Early Detection of Dendroctonus valens Infestation with Machine Learning Algorithms Based on Hyperspectral Reflectance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Spectral Measurements and Pre-Processing
2.3. Hyperspectral Statistical Analyses
2.4. Spectral Vegetation Indices (SVIs)
2.5. Classification Procedures
2.5.1. Random Forest (RF)
2.5.2. Support Vector Machine (SVM)
2.5.3. Artificial Neural Network (ANN)
2.5.4. Optimizing Hyperparameters of the Three Classifiers
2.5.5. Evaluation of Model Performance
2.6. Variable Importance
3. Results
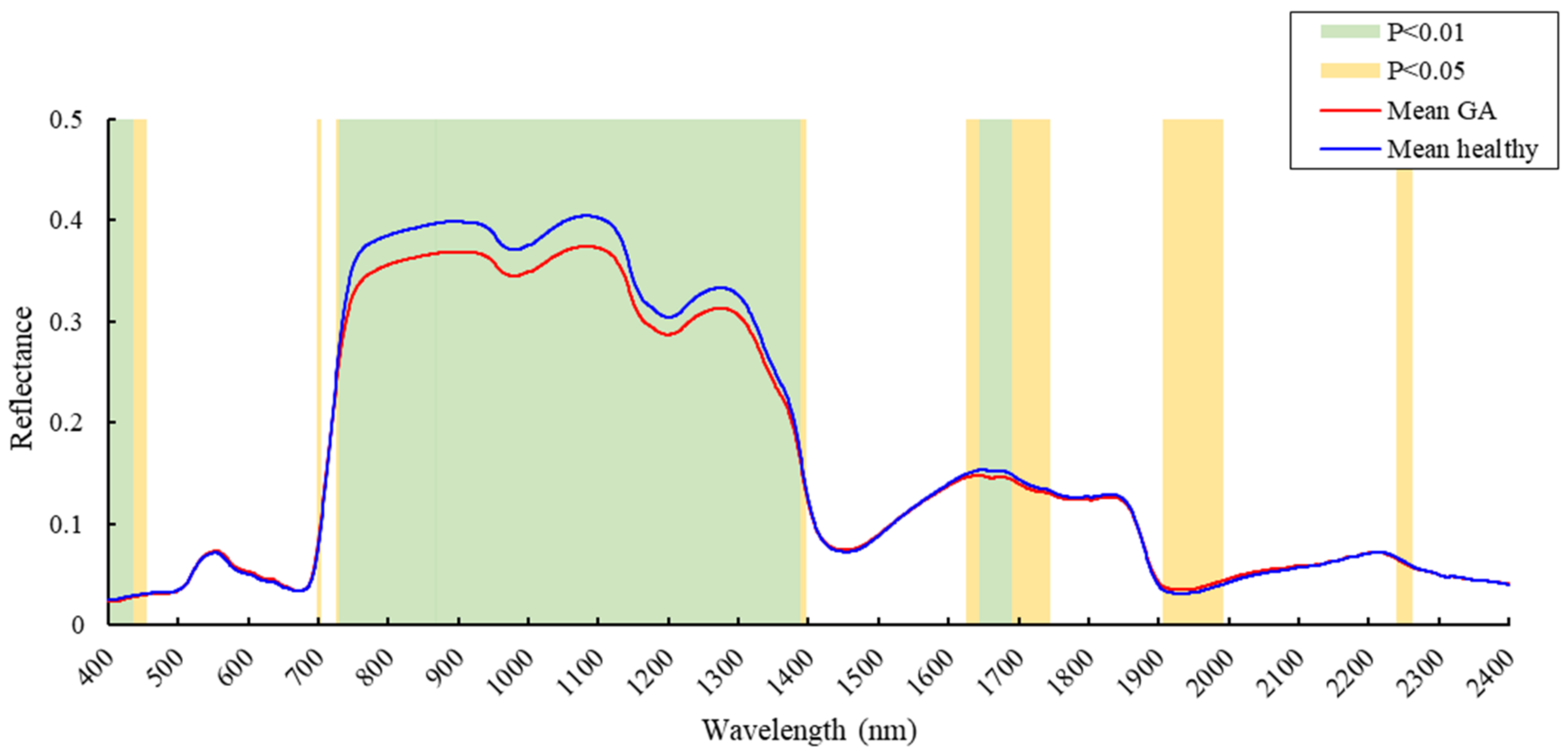
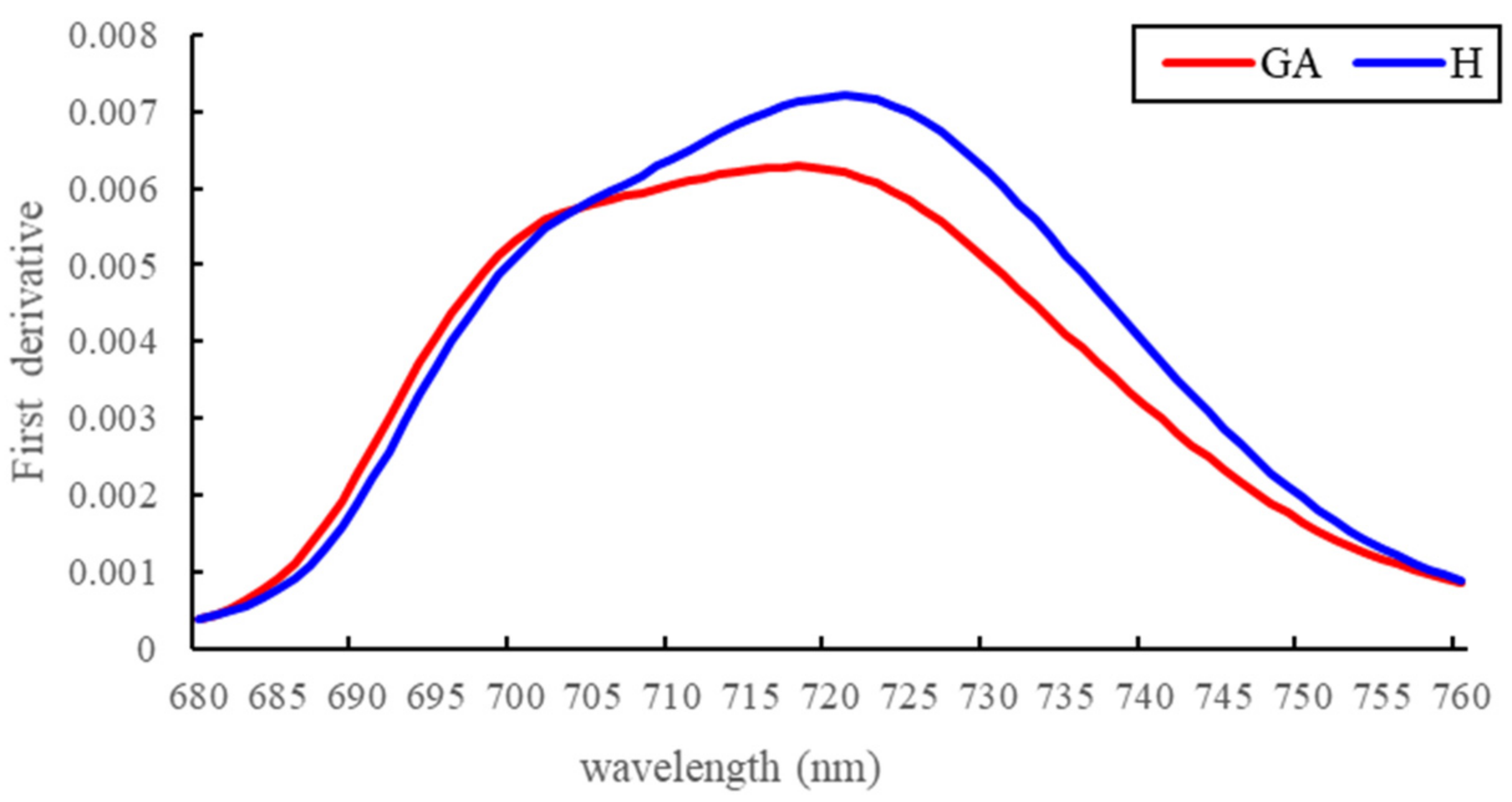
3.1. Spectral Reflectance Analysis
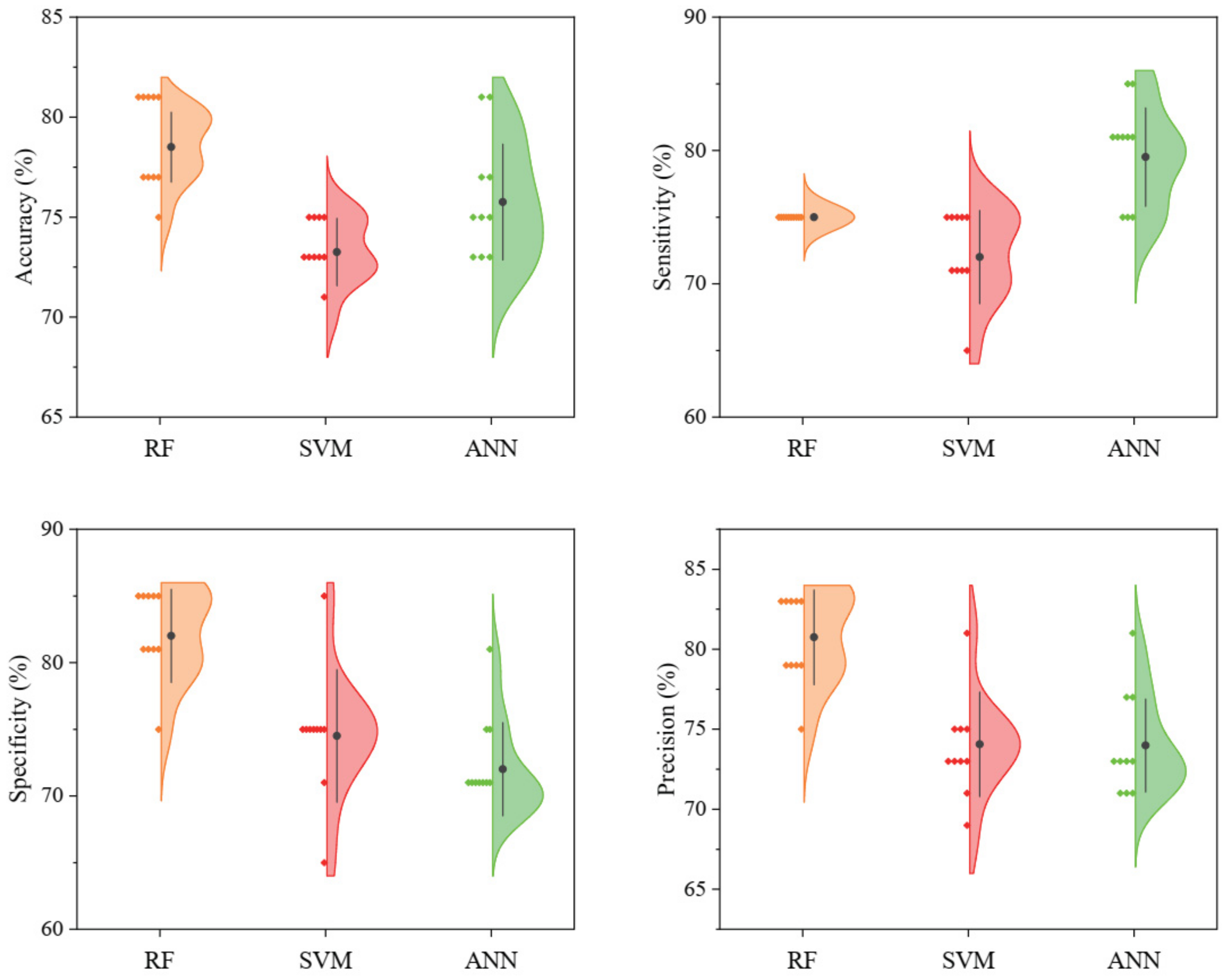
3.2. Classification Models
3.3. Important Variables
4. Discussion
4.1. Spectral Changes at the GA Stage
4.2. Classification Models and Variable Importance Evaluation
4.3. Implications for Remote Sensing
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yan, Z.; Sun, J.; Don, O.; Zhang, Z. The red turpentine beetle, Dendroctonus valens LeConte (Scolytidae): An exotic invasive pest of pine in China. Biodivers. Conserv. 2005, 14, 1735–1760. [Google Scholar] [CrossRef]
- Xu, H.; Li, Y.; Li, Z. The analysis of outbreak reason and spread direction of Dendroctonus valens. Plant Quar. 2006, 20, 278–280. [Google Scholar]
- Ge, X.; Jiang, C.; Chen, L.; Qiu, S.; Zhao, Y.; Wang, T.; Zong, S. Predicting the potential distribution in China of Dendroctonus valens Leconte. Environ. Environ. Entomol. 2018, 40, 758–768. [Google Scholar]
- Pan, J. Invasive Characteristics, Spatial Distribution and Sampling Technique of Dendroctonus valens Population. Master’s Thesis, Beijing Forestry University, Beijing, China, 2011. [Google Scholar]
- Zhang, L.; Chen, Q.; Zhang, X. Studies on the morphological characters and bionomics of Dendroctonus valens Leconte. Sci. Silvae Sin. 2002, 38, 95–99. [Google Scholar]
- Zhao, J.; Yang, Z.; Ren, X.; Liang, X. Biological characteristics and occurring law of Dendroctonus valens in China. Sci. Silvae Sin. 2008, 44, 99–105. [Google Scholar]
- Wulder, M.A.; Dymond, C.C.; White, J.; Leckie, D.G.; Carroll, A.L. Surveying mountain pine beetle damage of forests: A review of remote sensing opportunities. For. Ecol. Manag. 2006, 221, 27–41. [Google Scholar] [CrossRef]
- Foster, A.C.; Walter, J.A.; Shugart, H.H.; Sibold, J.; Negron, J. Spectral evidence of early-stage spruce beetle infestation in Engelmann spruce. For. Ecol. Manag. 2017, 384, 347–357. [Google Scholar] [CrossRef] [Green Version]
- Zhan, Z.; Yu, L.; Li, Z.; Ren, L.; Gao, B.; Wang, L.; Luo, Y. Combining GF-2 and Sentinel-2 images to detect tree mortality caused by red turpentine beetle during the early outbreak stage in North China. Forests 2020, 11, 172. [Google Scholar] [CrossRef] [Green Version]
- Hall, R.J.; Castilla, G.; White, J.C.; Cooke, B.J.; Skakun, R.S. Remote sensing of forest pest damage: A review and lessons learned from a Canadian perspective. Can. Entomol. 2016, 148, S296–S356. [Google Scholar] [CrossRef]
- Lausch, A.; Erasmi, S.; King, D.J.; Magdon, P.; Heurich, M. Understanding forest health with remote sensing-Part I—A review of spectral traits, processes and remote-sensing characteristics. Remote Sens. (Basel) 2016, 8, 1029. [Google Scholar] [CrossRef] [Green Version]
- Lausch, A.; Erasmi, S.; King, D.J.; Magdon, P.; Heurich, M. Understanding forest health with remote sensing-Part II—A review of approaches and data models. Remote Sens. 2017, 9, 129. [Google Scholar] [CrossRef] [Green Version]
- Stone, C.; Mohammed, C. Application of remote sensing technologies for assessing planted forests damaged by insect pests and fungal pathogens: A review. Curr. For. Rep. 2017, 3, 75–92. [Google Scholar] [CrossRef]
- Senf, C.; Seidl, R.; Hostert, P. Remote sensing of forest insect disturbances: Current state and future directions. Int. J. Appl. Earth Obs. Geoinf. 2017, 60, 49–60. [Google Scholar] [CrossRef] [Green Version]
- Goodwin, N.R.; Coops, N.C.; Wulder, M.A.; Gillanders, S.; Schroeder, T.A.; Nelson, T. Estimation of insect infestation dynamics using a temporal sequence of Landsat data. Remote Sens. Environ. 2008, 112, 3680–3689. [Google Scholar] [CrossRef]
- Liang, L.; Chen, Y.; Hawbaker, T.J.; Zhu, Z.; Gong, P. Mapping mountain pine beetle mortality through growth trend analysis of time-series Landsat data. Remote Sens. 2014, 6, 5696–5716. [Google Scholar] [CrossRef] [Green Version]
- Grabska, E.; Hawryło, P.; Socha, J. Continuous detection of small-scale changes in scots pine dominated stands using dense Sentinel-2 time series. Remote Sens. 2020, 12, 1298. [Google Scholar] [CrossRef] [Green Version]
- Gomez, D.; Ritger, H.; Pearce, C.; Eickwort, J.; Hulcr, J. Ability of remote sensing systems to detect bark beetle spots in the southeastern US. Forests 2020, 11, 1167. [Google Scholar] [CrossRef]
- Meddens, A.J.H.; Hicke, J.A.; Vierling, L.A.; Hudak, A.T. Evaluating methods to detect bark beetle-caused tree mortality using single-date and multi-date Landsat imagery. Remote Sens. Environ. 2013, 132, 49–58. [Google Scholar] [CrossRef]
- Abdullah, H.; Skidmore, A.K.; Darvishzadeh, R.; Heurich, M. Sentinel-2 accurately maps green-attack stage of European spruce bark beetle (Ips typographus, L.) compared with Landsat-8. Remote Sens. Ecol. Conserv. 2018, 5, 87–106. [Google Scholar] [CrossRef] [Green Version]
- Abdullah, H.; Darvishzadeh, R.; Skidmore, A.K.; Heurich, M. Sensitivity of Landsat-8 OLI and TIRS data to foliar properties of early stage bark beetle (Ips typographus, L.) infestation. Remote Sens. 2019, 11, 398. [Google Scholar] [CrossRef] [Green Version]
- Huo, L.; Persson, H.J.; Lindberg, E. Early detection of forest stress from European spruce bark beetle attack, and a new vegetation index: Normalized distance red & SWIR (NDRS). Remote Sens. Environ. 2021, 255, 112240. [Google Scholar] [CrossRef]
- Bárta, V.; Lukeš, P.; Homolová, L. Early detection of bark beetle infestation in Norway spruce forests of Central Europe using Sentinel-2. Int. J. Appl. Earth Obs. Geoinf. 2021, 100, 102335. [Google Scholar] [CrossRef]
- White, J.C.; Wulder, M.; Brooks, D.; Reich, R. Detection of red attack stage mountain pine beetle infestation with high spatial resolution satellite imagery. Remote Sens. Environ. 2005, 96, 340–351. [Google Scholar] [CrossRef]
- Coops, N.C.; Johnson, M.; Wulder, M.; White, J. Assessment of QuickBird high spatial resolution imagery to detect red attack damage due to mountain pine beetle infestation. Remote Sens. Environ. 2006, 103, 67–80. [Google Scholar] [CrossRef]
- Dennison, P.E.; Brunelle, A.R.; Carter, V.A. Assessing canopy mortality during a mountain pine beetle outbreak using GeoEye-1 high spatial resolution satellite data. Remote Sens. Environ. 2010, 114, 2431–2435. [Google Scholar] [CrossRef]
- Immitzer, M.; Atzberger, C. Early detection of bark beetle infestation in Norway Spruce (Picea abies L.) using WorldView-2 data. Photogramm. Fernerkund. Geoinf. 2014, 5, 351–367. [Google Scholar] [CrossRef]
- Mullen, K. Early Detection of Mountain Pine Beetle Damage in Ponderosa Pine Forests of the Black Hills Using Hyperspectral and WorldView-2 Data. Master’s Thesis, Minnesota State University, Mankato, MN, USA, 2016. [Google Scholar]
- Lausch, A.; Heurich, M.; Gordalla, D.; Dobner, H.-J.; Gwillym-Margianto, S.; Salbach, C. Forecasting potential bark beetle outbreaks based on spruce forest vitality using hyperspectral remote-sensing techniques at different scales. For. Ecol. Manag. 2013, 308, 76–89. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Ghosh, A.; Joshi, P.K.; Koch, B. Assessing the potential of hyperspectral imagery to map bark beetle-induced tree mortality. Remote Sens. Environ. 2014, 140, 533–548. [Google Scholar] [CrossRef]
- Niemann, K.O.; Quinn, G.; Stephen, R.; Visintini, F.; Parton, D. Hyperspectral remote sensing of mountain pine beetle with an emphasis on previsual assessment. Can. J. Remote Sens. 2015, 41, 191–202. [Google Scholar] [CrossRef]
- Näsi, R.; Honkavaara, E.; Lyytikäinen-Saarenmaa, P.; Blomqvist, M.; Litkey, P.; Hakala, T.; Viljanen, N.; Kantola, T.; Tanhuanpää, T.; Holopainen, M. Using UAV-based photogrammetry and hyperspectral imaging for mapping bark beetle damage at tree-level. Remote Sens. 2015, 7, 15467–15493. [Google Scholar] [CrossRef] [Green Version]
- Näsi, R.; Honkavaara, E.; Blomqvist, M.; Lyytikäinen-Saarenmaa, P.; Hakala, T.; Viljanen, N.; Kantola, T.; Holopainen, M. Remote sensing of bark beetle damage in urban forests at individual tree level using a novel hyperspectral camera from UAV and aircraft. Urban For. Urban Green. 2018, 30, 72–83. [Google Scholar] [CrossRef]
- Honkavaara, E.; Näsi, R.; Oliveira, R.; Viljanen, N.; Suomalainen, J.; Khoramshahi, E.; Hakala, T.; Nevalainen, O.; Markelin, L.; Vuorinen, M.; et al. Using multitemporal hyper- and multispectral Uav imaging for detecting bark beetle infestation on Norway spruce. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2020, XLIII-B3-2, 429–434. [Google Scholar] [CrossRef]
- Ahern, F.J. The effects of bark beetle stress on the foliar spectral reflectance of lodgepole pine. Int. J. Remote Sens. 1988, 9, 1451–1468. [Google Scholar] [CrossRef]
- Reichmuth, A.; Henning, L.; Pinnel, N.; Bachmann, M.; Rogge, D. Early detection of vitality changes of multi-temporal Norway spruce laboratory needle measurements—The ring-barking experiment. Remote Sens. 2018, 10, 57. [Google Scholar] [CrossRef] [Green Version]
- Cheng, T.; Rivard, B.; Sanchez-Azofeifa, A.; Feng, J.; Calvo-Polanco, M. Continuous wavelet analysis for the detection of green attack damage due to mountain pine beetle infestation. Remote Sens. Environ. 2010, 114, 899–910. [Google Scholar] [CrossRef]
- Abdullah, H.; Darvishzadeh, R.; Skidmore, A.K.; Groen, T.A.; Heurich, M. European spruce bark beetle (Ips typographus, L.) green attack affects foliar reflectance and biochemical properties. Int. J. Appl. Earth Obs. Geoinf. 2018, 64, 199–209. [Google Scholar] [CrossRef] [Green Version]
- Ismail, R.; Mutanga, O. Discriminating the early stages of Sirex noctilio infestation using classification tree ensembles and shortwave infrared bands. Int. J. Remote Sens. 2011, 32, 4249–4266. [Google Scholar] [CrossRef]
- Liu, Y.; Zhan, Z.; Ren, L.; Ze, S.; Yu, L.; Jiang, Q.; Luo, Y. Hyperspectral evidence of early-stage pine shoot beetle attack in Yunnan pine. For. Ecol. Manag. 2021, 497, 119505. [Google Scholar] [CrossRef]
- Yu, R.; Ren, L.; Luo, Y. Early detection of pine wilt disease in Pinus tabuliformis in North China using a field portable spectrometer and UAV-based hyperspectral imagery. For. Ecosyst. 2021, 8, 44. [Google Scholar] [CrossRef]
- Liu, Y. A comparative study on feature selection methods for drug discovery. J. Chem. Inf. Comput. Sci. 2004, 44, 1823–1828. [Google Scholar] [CrossRef] [Green Version]
- Kushmerick, N. Wrapper induction: Efficiency and expressiveness. Artif. Intell. 2000, 118, 15–68. [Google Scholar] [CrossRef] [Green Version]
- Burger, S.V. Introduction to Machine Learning with R; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2018. [Google Scholar]
- Beck, P.; Zarco-Tejada, P.J.; Strobl, P.; San Miguel, J. The Feasibility of Detecting Trees Affected by the Pine Wood Nematode Using Remote Sensing; Publications Office of the European Union: Luxembourg, 2015; pp. 1831–9424. [Google Scholar]
- Tian, Y.-C.; Gu, K.-J.; Chu, X.; Yao, X.; Cao, W.-X.; Zhu, Y. Comparison of different hyperspectral vegetation indices for canopy leaf nitrogen concentration estimation in rice. Plant Soil 2013, 376, 193–209. [Google Scholar] [CrossRef]
- Carter, G.A.; Miller, R.L. Early detection of plant stress by digital imaging within narrow stress-sensitive wavebands. Remote Sens. Environ. 1994, 50, 295–302. [Google Scholar] [CrossRef]
- Zhu, Y.; Yao, X.; Tian, Y.; Liu, X.; Cao, W. Analysis of common canopy vegetation indices for indicating leaf nitrogen accumulations in wheat and rice. Int. J. Appl. Earth Obs. Geoinf. 2008, 10, 1–10. [Google Scholar] [CrossRef]
- Blackburn, G.A. Spectral indices for estimating photosynthetic pigment concentrations: A test using senescent tree leaves. Int. J. Remote Sens. 1998, 19, 657–675. [Google Scholar] [CrossRef]
- Chappelle, E.W.; Kim, M.S.; McMurtrey, J.E., III. Ratio analysis of reflectance spectra (RARS): An algorithm for the remote estimation of the concentrations of chlorophyll a, chlorophyll b, and carotenoids in soybean leaves. Remote Sens. Environ. 1992, 39, 239–247. [Google Scholar] [CrossRef]
- Ceccato, P.; Flasse, S.; Tarantola, S.; Jacquemoud, S.; Grégoire, J.-M. Detecting vegetation leaf water content using reflectance in the optical domain. Remote Sens. Environ. 2001, 77, 22–33. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Lang, M.; Sowinska, M.; Heisel, F.; Miehé, J.A. Detection of vegetation stress via a new high resolution fluorescence imaging system. J. Plant Physiol. 1996, 148, 599–612. [Google Scholar] [CrossRef]
- Lee, Y.-J.; Yang, C.-M.; Chang, K.-W.; Shen, Y. A simple spectral index using reflectance of 735 nm to assess nitrogen status of rice canopy. Agron. J. 2008, 100, 205–212. [Google Scholar] [CrossRef]
- Vogelmann, J.E.; Rock, B.N.; Moss, D.M. Red edge spectral measurements from sugar maple leaves. Int. J. Remote Sens. 1993, 14, 1563–1575. [Google Scholar] [CrossRef]
- Gamon, J.A.; Peñuelas, J.; Field, C.B. A narrow-waveband spectral index that tracks diurnal changes in photosynthetic efficiency. Remote Sens. Environ. 1992, 41, 35–44. [Google Scholar] [CrossRef]
- Merton, R.; Huntington, J. Early simulation results of the ARIES-1 satellite sensor for multi-temporal vegetation research derived from AVIRIS. In Proceedings of the Eighth Annual JPL Airborne Earth Science Workshop, Pasadena, CA, USA, 9–11 February 1999. [Google Scholar]
- Gitelson, A.A.; Zur, Y.; Chivkunova, O.B.; Merzlyak, M.N. Assessing carotenoid content in plant leaves with reflectance spectroscopy. Photochem. Photobiol. 2002, 75, 272–281. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Gritz, Y.; Merzlyak, M.N. Relationships between leaf chlorophyll content and spectral reflectance and algorithms for non-destructive chlorophyll assessment in higher plant leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Gitelson, A.A.; Viña, A.; Ciganda, V.; Rundquist, D.C.; Arkebauer, T.J. Remote estimation of canopy chlorophyll content in crops. Geophys. Res. Lett. 2005, 32, L08403. [Google Scholar] [CrossRef] [Green Version]
- Zarco-Tejada, P.J.; Miller, J.R.; Noland, T.L.; Mohammed, G.H.; Sampson, P.H. Scaling-up and model inversion methods with narrowband optical indices for chlorophyll content estimation in closed forest canopies with hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2001, 39, 1491–1507. [Google Scholar] [CrossRef] [Green Version]
- Gao, B.-C. NDWI—A normalized difference water index for remote sensing of vegetation liquid water from space. Remote Sens. Environ. 1996, 58, 257–266. [Google Scholar] [CrossRef]
- Haubrock, S.-N.; Chabrillat, S.; Lemmnitz, C.; Kaufmann, H. Surface soil moisture quantification models from reflectance data under field conditions. Int. J. Remote Sens. 2008, 29, 3–29. [Google Scholar] [CrossRef]
- Serrano, L.; Peñuelas, J.; Ustin, S.L. Remote sensing of nitrogen and lignin in Mediterranean vegetation from AVIRIS data: Decomposing biochemical from structural signals. Remote Sens. Environ. 2002, 81, 355–364. [Google Scholar] [CrossRef]
- Wang, Q.; Li, P. Hyperspectral indices for estimating leaf biochemical properties in temperate deciduous forests: Comparison of simulated and measured reflectance data sets. Ecol. Indic. 2012, 14, 56–65. [Google Scholar] [CrossRef]
- Dash, J.; Curran, P.J. The MERIS terrestrial chlorophyll index. Int. J. Remote Sens. 2004, 25, 5403–5413. [Google Scholar] [CrossRef]
- Reyniers, M.; Walvoort, D.J.J.; De Baardemaaker, J. A linear model to predict with a multi-spectral radiometer the amount of nitrogen in winter wheat. Int. J. Remote Sens. 2006, 27, 4159–4179. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Pattey, E.; Zarco-Tejada, P.J.; Strachan, I.B. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Gao, J.; Liang, T.; Yin, J.; Ge, J.; Feng, Q.; Wu, C.; Hou, M.; Liu, J.; Xie, H. Estimation of alpine grassland forage nitrogen coupled with hyperspectral characteristics during different growth periods on the tibetan plateau. Remote Sens. 2019, 11, 2085. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Niu, Z.; Tang, Q.; Huang, W. Estimating chlorophyll content from hyperspectral vegetation indices: Modeling and validation. Agric. For. Meteorol. 2008, 148, 1230–1241. [Google Scholar] [CrossRef]
- Zhu, X. Playing with R: Data Analitical Thinking to Practice; China Renmin University Press: Beijing, China, 2018. [Google Scholar]
- Sothe, C.; De Almeida, C.M.; Schimalski, M.B.; La Rosa, L.E.C.; Castro, J.D.B.; Feitosa, R.Q.; Dalponte, M.; Lima, C.L.; Liesenberg, V.; Miyoshi, G.T.; et al. Comparative performance of convolutional neural network, weighted and conventional support vector machine and random forest for classifying tree species using hyperspectral and photogrammetric data. GISci. Remote Sens. 2020, 57, 369–394. [Google Scholar] [CrossRef]
- Gutierrez, D.D. Machine Learning and Data Science: An Introduction to Statistical Learning Methods with R; Technics Publications: Bernards, NJ, USA, 2015. [Google Scholar]
- Lesmeister, C. Mastering Machine Learning with R, 2nd ed.; Packt Publishing Ltd: Birmingham, UK, 2017. [Google Scholar]
- Rumpf, T.; Mahlein, A.-K.; Steiner, U.; Oerke, E.-C.; Dehne, H.-W.; Plümer, L. Early detection and classification of plant diseases with Support Vector Machines based on hyperspectral reflectance. Comput. Electron. Agric. 2010, 74, 91–99. [Google Scholar] [CrossRef]
- Variable Importance. Available online: https://topepo.github.io/caret/variable-importance.html (accessed on 10 March 2021).
- Curran, P.J. Remote sensing of foliar chemistry. Remote Sens. Environ. 1989, 30, 271–278. [Google Scholar] [CrossRef]
- Runesson, U.T. Considerations for Early Remote Detection of Mountain Pine Beetle in Green-Foliaged Lodgepole Pine. Ph.D. Thesis, University of British Columbia, Vancouver, BC, Canada, 1991. [Google Scholar]
- Abdullah, H.; Skidmore, A.K.; Darvishzadeh, R.; Heurich, M. Timing of red-edge and shortwave infrared reflectance critical for early stress detection induced by bark beetle (Ips typographus, L.) attack. Int. J. Appl. Earth Obs. Geoinf. 2019, 82, 101900. [Google Scholar] [CrossRef]
- Sims, D.A.; Gamon, J.A. Estimation of vegetation water content and photosynthetic tissue area from spectral reflectance: A comparison of indices based on liquid water and chlorophyll absorption features. Remote Sens. Environ. 2003, 84, 526–537. [Google Scholar] [CrossRef]
- Yu, L.; Zhan, Z.; Ren, L.; Zong, S.; Luo, Y.; Huang, H. Evaluating the potential of WorldView-3 data to classify different shoot damage ratios of pinus yunnanensis. Forests 2020, 11, 417. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.; Huang, J.; Zong, S.; Huang, H.; Luo, Y. Detecting shoot beetle damage on Yunnan pine using Landsat time-series data. Forests 2018, 9, 39. [Google Scholar] [CrossRef] [Green Version]





| Spectral Indices | Formulation or Depiction | Spectral Regions | Ref. |
|---|---|---|---|
| Blue index | BI = R450/R490 | Blue | [45] |
| Simple ratio indices | SR1 = R545/R538 | Green | [46] |
| SR2 = R694/R760 | Red edge | [47] | |
| Ratio vegetation index | RVI = R810/R660 | Red, NIR | [48] |
| Difference vegetation index | DVI = R810 − R680 | Red edge, NIR | [48] |
| Pigment-specific simple ratio | PSSRb= R800/R635 | Red, NIR | [49] |
| PSSRc = R800/R470 | Blue, NIR | [49] | |
| Pigment-specific normalized difference | PSNDa = (R800 − R680)/(R800 + R680) | Red edge, NIR | [49] |
| PSNDb = (R800 − R635)/(R800 + R635) | Red, NIR | [49] | |
| PSNDc = (R800 − R470)/(R800 + R470) | Blue, NIR | [49] | |
| Ratio analysis of reflectance spectra | RARS = R746/R513 | Green, red edge | [50] |
| Moisture stress index | MSI = R1600/R820 | NIR, SWIR | [51] |
| Lichtenthaler index | LIC = R440/R740 | Blue, red edge | [52] |
| First derivative of reflectance spectrum at 735 nm | D735 | Red edge | [53] |
| A ratio of first-derivative values | Voge = D715/D705 | Red edge | [54] |
| First-derivative difference index | DID = D1183 − D876 | NIR | [46] |
| Physiological reflectance index | PRI515 = (R515 − R531)/(R515 + R531) | Green | [55] |
| Photochemical reflectance index | PRIm2 = (R600 − R531)/(R600 + R531) | Green, orange | [45] |
| PRIm4 = (R570 − R531 − R670)/((R570 + R531 + R670) | Green, red | [45] | |
| PRIn = PRI570/[RDVI × (R700/R670)],PRI570 = (R570 − R531)/(R570 + R531), RDVI = (R800 − R670)/(R800 + R670)0.5 | Green, red, NIR | [45] | |
| Red-edge vegetation stress index | RVSI = (R714 − R752)/2 − R733 | Red edge | [56] |
| Carotenoid reflectance index | CRI550 = 1/R510 − 1/R550 | Green | [57] |
| RNIR•CRI550 | R770 × CRI550 | Green, NIR | [58] |
| Chlorophyll content index | CIred-edge = R840~870/R720~730 − 1 | Red edge, NIR | [59] |
| Curvature index | CUR = R675 × R690/R6832 | Red, red edge | [60] |
| Normalized difference vegetation index | NDVIg-b = (R537 − R440)/(R537 + R440) | Blue, green | [46] |
| Normalized difference water index | NDWI = (R860 − R1240)/(R860 + R1240) | NIR | [61] |
| Normalized soil moisture index | NSMI = (R1800 − R2119)/(R1800 + R2119) | SWIR | [62] |
| Normalized difference lignin index | NDLI = [log(1/R1754) − log(1/R1680)]/[log(1/R1754) + log(1/R1680)] | SWIR | [63] |
| Health index | HI = (R534 − R698)/(R534 + R698) − R704/2 | Green, red edge | [45] |
| Double-difference indices | DDn1 = 2R715 − R715−185 − R715+185 | Red, red edge, NIR | [64] |
| DDn2 = 2R1530 − R1530−525 − R1530+525 | NIR, SWIR | [64] | |
| DDn3 = 2R1235 − R1235−25 − R1235+25 | NIR | [64] | |
| MERIS terrestrial chlorophyll index | MTCI = (R754 − R709)/(R709 − R681) | Red edge | [65] |
| Optimal vegetation index | VIopt = 1.45 × (R8002 + 1)/(R670 + 0.45) | Red, NIR | [66] |
| Three-band spectral index | TBSI = (R605 − R521 − R682)/(R605 + R521 + R682) | Green, red, red edge | [46] |
| Modified triangular vegetation index | MTVI = 1.2 × [1.2 × (R800 − R550) − 2.5 × (R670 − R550)] | Green, red, NIR | [67] |
| Soil-adjusted vegetation index | SAVI = (1 + 0.5) × [(R800 − R670)/(R800 + R670 + 0.5)] | Red, NIR | [68] |
| Optimized SAVI | OSAVI1 = (1 + 0.16) × [(R800 − R670)/(R800 + R670 + 0.16)] | Red, NIR | [69] |
| OSAVI2 = (1 + 0.16) × [(R750 − R705)/(R750 + R705 + 0.16)] | Red edge | [69] |
| True Conditions | ||||
|---|---|---|---|---|
| Positive (GA) | Negative (H) | |||
| Test results | Positive (GA) | True positive (TP) | False positive (FP) | Precision = TP/(TP + FP) |
| Negative (H) | False negative (FN) | True negative (TN) | ||
| Sensitivity = TP/(TP + FN) | Specificity = TN/(FP + TN) | Accuracy = (TP + TN)/(TP + FP + TN + FN) | ||
| Classification Models | Hyperparameters | Evaluation Metrics (%) | ||||
|---|---|---|---|---|---|---|
| Accuracy (Kappa) | Sensitivity | Specificity | Precision | |||
| RF | mtry | 29 | 80 (0.6) | 75 | 85 | 83.33 |
| SVM | C | 21.08381 | 75 (0.5) | 75 | 75 | 75 |
| γ | 0.010476 | |||||
| ANN | size | 20 | 80 (0.6) | 80 | 80 | 80 |
| decay | 0.391524 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, B.; Yu, L.; Ren, L.; Zhan, Z.; Luo, Y. Early Detection of Dendroctonus valens Infestation with Machine Learning Algorithms Based on Hyperspectral Reflectance. Remote Sens. 2022, 14, 1373. https://doi.org/10.3390/rs14061373
Gao B, Yu L, Ren L, Zhan Z, Luo Y. Early Detection of Dendroctonus valens Infestation with Machine Learning Algorithms Based on Hyperspectral Reflectance. Remote Sensing. 2022; 14(6):1373. https://doi.org/10.3390/rs14061373
Chicago/Turabian StyleGao, Bingtao, Linfeng Yu, Lili Ren, Zhongyi Zhan, and Youqing Luo. 2022. "Early Detection of Dendroctonus valens Infestation with Machine Learning Algorithms Based on Hyperspectral Reflectance" Remote Sensing 14, no. 6: 1373. https://doi.org/10.3390/rs14061373
APA StyleGao, B., Yu, L., Ren, L., Zhan, Z., & Luo, Y. (2022). Early Detection of Dendroctonus valens Infestation with Machine Learning Algorithms Based on Hyperspectral Reflectance. Remote Sensing, 14(6), 1373. https://doi.org/10.3390/rs14061373







