Mapping Restoration Activities on Dirk Hartog Island Using Remotely Piloted Aircraft Imagery
Abstract
:1. Introduction
1.1. Remotely Piloted Aircraft Environmental Monitoring
1.2. Object-Based Image Analysis
1.3. Dirk Hartog Island History
1.4. Objectives
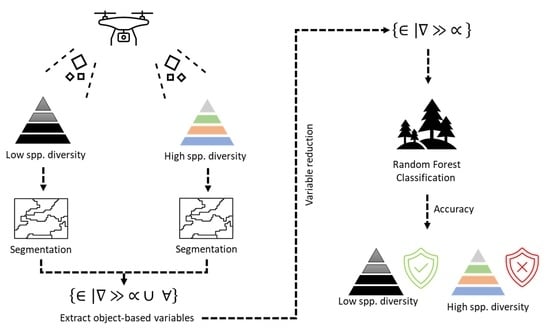
2. Materials and Methods
2.1. Study Area
2.2. Reference Data and Study Species
2.3. Object-Based Variables
2.4. Flight Height
2.5. Classification
3. Results
3.1. Object-Based Variables
3.2. Flight Height
3.3. Classification
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Garnett, S.; Latch, P.; Lindenmayer, D.; Woinarksi, J. Recovering Australian Threatened Species, 1st ed.; CSIRO: Clayton South, VIC, Australia, 2018. [Google Scholar]
- Ochoa, K.S.; Guo, Z. A framework for the management of agricultural resources with automated aerial imagery detection. Comput. Electron. Agric. 2019, 162, 53–69. [Google Scholar] [CrossRef]
- Buters, T.M.; Bateman, P.W.; Robinson, T.; Belton, D.; Dixon, K.W.; Cross, A.T. Methodological ambiguity and inconsistency constrain Unmanned Aerial Vehicles as a silver bullet for monitoring ecological restoration. Remote Sens. 2019, 11, 1180. [Google Scholar] [CrossRef] [Green Version]
- Lillesand, T.M.; Kiefer, R.W.; Chipman, J. Remote Sensing and Image Interpretation; John Wiley & Sons: New York, NY, USA, 2015. [Google Scholar]
- Getzin, S.; Wiegand, K.; Schöning, I. Assessing biodiversity in forests using very high-resolution images and unmanned aerial vehicles. Methods Ecol. Evol. 2011, 3, 397–404. [Google Scholar] [CrossRef]
- Robinson, T.P.; Wardell-Johnson, G.W.; Pracilio, G.; Brown, C.; Corner, R.; Klinken, R.D.v. Testing the discrimination and detection limits of WorldView-2 imagery on a challenging invasive plant target. Int. J. Appl. Earth Obs. Geoinf. 2016, 44, 23–30. [Google Scholar] [CrossRef]
- Baena, S.; Moat, J.; Whaley, O.; Boyd, D.S. Identifying species from the air: UAVs and the very high resolution challenge for plant conservation. PLoS ONE 2017, 12, e0188714. [Google Scholar] [CrossRef] [Green Version]
- Kumar, U.; Dasgupta, A.; Mukhopadhyay, C.; Ramachandra, T.V. Examining the Effect of Ancillary and Derived Geographical Data on Improvement of Per-Pixel Classification Accuracy of Different Landscapes. J. Indian Soc. Remote Sens. 2018, 46, 407–422. [Google Scholar] [CrossRef]
- Zhang, X.; Xiao, P.; Feng, X. An unsupervised evaluation method for remotely sensed imagery segmentation. IEEE 2012, 9, 156–160. [Google Scholar] [CrossRef]
- Bolyn, C.; Michez, A.; Gaucher, P.; Lejeune, P.; Bonnet, S. Forest mapping and species composition using supervised per pixel classification of Sentinel-2 imagery. Biotechnol. Agron. Société Environ. 2018, 22, 172–187. [Google Scholar] [CrossRef]
- Lu, D.; Weng, Q. Survey of image classification methods and techniques for improving classification performance. Int. J. Remote Sens. 2007, 28, 823–870. [Google Scholar] [CrossRef]
- Dronova, I. Object-based image analysis in wetland research: A review. Remote Sens. 2015, 7, 6380–6413. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C. Multiscale quantification of urban composition from EO-1/Hyperion data using object-based spectral unmixing. Int. J. Appl. Earth Obs. Geoinf. 2016, 47, 153–162. [Google Scholar] [CrossRef]
- Zhou, Z.; Ma, L.; Fu, T.; Zhang, G.; Yao, M.; Manchun. Change detection in coral reef environment using high-resolution images: Comparison of object-based and pixel-based paradigms. ISPRS Int. J. Geo-Inf. 2018, 7, 441. [Google Scholar] [CrossRef] [Green Version]
- Yuba, N.; Kawamura, K.; Yasuda, T.; Lim, J.; Yoshitoshi, R.; Watanabe, N.; Kurokawa, Y.; Maeda, T. Discriminating Pennisetum alopecuoides plants in a grazed pasture from unmanned aerial vehicles using object-based image analysis and random forest classifier. Grassl. Sci. 2020, 67, 73–82. [Google Scholar] [CrossRef]
- Ye, S.; Pontius, R.G.; Rakshit, R. A review of accuracy assessment for object-based image analysis: From per-pixel to per-polygon approaches. ISPRS J. Photogramm. Remote Sens. 2018, 141, 137–147. [Google Scholar] [CrossRef]
- Silveira, E.M.O.; Silv, S.H.G.; Acerbi-Junior, F.W.; Carvalho, M.C.; Carvalho, L.M.T.; Scolforo, J.R.S.; Wulder, M.A. Object-based random forest modelling of aboveground forest biomass outperforms a pixel-based approach in a heterogeneous and mountain tropical environment. Int. J. Appl. Earth Obs. Geoinf. 2019, 78, 175–188. [Google Scholar] [CrossRef]
- Wu, Q.; Zhong, R.; Zhao, W.; Fu, H.; Song, K. A comparison of pixel-based decision tree and object-based Support Vector Machine methods for land-cover classification based on aerial images and airborne lidar data. Int. J. Remote Sens. 2017, 38, 7176–7195. [Google Scholar] [CrossRef]
- Addink, E.A.; Jong, S.M.D.; Pebesma, E. The importance of scale in object-based mapping of vegetation parameters with hyperspectral imagery. Photogramm. Eng. Remote Sens. 2007, 73, 905–912. [Google Scholar] [CrossRef] [Green Version]
- Blaschke, T. Object based image analysis for remote sensing. ISPRS J. Photogramm. Remote Sens. 2010, 65, 2–16. [Google Scholar] [CrossRef] [Green Version]
- Baynes, A. The mammals of Shark Bay, Western Australia. In Research in Shark Bay, Report of the France-Australe Bicentenary Expedition Committee; Berry, P.F., Bradshaw, S.D., Wilson, B.R., Eds.; Western Australian Museum: Perth, Australia, 1990. [Google Scholar]
- McKenzie, N.L.; Hall, N.; Muir, W.P. Non-volant mammals of the southern Carnarvon Basin, Western Australia. Rec. West. Aust. Mus. Suppl. 2000, 479–510. [Google Scholar] [CrossRef]
- Algar, D.; Morris, K.; Asher, J.; Cowen, S. Dirk Hartog Island ‘Return to 1616’ Project—The first six years (2014 to 2019). Ecol. Manag. Restor. 2020, 21, 173–183. [Google Scholar] [CrossRef]
- Asher, J.; Morris, K. Dirk Hartog Island Biosecurity Implementation Plan—A Shared Responsibility; Department of Parks and Wildlife: Bentley, Australia, 2015. [Google Scholar]
- Diss, K. Dirk Hartog Island Turns back the Clock 400 Years to a Time before European Settlement. Available online: https://www.abc.net.au/news/2018-10-14/dirk-hartog-island-being-sent-400-years-back-in-time/10368926 (accessed on 19 October 2019).
- Heriot, S.; Asher, J.; Williams, M.R.; Moro, D. The eradication of ungulates (sheep and goats) from DirkHartog Island, Shark Bay World Heritage Area, Australia. Biol. Invasions 2019, 21, 1789–1805. [Google Scholar] [CrossRef]
- Cowen, S.; Rayner, K.; Sims, C.; Morris, K. Dirk Hartog Island National Park Ecological Restoration Project: Stage One-Trial Hare-Wallaby Translocations and Monitoring; Department of Biodiversity, Conservation and Attractions: Bentley, Australia, 2018. [Google Scholar]
- Valentine, L.E.; Bretz, M.; Ruthrof, K.X.; Fisher, R.; Hardy, G.E.S.J.; Fleming, P.A. Scratching beneath the surface: Bandicoot bioturbation contributes to ecosystem processes. Austral Ecol. 2017, 42, 265–276. [Google Scholar] [CrossRef] [Green Version]
- Palmer, B.J.; Valentine, L.E.; Page, M.; Hobbs, R.J. Translocations of digging mammals and their potential for ecosystem restoration: A review of goals and monitoring programmes. Mammal Rev. 2020, 50, 382–398. [Google Scholar] [CrossRef]
- Tregoning, P. Is the Australian Plate deforming? A space geodetic perspective. Geol. Soc. Am. 2003, 372, 41–48. [Google Scholar] [CrossRef]
- Guern, P.L.; Davaud, E. Recognition of ancient carbonate wind deposits: Lessons from a modern analogue, Chrissi Island, Crete. Sedimentology 2005, 52, 915–926. [Google Scholar] [CrossRef]
- Bowder, S. Before Dirk Hartog: Prehistoric archaeoidgical research in Shark Bay, Western Australia. Aust. Archaeol. 1990, 30, 46–57. [Google Scholar] [CrossRef]
- Harvey, A.; Johnson, M.E.; Harvey, R. Heterozoan carbonate-enriched beach sand and coastal dunes—with particular reference to rhodoliths, Dirk Hartog Island, Shark Bay, Western Australia. Facies 2018, 64, 1–18. [Google Scholar] [CrossRef]
- Bureau of Meteorology. Climate Data Online. Available online: http://www.bom.gov.au/climate/data/ (accessed on 2 October 2019).
- Maryan, B. Herpetofauna of Dirk Hartog Island Shark Bay area, Western Australia. Herpetofauna 1996, 26, 8–11. [Google Scholar]
- VC Technology. Litchi, Version 4.14.0; VC Technology Ltd.: London, UK, 2019. [Google Scholar]
- Agisoft. Photoscan-pro, Version 1.4.2; Agisoft LLC: St Petersburg, Russia, 2018. [Google Scholar]
- ESRI. ArcGIS Pro; Environmental Systems Research Institute: Redlands, CA, USA, 2019. [Google Scholar]
- The R Foundation. R, Version 3.6.1; The R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- R Studio. RStudio, Version 1.2.1335; R Studio, Inc.: Boston, MA, USA, 2019. [Google Scholar]
- Trimble. eCognition, Version 9.5.1; Trimble Germany GmbH: Munich, Germany, 2019. [Google Scholar]
- Louhaichi, M.; Borman, M.M.; Johnson, D.E. Spatially Located Platform and Aerial Photography for Documentation of Grazing Impacts on Wheat. Geocarto Int. 2001, 16, 65–70. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Peerbhay, K.Y.; Mutanga, O.; Ismail, R. Investigating the Capability of Few Strategically Placed Worldview-2 Multispectral Bands to Discriminate Forest Species in KwaZulu-Natal, South Africa. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 307–316. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Cohen, J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- CASA. Part 101 (Unmanned Aircraft and Rockets) Manual of Standards 2019 (as Amended); Compilation No. 3; Civil Aviation Safety Authority: Canberra, Australia, 2020. [Google Scholar]
- Landis, J.R.; Koch, G.G. The Measurement of Observer Agreement for Categorical Data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef] [Green Version]
- Chianucci, F.; Disperati, L.; Guzzi, D.; Bianchini, D.; Nardino, V.; Lastri, C.; Rindinella, A.; Corona, P. Estimation of canopy attributes in beech forests using true colour digital images from a small fixed-wing UAV. Int. J. Appl. Earth Obs. Geoinf. 2016, 47, 60–68. [Google Scholar] [CrossRef] [Green Version]
- Emery, S.M. Succession: A Closer Look. Nat. Educ. Knowl. 2010, 3, 45. [Google Scholar]
- Nogueira, K.; Santos, J.A.d.; Menini, N.; Silva, T.S.F.; Morellato, L.P.C.; Torres, R.d.S. Spatio-temporal vegetation pixel classification by using convolutional networks. IEEE Geosci. Remote Sens. Lett. 2019, 1, 1–5. [Google Scholar] [CrossRef]
- Alberto, R.T.; Serrano, S.C.; Damian, G.B.; Camaso, E.E.; Celestino, A.B.; Hernando, P.J.C.; Isip, M.F.; Orge, K.M.; Quinto, M.J.C.; Tagaca, R.C. Object based agricultural land cover classification map of shadowed areas from aerial image and LiDAR data using Support Vector Machine. In Proceedings of the XXIII ISPRS Congress, Prague, Czech Republic, 12–19 July 2016; pp. 45–50. [Google Scholar]
- Babar, M.A.; Ginkel, M.v.; Klatt, A.R.; Prasad, B.; Reynolds, M.P. The potential of using spectral reflectance indices to estimate yield in wheat grown under reduced irrigation. Euphytica 2006, 150, 155–172. [Google Scholar] [CrossRef]
- Niphadkar, M.; Nagendra, H.; Tarantino, C.; Adamo, M.; Blonda, P. Comparing Pixel and Object-Based Approaches to Map an Understorey Invasive Shrub in Tropical Mixed Forests. Front. Plant Sci. 2017, 8, 892. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Luo, M.; Xu, L.; Zhou, X.; Ren, J.; Zhou, J. Object-based random forest classification of land cover from remotely sensed imagery for industrial and mining reclamation. In Proceedings of the ISPRS TC III Mid Term Symposium ‘Developments, Technologies and Applications in Remote Sensing’, Beijing, China, 7–10 May 2018. [Google Scholar]
- Jiao, L.; Liu, Y. Analyzing the shape characteristics of land use classes in remote sensing imagery. In Proceedings of the XXII ISPRS Congress, Melbourne, VIC, Australia, 25 August–1 September 2012; pp. 135–140. [Google Scholar]
- Memarian, H.; Balasundram, S.K.; Khosla, R. Comparison between pixel- and object-based image classification of a tropical landscape using Système Pour l’Observation de la Terre-5 imagery. J. Appl. Remote Sens. 2013, 7, 073512. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Liu, X.; Chen, Y.; Liang, X. Land-cover mapping using Random Forest classification and incorporating NDVI time-series and texture: A case study of central Shandong. Int. J. Remote Sens. 2018, 39, 8703–8723. [Google Scholar] [CrossRef]
- Lu, D.; Hetrick, S.; Moran, E. Impervious surface mapping with Quickbird imagery. Int. J. Remote Sens. 2011, 32, 2519–2533. [Google Scholar] [CrossRef] [Green Version]
- Kelly, M.; Blanchard, S.D.; Kersten, E.; Koy, K. Terrestrial remotely sensed imagery in support of public health: New avenues of research using object-based image analysis. Remote Sens. 2011, 3, 2321–2345. [Google Scholar] [CrossRef] [Green Version]
- Kiiveri, H.T.; Caccetta, P.; Evans, F. Use of conditional probability networks for environmental monitoring. Int. J. Remote Sens. 2010, 22, 1173–1190. [Google Scholar] [CrossRef]
- Zhou, K.; Cheng, T.; Zhu, Y.; Cao, W.; Ustin, S.L.; Zheng, H.; Yao, X.; Tian, Y. Assessing the impact of spatial resolution on the estimation of leaf nitrogen concentration over the full season of paddy rice using near-surface imaging spectroscopy data. Front. Plant Sci. 2018, 9, 964. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Candón, D.; Castro, A.I.D.; López-Granados, F. Assessing the accuracy of mosaics from unmanned aerial vehicle (UAV) imagery for precision agriculture purposes in wheat. Precis. Agric. 2014, 15, 44–56. [Google Scholar] [CrossRef] [Green Version]
- Anderson, K.; Gaston, K.J. Lightweight unmanned aerial vehicles will revolutionize spatial ecology. Front. Ecol. Environ. 2013, 11, 138–146. [Google Scholar] [CrossRef] [Green Version]
- Laliberte, A.S.; Rango, A. Incorporation of texture, intensity, hue, and saturation for rangeland monitoring with unmanned aircraft imagery. In Proceedings of the ISPRS XXXVIII-4/C1 GEOgraphic Object Based Image Analysis (GEOBIA) for the 21st Century ‘Pixels, Objects, Intelligence’, Calgary, Canada, 5–8 August 2008. [Google Scholar]
- El-naggar, A.M. Determination of optimum segmentation parameter values for extracting building from remote sensing images. Alex. Eng. J. 2018, 57, 3089–3097. [Google Scholar] [CrossRef]
- Dalponte, M.; Ørka, H.; Gobakken, T.; Gianelle, D.; Næsset, E. Tree species classification in boreal forests with hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2013, 51, 2632–2645. [Google Scholar] [CrossRef]
- Raczko, E.; Zagajewski, B. Comparison of support vector machine, random forest and neural network classifiers for tree species classification on airborne hyperspectral APEX images. Eur. J. Remote Sens. 2017, 50, 144–154. [Google Scholar] [CrossRef] [Green Version]
- Paczkowska, G. Acanthocarpus Preissii Lehm. Available online: https://florabase.dpaw.wa.gov.au/browse/profile/1208 (accessed on 27 October 2019).
- Paczkowska, G. Threlkeldia Diffusa R.Br. Coast Bonefruit. Available online: https://florabase.dpaw.wa.gov.au/browse/profile/2644 (accessed on 27 October 2019).
- Coleman, H. Triodia Plurinervata N.T.Burb. Available online: https://florabase.dpaw.wa.gov.au/browse/profile/694 (accessed on 27 October 2019).
- Paczkowska, G. Cenchrus Ciliaris L. Buffel Grass. Available online: https://florabase.dpaw.wa.gov.au/browse/profile/258 (accessed on 27 October 2019).
- Snavely, R.A.; Uyeda, K.A.; Stow, D.A.; O’Leary, J.F.; Lambert, J. Mapping vegetation community types in a highly disturbed landscape: Integrating hierarchical object-based image analysis with lidar-derived canopy height data. Int. J. Remote Sens. 2019, 40, 4384–4400. [Google Scholar] [CrossRef]
- Spooner, A. Atriplex Vesicaria Benth. Bladder Saltbush. Available online: https://florabase.dpaw.wa.gov.au/browse/profile/2481 (accessed on 26 October 2019).
- Paczkowska, G. Exocarpos aphyllus R.Br. Leafless Ballart. Available online: https://florabase.dpaw.wa.gov.au/browse/profile/10977 (accessed on 27 October 2019).
- Mallen-Cooper, M.; Nakagawa, S.; Eldridge, D.J. Global meta-analysis of soil-disturbing vertebrates reveals strong effects on ecosystem patterns and processes. Glob. Ecol. Biogeogr. 2019, 28, 661–679. [Google Scholar] [CrossRef]
- Travers, S.K.; Eldridge, D.J.; Koen, T.B.; Soliveres, S. Animal foraging pit soil enhances the performance of a native grass under stressful conditions. Plant Soil 2012, 352, 341–351. [Google Scholar] [CrossRef]
- Grossman, B.F.; Hayward, M.W.; Gibb, H. An experimental test of multi-scalar impacts of digging mammal reintroductions on invertebrate burrows. Soil Biol. Biochem. 2019, 132, 101–110. [Google Scholar] [CrossRef]
- Louw, M.A.; Roux, P.C.l.; Meyer-Milne, E.; Haussmann, N.S. Mammal burrowing in discrete landscape patches further increases soil and vegetation heterogeneity in an arid environment. J. Arid Environ. 2017, 141, 68–75. [Google Scholar] [CrossRef]





| Variable Group | Variable | Study Site 1 VIP Score | Study Site 2 VIP Score |
|---|---|---|---|
| Mean red band | 1.76 | 1.17 | |
| Reflectance | Mean green band | 1.58 | 1.10 |
| Mean blue band | 1.79 | 1.24 | |
| Spectral index–Green Leaf Algorithm (GLA) | Mean GLA | 0.25 | 0.80 |
| Median GLA | 0.06 | 0.56 | |
| Maximum GLA | 1.58 | 0.98 | |
| GLA 90th percentile value | 1.53 | 0.97 | |
| Height–Canopy Height Model (CHM) | Mean CHM | 0.74 | 0.80 |
| Median CHM | 1.18 | 1.43 | |
| Minimum CHM | 0.88 | 1.01 | |
| Maximum CHM | 1.35 | 1.68 | |
| CHM 90th percentile value | 1.30 | 1.61 | |
| Texture | Mean | 0.11 | 0.79 |
| Correlation | 0.65 | 0.68 | |
| Contrast | 0.35 | 0.76 | |
| Homogeneity | 0.52 | 1.24 | |
| Entropy | 0.51 | 0.74 | |
| Shape | Roundness | 0.02 | 0.53 |
| Compactness | 0.00 | 0.51 | |
| Length/width | 0.04 | 0.27 | |
| Area per pixel | 0.03 | 0.75 |
| A. ligulata | A. vesicaria | Ground | T. plurinervata | S. spinescens | A. preissii | A. cuneiformis | C. ciliaris | E. aphyllus | T. diffusa | P. phillyreoides | Comm Error (%) | ||
| (A) | A. ligulata | 268 | 31 | 54 | 3 | - | 23 | 123 | 2 | 89 | 9 | 4 | 55.8 |
| A. vesicaria | 28 | 59 | 129 | 11 | - | 26 | 33 | 1 | 10 | 26 | 2 | 81.8 | |
| Ground | 12 | 22 | 838 | 7 | - | 16 | 18 | 2 | 14 | 11 | 1 | 10.9 | |
| T. plurinervata | 4 | 8 | 48 | 14 | - | 12 | 17 | 1 | 0 | 19 | 0 | 88.6 | |
| S. spinescens | - | - | - | - | - | - | - | - | - | - | - | ||
| A. preissii | 17 | 30 | 60 | 2 | - | 62 | 35 | 0 | 1 | 10 | 3 | 71.8 | |
| A. cuneiformis | 55 | 12 | 53 | 2 | - | 10 | 412 | 2 | 27 | 9 | 1 | 29.2 | |
| C. ciliaris | 1 | 4 | 37 | 5 | - | 5 | 6 | 17 | 1 | 13 | 0 | 80.9 | |
| E. aphyllus | 162 | 6 | 1 | 1 | - | 9 | 53 | 1 | 155 | 4 | 2 | 60.7 | |
| T. diffusa | 2 | 14 | 58 | 9 | - | 12 | 30 | 2 | 3 | 11 | 0 | 90.5 | |
| P. phillyreoides | 10 | 4 | 0 | 0 | - | 6 | 18 | 0 | 0 | 4 | 1 | 97.7 | |
| Om error (%) | 52.1 | 68.9 | 34.4 | 74.1 | - | 65.8 | 44.7 | 37.0 | 48.3 | 90.5 | 92.9 | ||
| Overall accuracy (%) | 53.0 | ||||||||||||
| (B) | A. ligulata | 1274 | 26 | 25 | 80 | 248 | 22.9 | ||||||
| A. vesicaria | 34 | 70 | 0 | 28 | 139 | 74.2 | |||||||
| Ground | 4 | 0 | 212 | 7 | 0 | 3.6 | |||||||
| T. plurinervata | 16 | 1 | 34 | 317 | 2 | 14.3 | |||||||
| S. spinescens | 145 | 64 | 18 | 50 | 294 | 48.51 | |||||||
| Om error (%) | 13.3 | 56.5 | 26.6 | 24.2 | 56.9 | ||||||||
| Overall accuracy (%) | 70.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wilson, L.; van Dongen, R.; Cowen, S.; Robinson, T.P. Mapping Restoration Activities on Dirk Hartog Island Using Remotely Piloted Aircraft Imagery. Remote Sens. 2022, 14, 1402. https://doi.org/10.3390/rs14061402
Wilson L, van Dongen R, Cowen S, Robinson TP. Mapping Restoration Activities on Dirk Hartog Island Using Remotely Piloted Aircraft Imagery. Remote Sensing. 2022; 14(6):1402. https://doi.org/10.3390/rs14061402
Chicago/Turabian StyleWilson, Lucy, Richard van Dongen, Saul Cowen, and Todd P. Robinson. 2022. "Mapping Restoration Activities on Dirk Hartog Island Using Remotely Piloted Aircraft Imagery" Remote Sensing 14, no. 6: 1402. https://doi.org/10.3390/rs14061402
APA StyleWilson, L., van Dongen, R., Cowen, S., & Robinson, T. P. (2022). Mapping Restoration Activities on Dirk Hartog Island Using Remotely Piloted Aircraft Imagery. Remote Sensing, 14(6), 1402. https://doi.org/10.3390/rs14061402