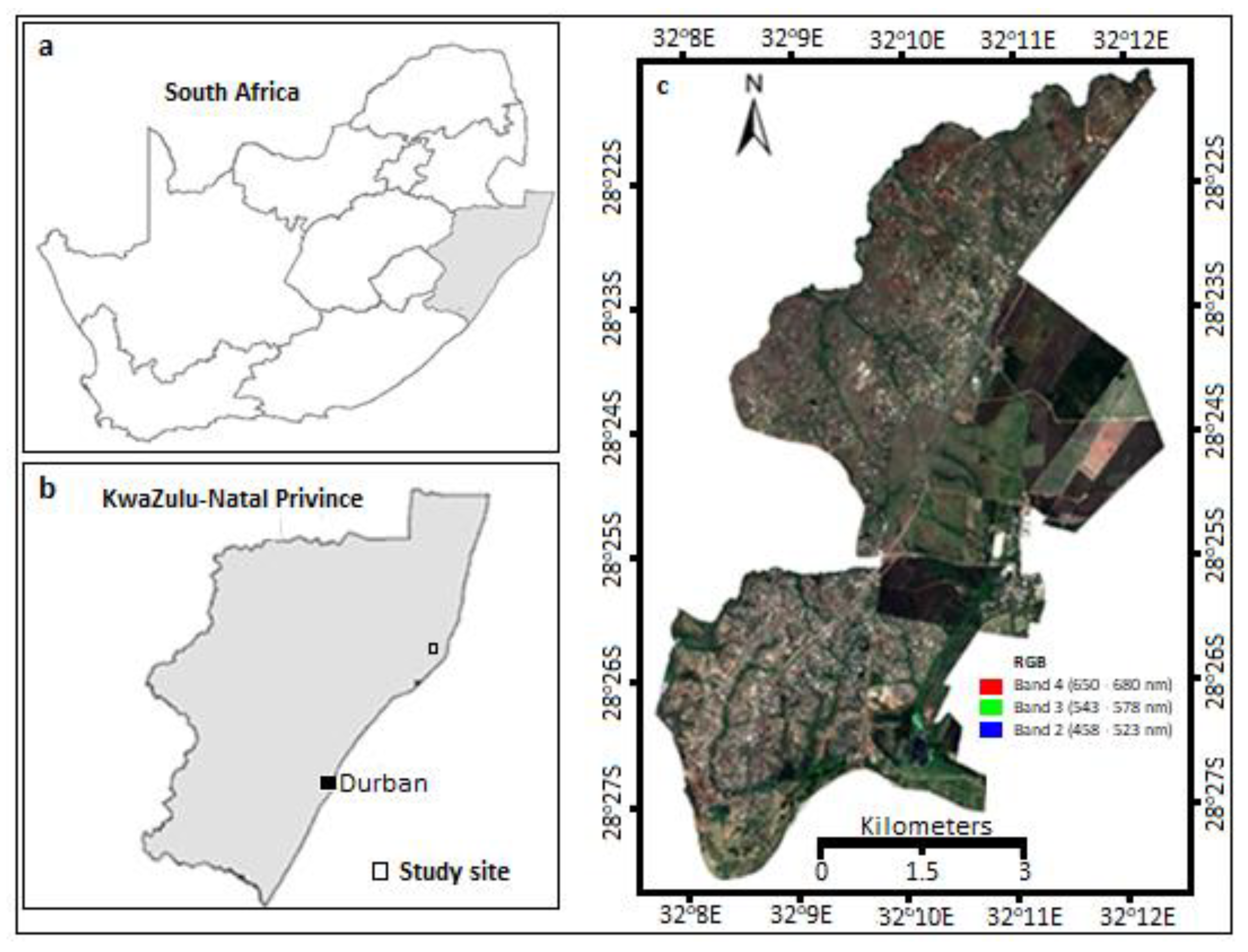
2.1. The Study Area
2.2. Reference Data
2.3. Acquistion of Multi-Date Sentinel-2 Images and Pre-Processing
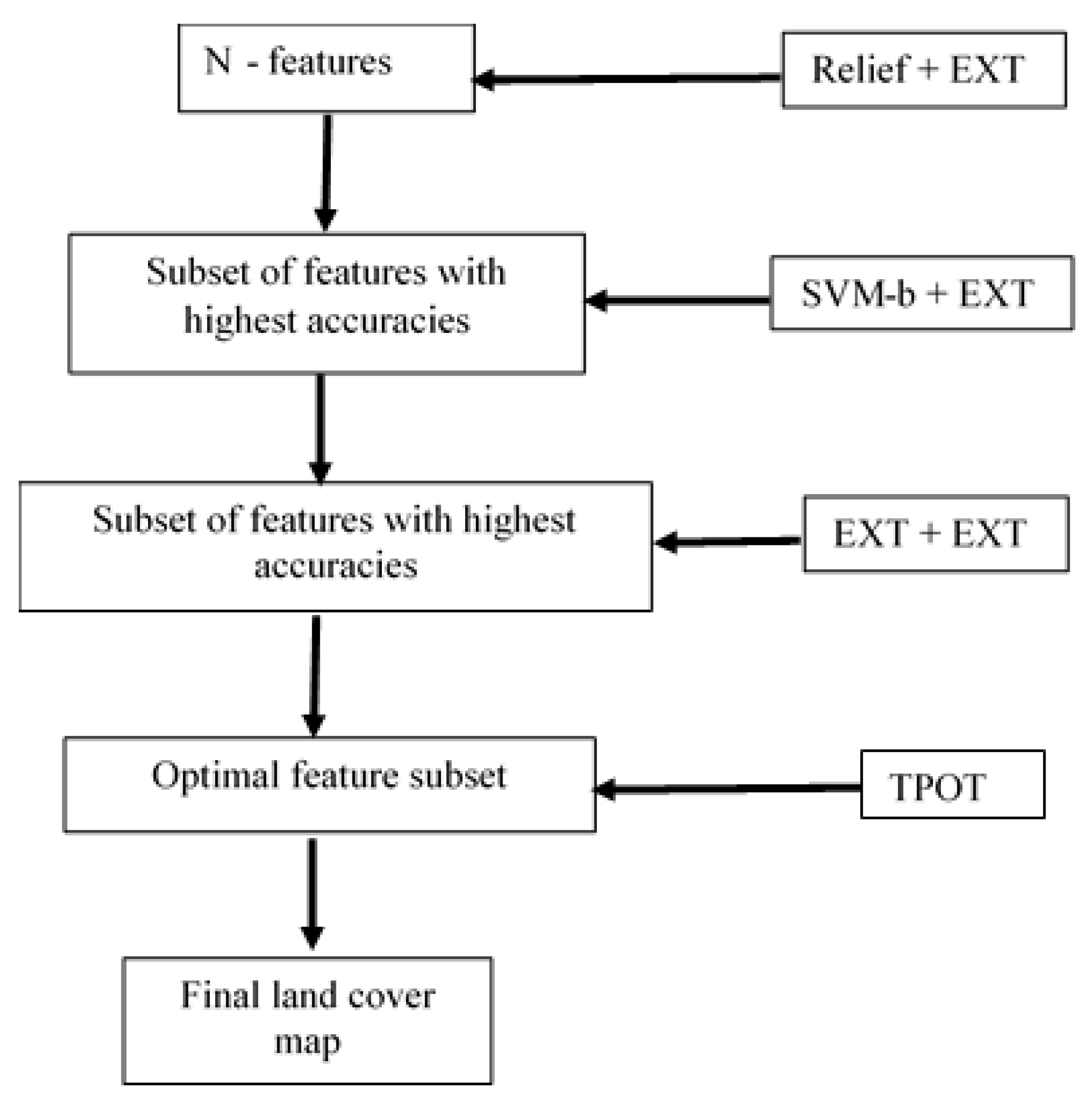
2.4. Feature Selection and Classification Methods
2.4.1. ReliefF
2.4.2. Support Vector Machine—Backward
2.4.3. ExtraTrees Classifier
2.4.4. The Tree-Based Pipeline Optimization Tool (TPOT)
2.4.5. The ReliefF-Svmb-EXT-TPOT System
2.4.6. Model Assessment Metrics
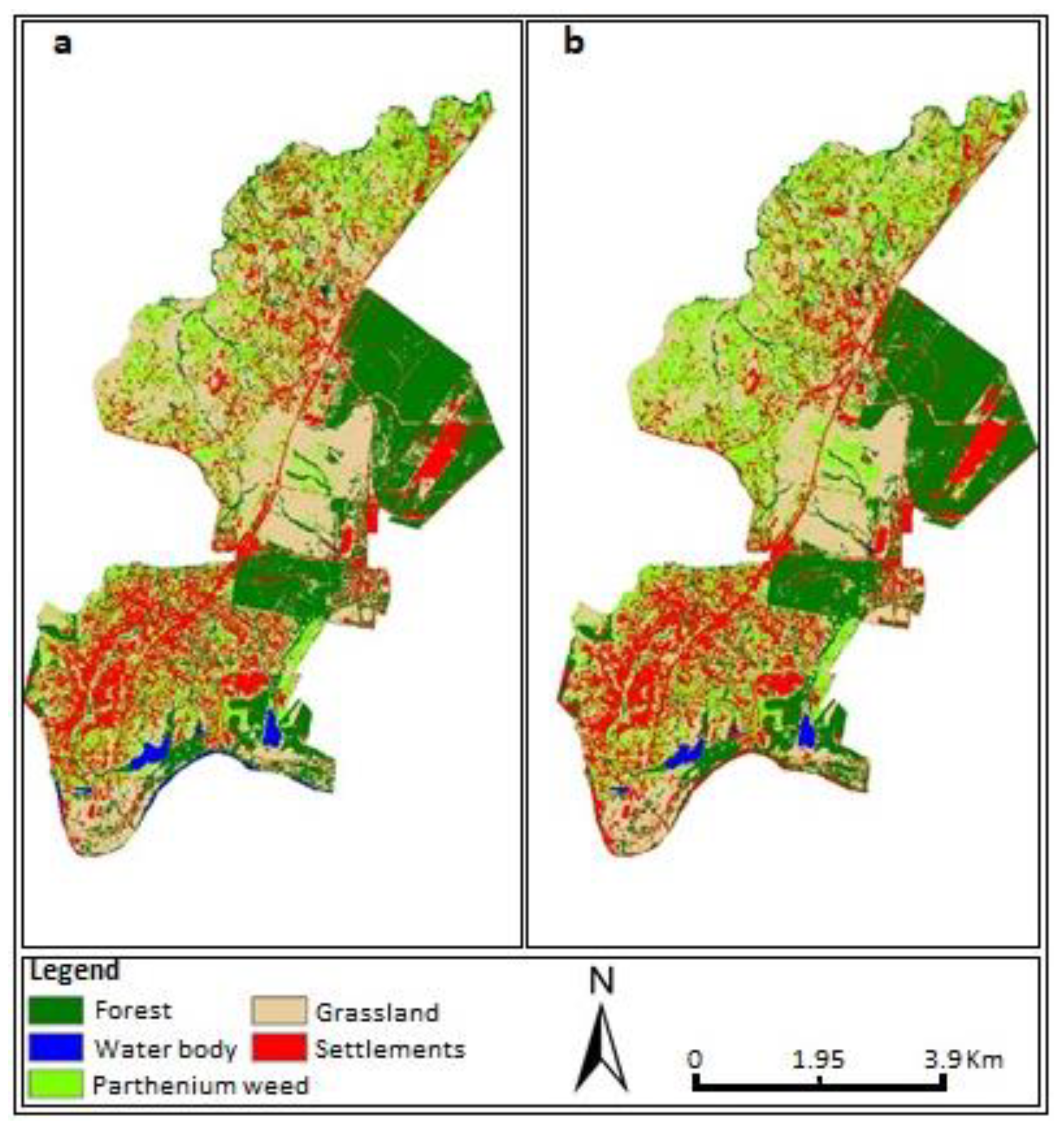
3. Results
4. Discussion
5. Conclusions
- (a)
- The TPOT can work well on a high-dimensional dataset, such as multi-date Sentinel-2 imagery, but at a higher computational cost;
- (b)
- Combining a hybrid feature selection method with the TPOT decreases the computational costs of the TPOT on a high-dimensional dataset, with a slight increase in the classification accuracies.
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| Models | Recommended Pipelines |
|---|---|
| TPOT | ExtraTreesClassifier(SelectPercentile(PCA(RobustScaler(input_matrix), iterated_power = 8, svd_solver = randomized), percentile = 51), bootstrap = False, criterion = entropy, max_features = 0.15000000000000002, min_samples_leaf = 1, min_samples_split = 4, n_estimators = 100) |
| ReliefF-Svmb-EXT-TPOT | XGBClassifier(MLPClassifier(PCA(ZeroCount(StandardScaler(input_matrix)), iterated_power = 8, svd_solver = randomized), alpha = 0.01, learning_rate_init = 0.1), learning_rate = 0.1, max_depth = 4, min_child_weight = 2, n_estimators = 100, n_jobs = 1, subsample = 0.7500000000000001, verbosity = 0) |
References
- Swati, G.; Halder, S.; Ganguly, A.; Chatterjee, P. Review on Parthenium hysterphorus as a potential energy source. Renew. Sustain. Energy Rev. 2013, 20, 420–429. [Google Scholar] [CrossRef]
- Lawrence, R.L.; Wood, S.; Sheley, R. Mapping invasive plants using hyperspectral imagery and Breiman Cutler classifications (RandomForest). Remote Sens. Environ. 2006, 100, 356–362. [Google Scholar] [CrossRef]
- Vuolo, F.; Neuwirth, M.; Immitzer, M.; Atzberger, C.; Ng, W. How much does multi-temporal Sentinel-2 data improve crop type classification? Int. J. Appl. Earth Obs. Geoinf. 2018, 72, 122–130. [Google Scholar] [CrossRef]
- Tottrup, C. Improving tropical forest mapping using multi-date Landsat TM data and pre-classification image smoothing. Int. J. Remote Sens. 2004, 25, 717–730. [Google Scholar] [CrossRef]
- Olson, R.S.; Moore, J.H. TPOT: A tree-based pipeline optimization tool for automating machine learning. JMLR Workshop Conf. Proc. 2016, 64, 66–74. [Google Scholar]
- Luo, G. A review of automatic selection methods for machine learning algorithms and hyper-parameter values. Netw. Modeling Anal. Health Inform. Bioinform. 2016, 5, 18. [Google Scholar] [CrossRef]
- Sohn, A.; Olson, R.; Moore, J. Toward the automated analysis of complex diseases in genome-wide association studies using genetic programming. In Proceedings of the Genetic and Evolutionary Computation Conference, Berlin, Germany, 15–19 July 2017; ACM: New York, NY, USA, 2017; pp. 489–496. [Google Scholar]
- Elshawi, M.; Sakr, S. Automated machine learning: State-of-the-art and open challenges. arXiv 2019, arXiv:1906.02287. [Google Scholar]
- Venkatesh, B.; Anuradha, J. A Hybrid Feature Selection Approach for Handling a High-Dimensional Data. Innovations in Computer Science and Engineering; Springer: Berlin/Heidelberg, Germany, 2019; pp. 365–373. [Google Scholar]
- Kganyago, M.; Odindi, J.; Mhangara, P. Selecting a subset of spectral bands for mapping invasive alien plants: A case of discriminating Parthenium hysterophorus using field spectroscopy data. Int. J. Remote Sens. 2017, 38, 5608–5625. [Google Scholar] [CrossRef]
- Rouhi, A.; Nezamabadi-pour, H. A hybrid feature selection approach based on ensemble method for high-dimensional data. In Proceedings of the 2017 2nd Conference on Swarm Intelligence and Evolutionary Computation (CSIEC), Kerman, Iran, 7–9 March 2017. [Google Scholar]
- Kiala, Z.; Mutanga, O.; Odindi, J.; Peerbhay, K. Feature Selection on Sentinel-2 Multispectral Imagery for Mapping a Landscape Infested by Parthenium Weed. Remote Sens. 2019, 11, 1892. [Google Scholar] [CrossRef] [Green Version]
- Robnik-Šikonja, M.; Kononenko, I. Theoretical and empirical analysis of ReliefF and RReliefF. Mach. Learn. 2003, 53, 23–69. [Google Scholar] [CrossRef] [Green Version]
- Geurts, P.; Ernst, D.; Wehenkel, L. Extremely randomized trees. Mach. Learn. 2006, 63, 3–42. [Google Scholar] [CrossRef] [Green Version]
- Henry, M.C. Comparison of single-and multi-date Landsat data for mapping wildfire scars in Ocala National Forest, Florida. Photogramm. Eng. Remote Sens. 2008, 74, 881–891. [Google Scholar] [CrossRef] [Green Version]
- Adelabu, S.; Mutanga, O.; Adam, E. Testing the reliability and stability of the internal accuracy assessment of random forest for classifying tree defoliation levels using different validation methods. Geocarto Int. 2015, 30, 810–821. [Google Scholar] [CrossRef]
- Congedo, L. Semi-automatic classification plugin documentation. Release 2016, 4, 29. [Google Scholar]
- QGIS Development Team. QGIS Geographic Information System, 2014. Open Source Geospatial Foundation Project. Available online: http://qgis.osgeo.org (accessed on 10 June 2018).
- Farrell, A.; Wan, G.; Rush, S.; Martin, J.; Belant, J.; Butler, A.; Godwin, D. Machine learning of large-scale spatial distributions of wild turkeys with high-dimensional environmental data. Ecol. Evol. 2019, 9, 5938–5949. [Google Scholar] [CrossRef]
- Zhu, Z.; Ong, Y.; Dash, M. Wrapper–filter feature selection algorithm using a memetic framework. IEEE Trans. Syst. Man Cybern. Part 2007, 37, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Kohavi, R.; John, G.H. Wrappers for feature subset selection. Artif. Intell. 1997, 97, 273–324. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Cheng, K.; Wang, S.; Morstatter, F.; Trevino, R.; Tang, J.; Liu, H. Feature selection: A data perspective. ACM Comput. Surv. 2017, 50, 94. [Google Scholar] [CrossRef] [Green Version]
- Fortin, F.-A.; Rainville, F.; Gardner, M.; Parizeau, M.; Gagne, C. DEAP: Evolutionary algorithms made easy. J. Mach. Learn. Res. 2012, 13, 2171–2175. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Lunetta, R.S.; Lyon, J.G. Remote Sensing and GIS Accuracy Assessmen; CRC Press: Boca Raton, FL, USA, 2004. [Google Scholar]
- Royimani, L.; Mutanga, O.; Odindi, J.; Kiala, K.; Sibanda, M.; Dube, T. Distribution of Parthenium hysterophoru L. with variation in rainfall using multi-year SPOT data and random forest classification. Remote Sens. Appl. Soc. Environ. 2018, 13, 215–223. [Google Scholar] [CrossRef]
- Kganyago, M.; Odindi, J.; Mhangara, P. Evaluating the capability of Landsat 8 OLI and SPOT 6 for discriminating invasive alien species in the African Savanna landscape. Int. J. Appl. Earth Obs. Geoinf. 2018, 67, 10–19. [Google Scholar] [CrossRef]
- Casady, G.M.; Hanley, R.; Seelan, S. Detection of leafy spurge (Euphorbia esula) using multi-date high-resolution satellite imagery. Weed Technol. 2005, 19, 462–467. [Google Scholar] [CrossRef]
- Thejas, G.; Joshi, S.; Iyengar, S.; Sunitha, N.; Badrinath, P. Mini-Batch Normalized Mutual Information: A Hybrid Feature Selection Method. IEEE Access 2019, 7, 116875–116885. [Google Scholar] [CrossRef]
- Lusa, L. Gradient boosting for high-dimensional prediction of rare events. Comput. Stat. Data Anal. 2017, 113, 19–37. [Google Scholar]
- Samat, A.; Persello, C.; Liu, S.; Li, E.; Miao, Z.; Abuduwaili, J. Classification of VHR multispectral images using extratrees and maximally stable extremal region-guided morphological profile. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2018, 11, 3179–3195. [Google Scholar] [CrossRef]
- Peralta, B.; Soto, A. Embedded local feature selection within mixture of experts. Inf. Sci. 2014, 269, 176–187. [Google Scholar] [CrossRef]
- Hsu, C.-W.; Chung, C.; Lin, C. A practical guide to support vector classification. In Technical Report; Department of Computer Science and Information Engineering, University of National Taiwan: Taipei, Taiwan, 2003; pp. 1–12. [Google Scholar]
- Lin, X.; Yang, F.; Zhou, L.; Yin, P.; Kong, H.; Wing, W.; Lu, X.; Jia, L.; Wang, Q.; Xu, G. A support vector machine-recursive feature elimination feature selection method based on artificial contrast variables and mutual information. J. Chromatogr. B 2012, 910, 149–155. [Google Scholar] [CrossRef]
| Class | Number of GPS Points |
|---|---|
| Forest | 100 |
| Water body | 70 |
| Parthenium weed | 90 |
| Grassland | 92 |
| Settlement | 95 |
| Band | Spectral Band/Region | Pixel Size (m) | Wavelength Range (nm) |
|---|---|---|---|
| 1 | Coastal aerosol | 60 | 430–457 |
| 2 | Blue | 10 | 448–546 |
| 3 | Green | 10 | 538–583 |
| 4 | Red | 10 | 646–684 |
| 5 | Vegetation red edge | 20 | 694–713 |
| 6 | Vegetation red edge | 20 | 731–749 |
| 7 | Vegetation red edge | 20 | 769–797 |
| 8 | NIR | 10 | 763–908 |
| 8a | Vegetation red edge | 20 | 848–881 |
| 9 | Water vapour | 60 | 932–958 |
| 10 | SWIR-Cirrus | 60 | 1336–1411 |
| 11 | SWIR | 20 | 1542–1685 |
| 12 | SWIR | 20 | 2081–2323 |
| Parthenium Weed | Forest | Water Body | Grassland | Settlements | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Methods | PA | UA | PA | UA | PA | UA | PA | UA | PA | UA | OA |
| TPOT—Hybrid | 88% | 78% | 100% | 97% | 100% | 100% | 84% | 93% | 93% | 97% | 92.6% |
| TPOT—alone | 85% | 81% | 97% | 93% | 100% | 95% | 89% | 89% | 91% | 100% | 91.9% |
| (a) | Reference Data | ||||||
|---|---|---|---|---|---|---|---|
| Forest | Water Body | Parthenium Weed | Grassland | Settlements | Total | ||
| Classified data | Forest | 29 | 0 | 0 | 1 | 0 | 30 |
| Water body | 0 | 21 | 0 | 0 | 0 | 21 | |
| Parthenium weed | 0 | 0 | 21 | 4 | 2 | 27 | |
| Grassland | 0 | 0 | 2 | 26 | 0 | 28 | |
| Settlements | 0 | 0 | 1 | 0 | 28 | 29 | |
| Total | 29 | 21 | 24 | 31 | 30 | 135 | |
| (b) | Reference Data | ||||||
| Forest | Water Body | Parthenium Weed | Grassland | Settlements | Total | ||
| Classified data | Forest | 28 | 0 | 1 | 1 | 0 | 30 |
| Water body | 0 | 20 | 0 | 0 | 1 | 21 | |
| Parthenium weed | 1 | 0 | 22 | 2 | 2 | 27 | |
| Grassland | 0 | 0 | 3 | 25 | 0 | 28 | |
| Settlements | 0 | 0 | 0 | 0 | 29 | 29 | |
| Total | 29 | 20 | 26 | 28 | 32 | 135 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiala, Z.; Odindi, J.; Mutanga, O. Determining the Capability of the Tree-Based Pipeline Optimization Tool (TPOT) in Mapping Parthenium Weed Using Multi-Date Sentinel-2 Image Data. Remote Sens. 2022, 14, 1687. https://doi.org/10.3390/rs14071687
Kiala Z, Odindi J, Mutanga O. Determining the Capability of the Tree-Based Pipeline Optimization Tool (TPOT) in Mapping Parthenium Weed Using Multi-Date Sentinel-2 Image Data. Remote Sensing. 2022; 14(7):1687. https://doi.org/10.3390/rs14071687
Chicago/Turabian StyleKiala, Zolo, John Odindi, and Onisimo Mutanga. 2022. "Determining the Capability of the Tree-Based Pipeline Optimization Tool (TPOT) in Mapping Parthenium Weed Using Multi-Date Sentinel-2 Image Data" Remote Sensing 14, no. 7: 1687. https://doi.org/10.3390/rs14071687
APA StyleKiala, Z., Odindi, J., & Mutanga, O. (2022). Determining the Capability of the Tree-Based Pipeline Optimization Tool (TPOT) in Mapping Parthenium Weed Using Multi-Date Sentinel-2 Image Data. Remote Sensing, 14(7), 1687. https://doi.org/10.3390/rs14071687









