Toward Automated Machine Learning-Based Hyperspectral Image Analysis in Crop Yield and Biomass Estimation
Abstract
1. Introduction
2. Materials and Methods
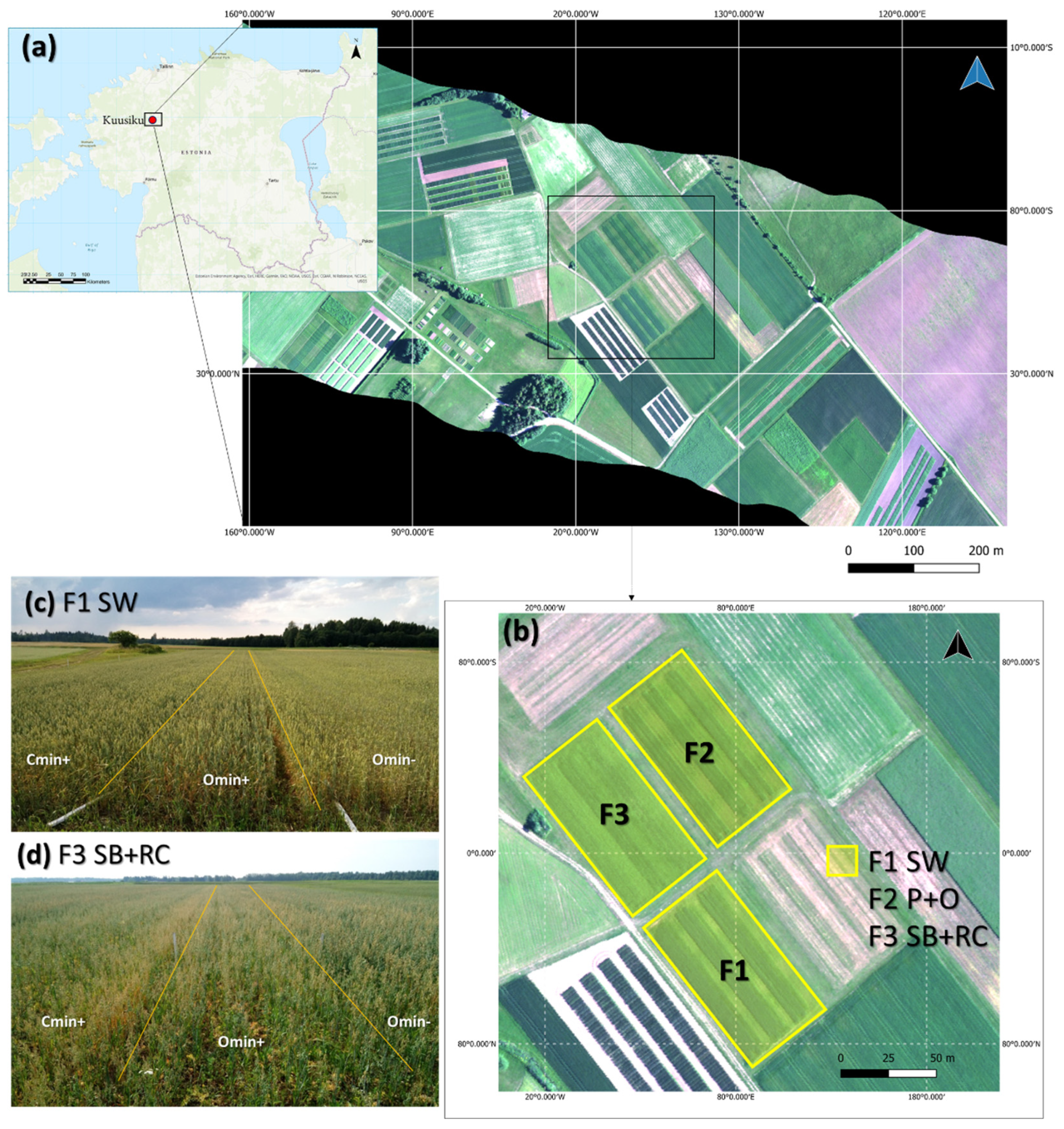
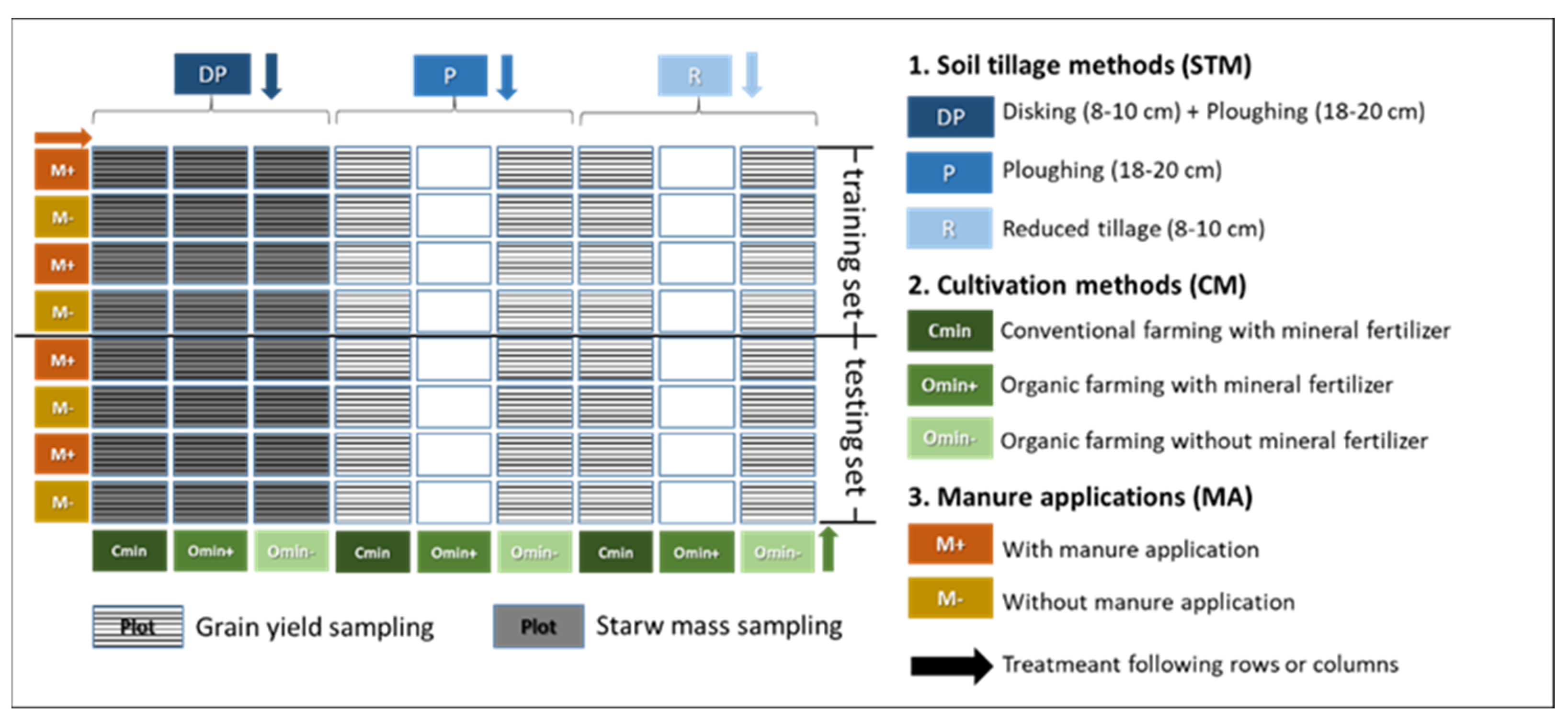
2.1. Research Site and Experiment Layout
2.2. Hyperspectral Image Data Collection
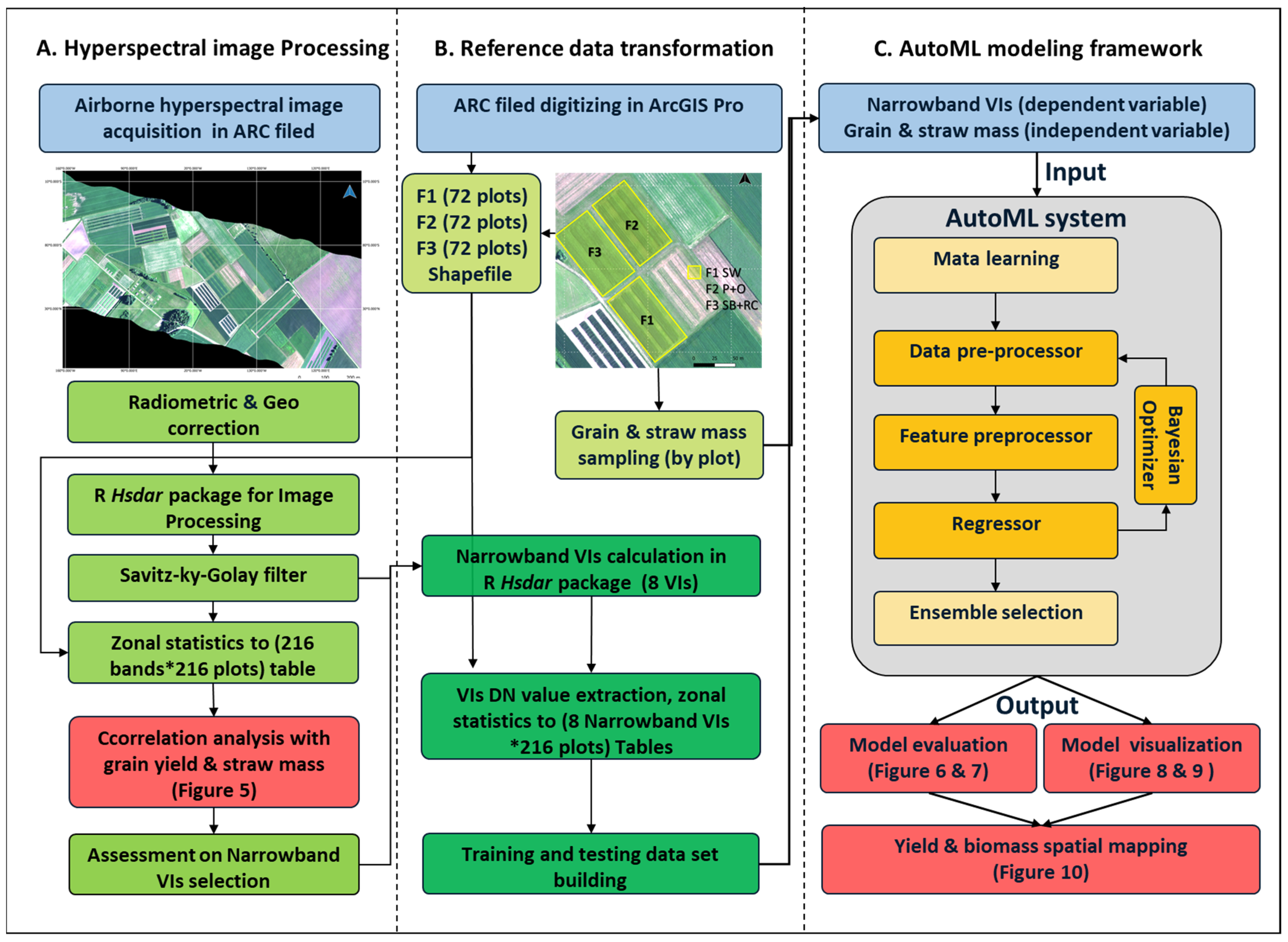
2.3. Hyperspectral Image Processing
2.4. Narrowband Vegetation Index
2.5. AutoML Regression with Auto-Sklearn
2.6. Model Evaluation
3. Results
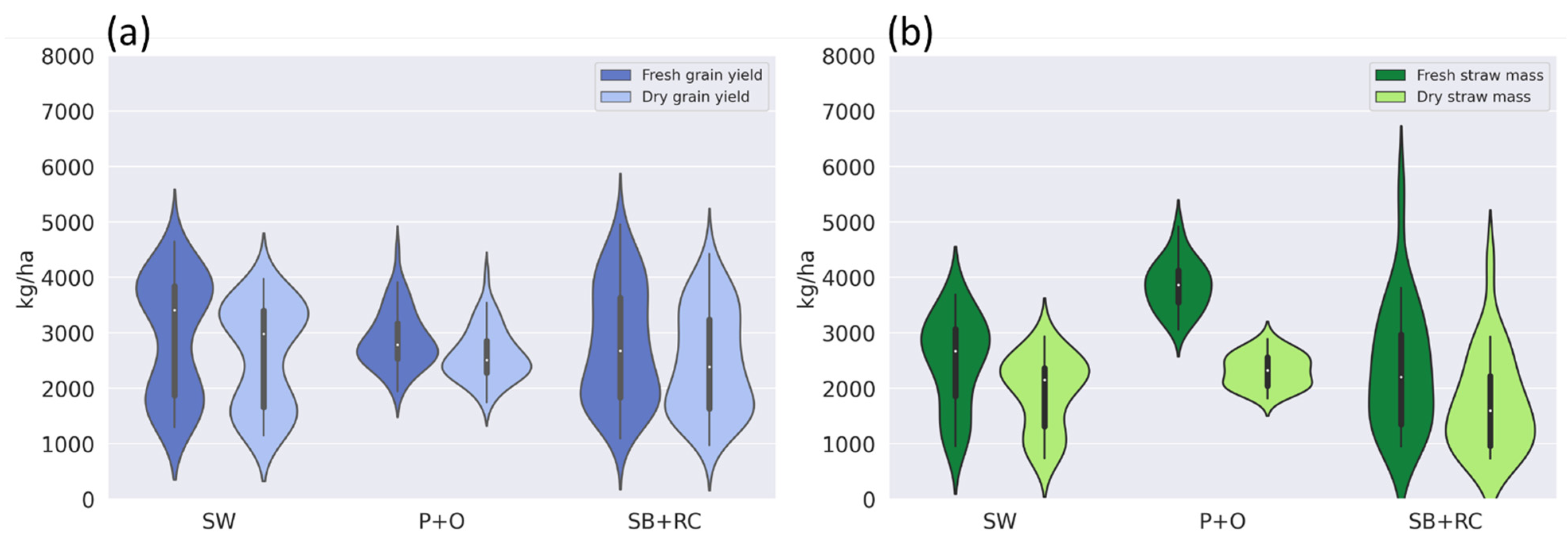
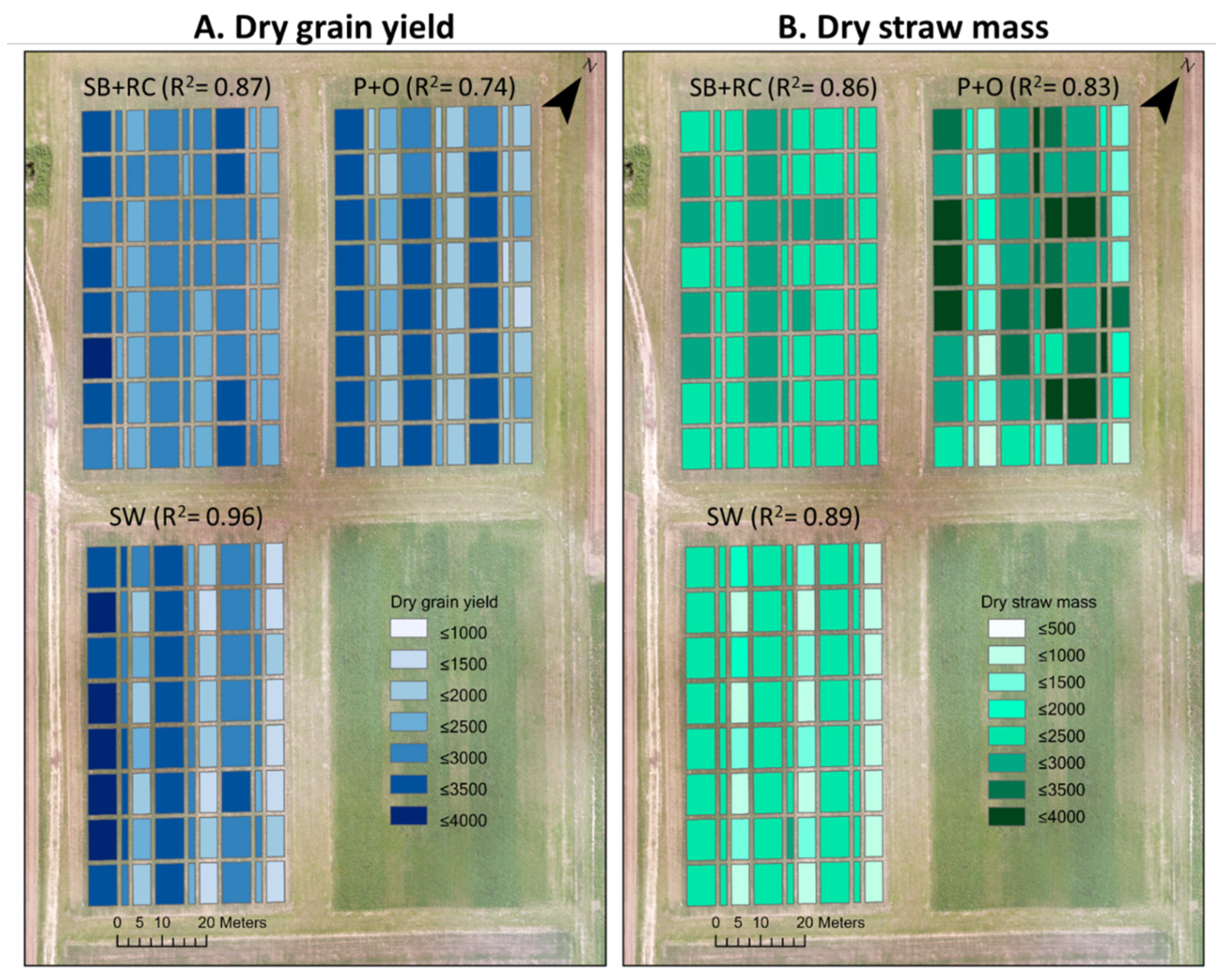
3.1. The Field Observation DM Data Analysis
3.2. The Hyperspectral Reflectance Signature under Various Agriculture Management Practises
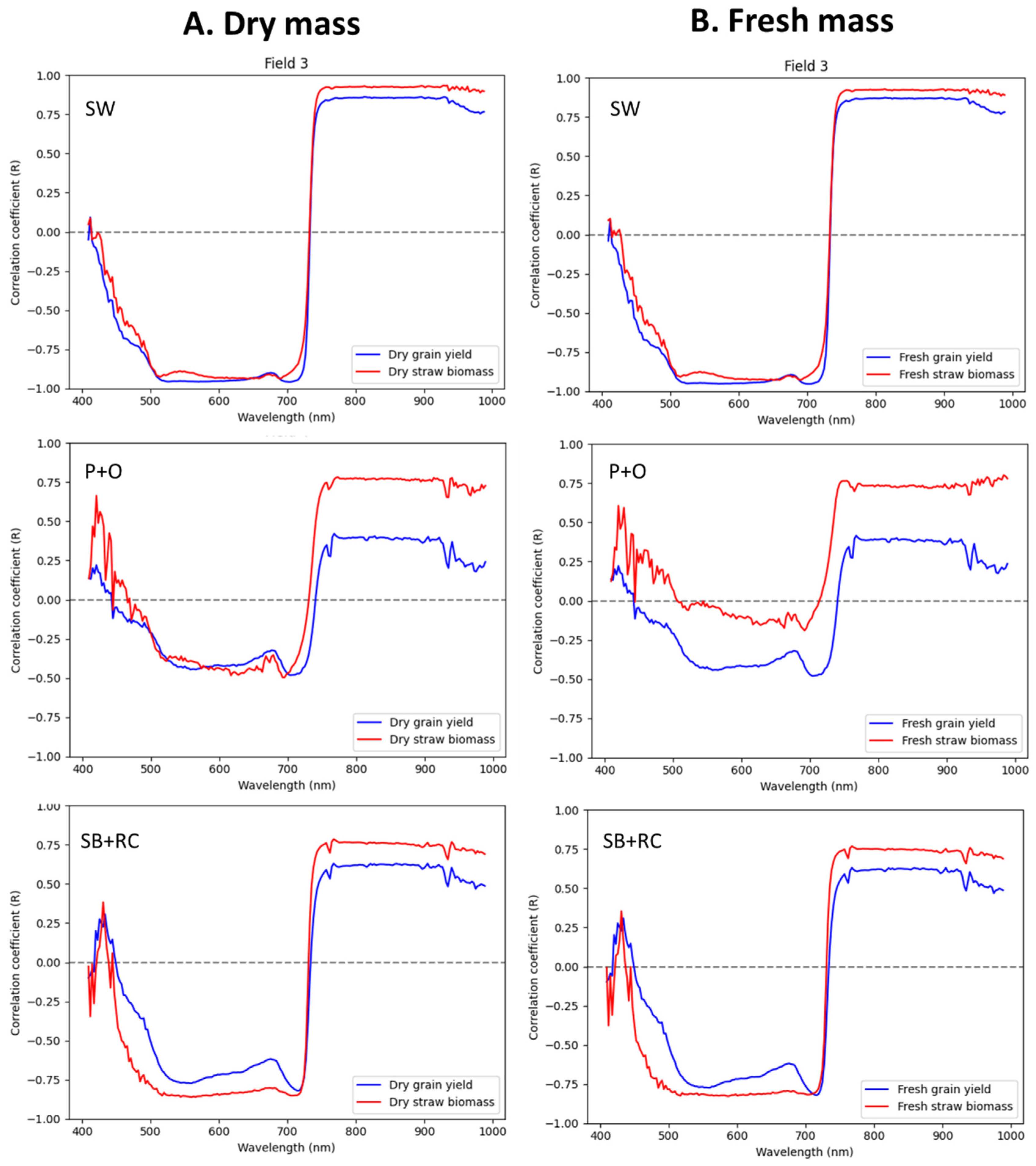
3.3. Characterization of the Correlation Coefficient with Averaged Radiance Hyperspectral Data and Field Observation
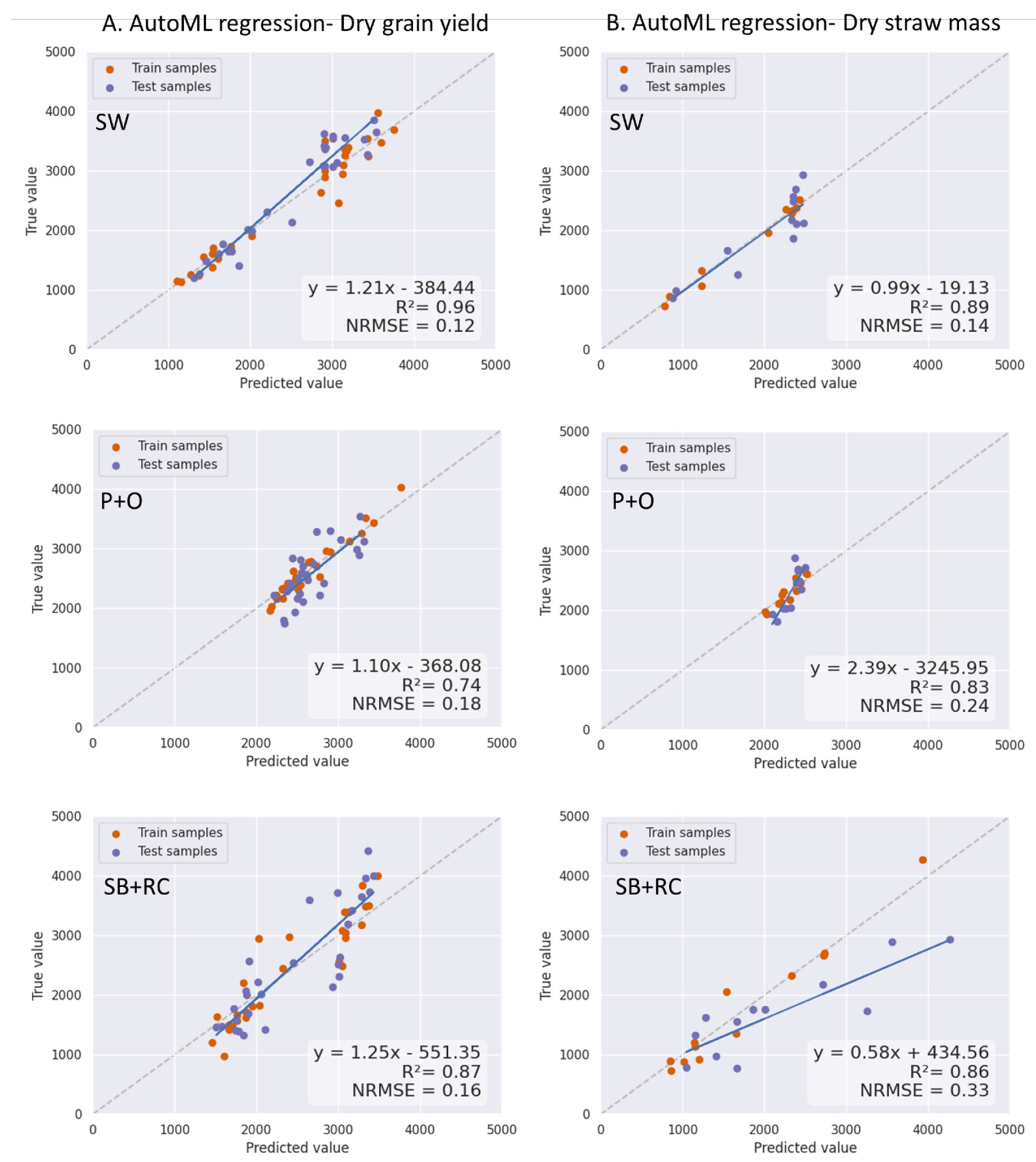
3.4. The AutoML Model Prediction and Evaluation
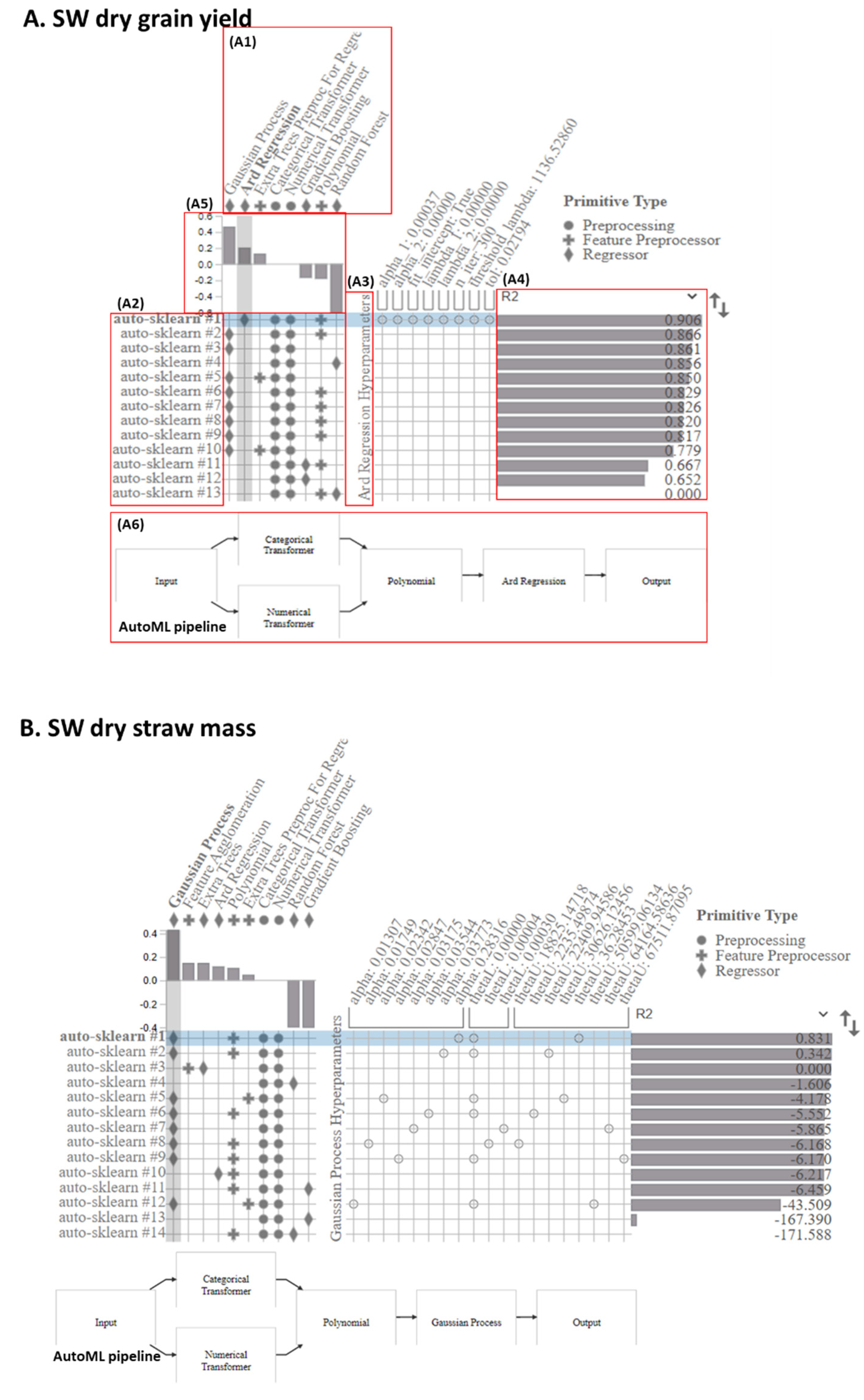
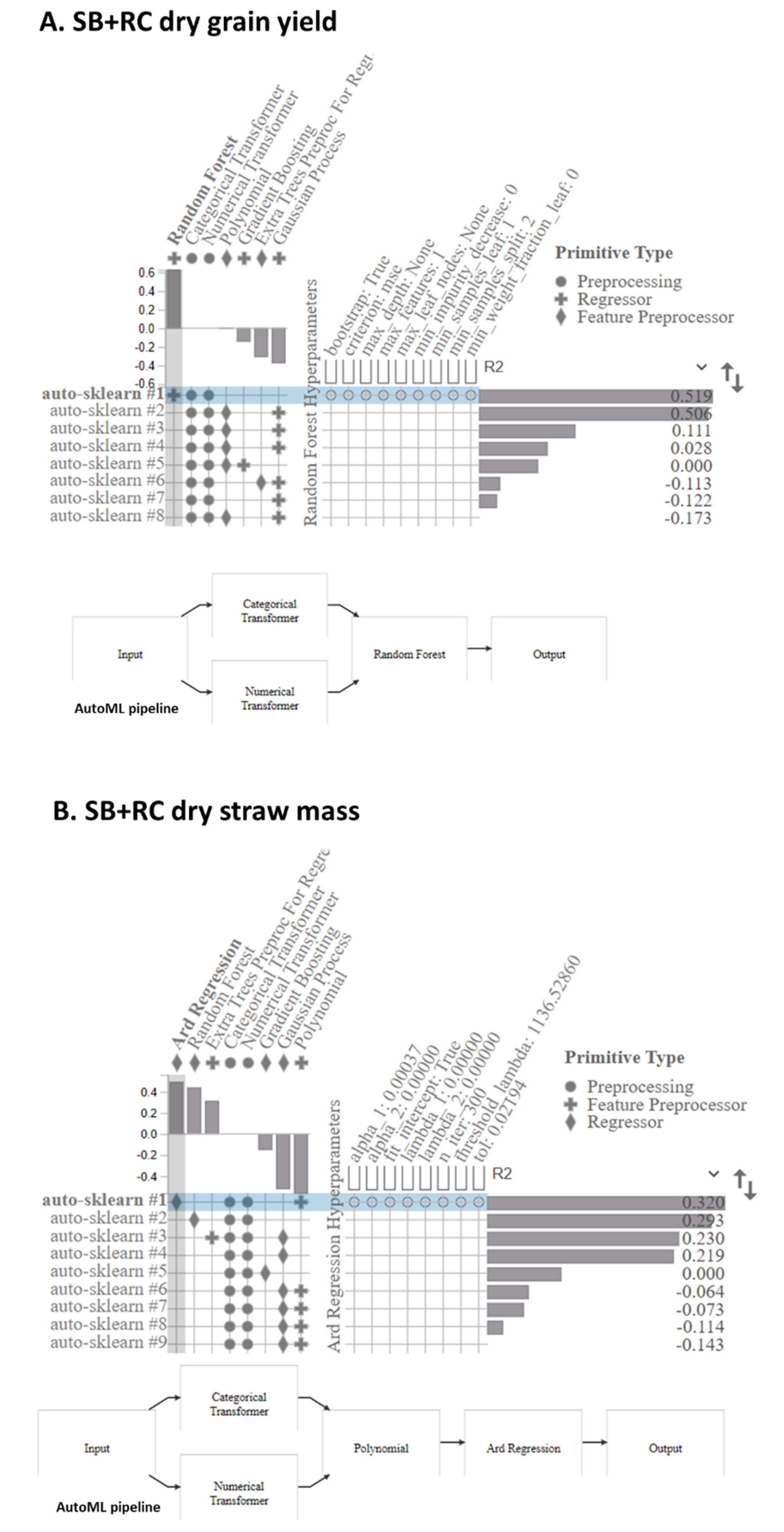
3.5. The AutoML Model Pipeline Visualization
3.6. The Field Observation DM Data Analysis
4. Discussion
4.1. The Effect of Hyperspectral Signatures and the Correlation between Crop Yield and Straw Mass
4.2. The Hyperspectral Narrowband VIs and AutoML Modelling
4.3. The AutoML Method’s Applicability and Impact in Hyperspectral Imaging
4.4. The Limitations in This Study
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Pavón-Pulido, N.; López-Riquelme, J.A.; Torres, R.; Morais, R.; Pastor, J.A. New Trends in Precision Agriculture: A Novel Cloud-Based System for Enabling Data Storage and Agricultural Task Planning and Automation. Precis. Agric. 2017, 18, 1038–1068. [Google Scholar] [CrossRef]
- Karthikeyan, L.; Chawla, I.; Mishra, A.K. A Review of Remote Sensing Applications in Agriculture for Food Security: Crop Growth and Yield, Irrigation, and Crop Losses. J. Hydrol. 2020, 586, 124905. [Google Scholar] [CrossRef]
- Wen, W.; Timmermans, J.; Chen, Q.; van Bodegom, P.M. A Review of Remote Sensing Challenges for Food Security with Respect to Salinity and Drought Threats. Remote Sens. 2021, 13, 6. [Google Scholar] [CrossRef]
- Yang, M.D.; Huang, K.S.; Kuo, Y.H.; Tsai, H.P.; Lin, L.M. Spatial and Spectral Hybrid Image Classification for Rice Lodging Assessment through UAV Imagery. Remote Sens. 2017, 9, 583. [Google Scholar] [CrossRef]
- Mandal, A.; Majumder, A.; Dhaliwal, S.S.; Toor, A.S.; Mani, P.K.; Naresh, R.K.; Gupta, R.K.; Mitran, T. Impact of Agricultural Management Practices on Soil Carbon Sequestration and Its Monitoring through Simulation Models and Remote Sensing Techniques: A Review. Crit. Rev. Environ. Sci. Technol. 2020, 52, 1–49. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Tremblay, N.; Zarco-Tejada, P.J.; Dextraze, L. Integrated Narrow-Band Vegetation Indices for Prediction of Crop Chlorophyll Content for Application to Precision Agriculture. Remote Sens. Environ. 2002, 81, 416–426. [Google Scholar] [CrossRef]
- Sahoo, R.N.; Ray, S.S.; Manjunath, K.R. Hyperspectral Remote Sensing of Agriculture. Curr. Sci. 2015, 108, 848–859. [Google Scholar] [CrossRef]
- Windfuhr, M.; Jonsén, J. Food Sovereignty towards Democracy in Localized Food Systems; FAO: Rome, Italy, 2005; Volume 339. [Google Scholar]
- Gómez, D.; Salvador, P.; Sanz, J.; Casanova, J.L. Potato Yield Prediction Using Machine Learning Techniques and Sentinel 2 Data. Remote Sens. 2019, 11, 1745. [Google Scholar] [CrossRef]
- Triplett, G.B.; Dick, W.A. No-Tillage Crop Production: A Revolution in Agriculture! Agron. J. 2008, 100, 153–165. [Google Scholar] [CrossRef]
- Karlen, D.L.; Kovar, J.L.; Cambardella, C.A.; Colvin, T.S. Thirty-Year Tillage Effects on Crop Yield and Soil Fertility Indicators. Soil Tillage Res. 2013, 130, 24–41. [Google Scholar] [CrossRef]
- Ashapure, A.; Jung, J.; Yeom, J.; Chang, A.; Maeda, M.; Maeda, A.; Landivar, J. A Novel Framework to Detect Conventional Tillage and No-Tillage Cropping System Effect on Cotton Growth and Development Using Multi-Temporal UAS Data. ISPRS J. Photogramm. Remote Sens. 2019, 152, 49–64. [Google Scholar] [CrossRef]
- Telles, T.S.; Reydon, B.P.; Maia, A.G. Effects of No-Tillage on Agricultural Land Values in Brazil. Land Use Policy 2018, 76, 124–129. [Google Scholar] [CrossRef]
- Fanigliulo, R.; Antonucci, F.; Figorilli, S.; Pochi, D.; Pallottino, F.; Fornaciari, L.; Grilli, R.; Costa, C. Light Drone-Based Application to Assess Soil Tillage Quality Parameters. Sensors 2020, 20, 728. [Google Scholar] [CrossRef] [PubMed]
- Desta, B.T.; Gezahegn, A.M.; Tesema, S.E. Impacts of Tillage Practice on the Productivity of Durum Wheat in Ethiopia. Cogent Food Agric. 2021, 7, 1869382. [Google Scholar] [CrossRef]
- Crews, T.E.; Peoples, M.B. Legume versus Fertilizer Sources of Nitrogen: Ecological Tradeoffs and Human Needs. Agric. Ecosyst. Environ. 2004, 102, 279–297. [Google Scholar] [CrossRef]
- Zikeli, S.; Gruber, S.; Teufel, C.F.; Hartung, K.; Claupein, W. Effects of Reduced Tillage on Crop Yield, Plant Available Nutrients and Soil Organic Matter in a 12-Year Long-Term Trial under Organic Management. Sustainability 2013, 5, 3876–3894. [Google Scholar] [CrossRef]
- Yang, X.M.; Drury, C.F.; Reynolds, W.D.; Reeb, M.D. Legume Cover Crops Provide Nitrogen to Corn during a Three-Year Transition to Organic Cropping. Agron. J. 2019, 111, 3253–3264. [Google Scholar] [CrossRef]
- Li, K.Y.; Burnside, N.G.; de Lima, R.S.; Peciña, M.V.; Sepp, K.; der Yang, M.; Raet, J.; Vain, A.; Selge, A.; Sepp, K. The Application of an Unmanned Aerial System and Machine Learning Techniques for Red Clover-Grass Mixture Yield Esti-Mation under Variety Performance Trials. Remote Sens. 2021, 13, 1994. [Google Scholar] [CrossRef]
- Loide, V. The Results of an NPK-Fertilisation Trial of Long-Term Crop Rotation on Carbonate-Rich Soil in Estonia. Acta Agric. Scand. Sect. B Soil Plant Sci. 2019, 69, 596–605. [Google Scholar] [CrossRef]
- Gianelle, D.; Vescovo, L.; Marcolla, B.; Manca, G.; Cescatti, A. Ecosystem Carbon Fluxes and Canopy Spectral Reflectance of a Mountain Meadow. Int. J. Remote Sens. 2009, 30, 435–449. [Google Scholar] [CrossRef]
- Seitz, S.; Goebes, P.; Puerta, V.L.; Pereira, E.I.P.; Wittwer, R.; Six, J.; van der Heijden, M.G.A.; Scholten, T. Conservation Tillage and Organic Farming Reduce Soil Erosion. Agron. Sustain. Dev. 2019, 39, 4. [Google Scholar] [CrossRef]
- Doyle, C.J.; Topp, C.F.E. The Economic Opportunities for Increasing the Use of Forage Legumes in North European Livestock Systems under Both Conventional and Organic Management. Renew. Agric. Food Syst. 2004, 19, 15–22. [Google Scholar] [CrossRef]
- Laidig, F.; Piepho, H.P.; Drobek, T.; Meyer, U. Genetic and Non-Genetic Long-Term Trends of 12 Different Crops in German Official Variety Performance Trials and on-Farm Yield Trends. Theor. Appl. Genet. 2014, 127, 2599–2617. [Google Scholar] [CrossRef] [PubMed]
- Lollato, R.P.; Roozeboom, K.; Lingenfelser, J.F.; da Silva, C.L.; Sassenrath, G. Soft Winter Wheat Outyields Hard Winter Wheat in a Subhumid Environment: Weather Drivers, Yield Plasticity, and Rates of Yield Gain. Crop Sci. 2020, 60, 1617–1633. [Google Scholar] [CrossRef]
- Andrade, J.F.; Rattalino Edreira, J.I.; Mourtzinis, S.; Conley, S.P.; Ciampitti, I.A.; Dunphy, J.E.; Gaska, J.M.; Glewen, K.; Holshouser, D.L.; Kandel, H.J.; et al. Assessing the Influence of Row Spacing on Soybean Yield Using Experimental and Producer Survey Data. Field Crops Res. 2019, 230, 98–106. [Google Scholar] [CrossRef]
- Munaro, L.B.; Hefley, T.J.; DeWolf, E.; Haley, S.; Fritz, A.K.; Zhang, G.; Haag, L.A.; Schlegel, A.J.; Edwards, J.T.; Marburger, D.; et al. Exploring Long-Term Variety Performance Trials to Improve Environment-Specific Genotype × Management Recommendations: A Case-Study for Winter Wheat. Field Crops Res. 2020, 255, 107848. [Google Scholar] [CrossRef]
- Frazier, A.E. Landscape Heterogeneity and Scale Considerations for Super-Resolution Mapping. Int. J. Remote Sens. 2015, 36, 2395–2408. [Google Scholar] [CrossRef]
- Ge, Y.; Chen, Y.; Stein, A.; Li, S.; Hu, J. Enhanced Subpixel Mapping with Spatial Distribution Patterns of Geographical Objects. IEEE Trans. Geosci. Remote Sens. 2016, 54, 2356–2370. [Google Scholar] [CrossRef]
- Feng, W.; Yao, X.; Zhu, Y.; Tian, Y.C.; Cao, W.X. Monitoring Leaf Nitrogen Status with Hyperspectral Reflectance in Wheat. Eur. J. Agron. 2008, 28, 394–404. [Google Scholar] [CrossRef]
- Thorp, K.R.; Wang, G.; Bronson, K.F.; Badaruddin, M.; Mon, J. Hyperspectral Data Mining to Identify Relevant Canopy Spectral Features for Estimating Durum Wheat Growth, Nitrogen Status, and Grain Yield. Comput. Electron. Agric. 2017, 136, 1–12. [Google Scholar] [CrossRef]
- Tsouros, D.C.; Bibi, S.; Sarigiannidis, P.G. A Review on UAV-Based Applications for Precision Agriculture. Information 2019, 10, 349. [Google Scholar] [CrossRef]
- Raeva, P.L.; Šedina, J.; Dlesk, A. Monitoring of Crop Fields Using Multispectral and Thermal Imagery from UAV. Eur. J. Remote Sens. 2019, 52, 192–201. [Google Scholar] [CrossRef]
- Baret, F.; Guyot, G.; Major, D.J. TSAVI: A Vegetation Index Which Minimizes Soil Brightness Effects on LAI and APAR Estimation. In Proceedings of the Digest—International Geoscience and Remote Sensing Symposium (IGARSS), Vancouver, BC, Canada, 10–14 July 1989; Volume 3. [Google Scholar]
- Qi, J.; Chehbouni, A.; Huete, A.R.; Kerr, Y.H.; Sorooshian, S. A Modified Soil Adjusted Vegetation Index. Remote Sens. Environ. 1994, 48, 119–126. [Google Scholar] [CrossRef]
- Rhyma, P.P.; Norizah, K.; Hamdan, O.; Faridah-Hanum, I.; Zulfa, A.W. Integration of Normalised Different Vegetation Index and Soil-Adjusted Vegetation Index for Mangrove Vegetation Delineation. Remote Sens. Appl. Soc. Environ. 2020, 17, 100280. [Google Scholar] [CrossRef]
- Zhou, G. Determination of Green Aboveground Biomass in Desert Steppe Using Litter-Soil-Adjusted Vegetation Index. Eur. J. Remote Sens. 2014, 47, 611–625. [Google Scholar] [CrossRef]
- Mutanga, O.; Skidmore, A.K. Narrow Band Vegetation Indices Overcome the Saturation Problem in Biomass Estimation. Int. J. Remote Sens. 2004, 25, 3999–4014. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Ustin, S.L.; Whiting, M.L. Temporal and Spatial Relationships between Within-Field Yield Variability in Cotton and High-Spatial Hyperspectral Remote Sensing Imagery. Agron. J. 2005, 97, 641–653. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Pattey, E.; Zarco-Tejada, P.J.; Strachan, I.B. Hyperspectral Vegetation Indices and Novel Algorithms for Predicting Green LAI of Crop Canopies: Modeling and Validation in the Context of Precision Agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Monteiro, P.F.C.; Filho, R.A.; Xavier, A.C.; Monteiro, R.O.C. Assessing Biophysical Variable Parameters of Bean Crop with Hyperspectral Measurements. Sci. Agric. 2012, 69, 87–94. [Google Scholar] [CrossRef]
- Xavier, A.C.; Theodor Rudorff, B.F.; Moreira, M.A.; Alvarenga, B.S.; de Freitas, J.G.; Salomon, M.V. Hyperspectral Field Reflectance Measurements to Estimate Wheat Grain Yield and Plant Height. Sci. Agric. 2006, 63, 130–138. [Google Scholar] [CrossRef][Green Version]
- Stagakis, S.; Markos, N.; Sykioti, O.; Kyparissis, A. Monitoring Canopy Biophysical and Biochemical Parameters in Ecosystem Scale Using Satellite Hyperspectral Imagery: An Application on a Phlomis Fruticosa Mediterranean Ecosystem Using Multiangular CHRIS/PROBA Observations. Remote Sens. Environ. 2010, 114, 977–994. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Miller, J.R.; Noland, T.L.; Mohammed, G.H.; Sampson, P.H. Scaling-up and Model Inversion Methods with Narrowband Optical Indices for Chlorophyll Content Estimation in Closed Forest Canopies with Hyperspectral Data. IEEE Trans. Geosci. Remote Sens. 2001, 39, 1491–1507. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J. Hyperspectral Remote Sensing of Closed Forest Canopies: Estimation of Chlorophyll Fluorescence and Pigment Content; York University: Toronto, ON, Canada, 2000. [Google Scholar]
- Schmidt, K.S.; Skidmore, A.K. Spectral Discrimination of Vegetation Types in a Coastal Wetland. Remote Sens. Environ. 2003, 85, 92–108. [Google Scholar] [CrossRef]
- Feng, J.; Jiao, L.; Liu, F.; Sun, T.; Zhang, X. Unsupervised Feature Selection Based on Maximum Information and Minimum Redundancy for Hyperspectral Images. Pattern Recognit. 2016, 51, 295–309. [Google Scholar] [CrossRef]
- Yang, M.D.; Huang, K.H.; Tsai, H.P. Integrating MNF and HHT Transformations into Artificial Neural Networks for Hyperspectral Image Classification. Remote Sens. 2020, 12, 2327. [Google Scholar] [CrossRef]
- Bajcsy, P.; Groves, P. Methodology for Hyperspectral Band Selection. Photogramm. Eng. Remote Sens. 2004, 70, 793–802. [Google Scholar] [CrossRef]
- Heiskanen, J.; Rautiainen, M.; Stenberg, P.; Mõttus, M.; Vesanto, V.H. Sensitivity of Narrowband Vegetation Indices to Boreal Forest LAI, Reflectance Seasonality and Species Composition. ISPRS J. Photogramm. Remote Sens. 2013, 78, 1–14. [Google Scholar] [CrossRef]
- Näsi, R.; Viljanen, N.; Kaivosoja, J.; Alhonoja, K.; Hakala, T.; Markelin, L.; Honkavaara, E. Estimating Biomass and Nitrogen Amount of Barley and Grass Using UAV and Aircraft Based Spectral and Photogrammetric 3D Features. Remote Sens. 2018, 10, 1082. [Google Scholar] [CrossRef]
- Li, C.; Ma, C.; Pei, H.; Feng, H.; Shi, J.; Wang, Y.; Chen, W.; Li, Y.; Feng, X.; Shi, Y. Estimation of Potato Biomass and Yield Based on Machine Learning from Hyperspectral Remote Sensing Data. J. Agric. Sci. Technol. B 2020, 10, 195–213. [Google Scholar] [CrossRef]
- Choudhury, M.R.; Das, S.; Christopher, J.; Apan, A.; Chapman, S.; Menzies, N.W.; Dang, Y.P. Improving Biomass and Grain Yield Prediction of Wheat Genotypes on Sodic Soil Using Integrated High-Resolution Multispectral, Hyperspectral, 3d Point Cloud, and Machine Learning Techniques. Remote Sens. 2021, 13, 3482. [Google Scholar] [CrossRef]
- Nawar, S.; Corstanje, R.; Halcro, G.; Mulla, D.; Mouazen, A.M. Delineation of Soil Management Zones for Variable-Rate Fertilization: A Review. Adv. Agron. 2017, 143, 175–245. [Google Scholar]
- Chlingaryan, A.; Sukkarieh, S.; Whelan, B. Machine Learning Approaches for Crop Yield Prediction and Nitrogen Status Estimation in Precision Agriculture: A Review. Comput. Electron. Agric. 2018, 151, 61–69. [Google Scholar]
- Zhang, W.; Zhang, Z.; Chao, H.C.; Guizani, M. Toward Intelligent Network Optimization in Wireless Networking: An Auto-Learning Framework. IEEE Wirel. Commun. 2019, 26, 76–82. [Google Scholar] [CrossRef]
- Colombo, R.; Bellingeri, D.; Fasolini, D.; Marino, C.M. Retrieval of Leaf Area Index in Different Vegetation Types Using High Resolution Satellite Data. Remote Sens. Environ. 2003, 86, 120–131. [Google Scholar] [CrossRef]
- He, X.; Zhao, K.; Chu, X. AutoML: A Survey of the State-of-the-Art. Knowl.-Based Syst. 2021, 212, 106622. [Google Scholar] [CrossRef]
- Mendoza, H.; Klein, A.; Feurer, M.; Springenberg, J.T.; Hutter, F. Towards Automatically-Tuned Neural Networks. In Proceedings of the Workshop on Automatic Machine Learning, New York, NY, USA, 24 June 2016. [Google Scholar]
- Yao, Q.; Wang, M.; Chen, Y.; Dai, W.; Hu, Y.Q.; Li, Y.F.; Tu, W.W.; Yang, Q.; Yu, Y. Taking the Human out of Learning Applications: A Survey on Automated Machine Learning. arXiv 2018, arXiv:1810.13306. [Google Scholar]
- Thornton, C.; Hutter, F.; Hoos, H.H.; Leyton-Brown, K. Auto-WEKA: Combined Selection and Hyperparameter Optimization of Classification Algorithms. In Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Chicago, IL, USA, 11–14 August 2013; pp. 847–855. [Google Scholar] [CrossRef]
- Feurer, M.; Klein, A.; Eggensperger, K.; Springenberg, J.T.; Blum, M.; Hutter, F. Efficient and Robust Automated Machine Learning. In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 7–12 December 2015; Volume 2015. [Google Scholar]
- Remeseiro, B.; Bolon-Canedo, V. A Review of Feature Selection Methods in Medical Applications. Comput. Biol. Med. 2019, 112, 103375. [Google Scholar] [CrossRef]
- Babaeian, E.; Paheding, S.; Siddique, N.; Devabhaktuni, V.K.; Tuller, M. Estimation of Root Zone Soil Moisture from Ground and Remotely Sensed Soil Information with Multisensor Data Fusion and Automated Machine Learning. Remote Sens. Environ. 2021, 260, 112434. [Google Scholar] [CrossRef]
- Koh, J.C.O.; Spangenberg, G.; Kant, S. Automated Machine Learning for High-throughput Image-based Plant Phenotyping. Remote Sens. 2021, 13, 858. [Google Scholar] [CrossRef]
- Komer, B.; Bergstra, J.; Eliasmith, C. Hyperopt-Sklearn: Automatic Hyperparameter Configuration for Scikit-Learn. In Proceedings of the 13th Python in Science Conference, Austin, TX, USA, 6–12 July 2014. [Google Scholar]
- Lehnert, L.W.; Meyer, H.; Obermeier, W.A.; Silva, B.; Regeling, B.; Thies, B.; Bendix, J. Hyperspectral Data Analysis in R: The Hsdar Package. J. Stat. Softw. 2019, 89, 1–23. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- FAO. World Reference Base for Soil Resources; World Soil Resources Report 103; FAO: Rome, Italy, 2006; ISBN 9251055114. [Google Scholar]
- The Mathworks Inc. MATLAB (R2019a), Version 9.6.0.1072779; The MathWorks Inc.: Natick, MA, USA, 2019.
- Beleites, C. Unstable Laser Emission Vignette for the Data Set Laser of the R Package HyperSpec. Spectrosc. Laser 2021, 75, 1–6. [Google Scholar]
- Blackburn, G.A. Spectral Indices for Estimating Photosynthetic Pigment Concentrations: A Test Using Senescent Tree Leaves. Int. J. Remote Sens. 1998, 19, 657–675. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and Photographic Infrared Linear Combinations for Monitoring Vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Fernández-Manso, A.; Fernández-Manso, O.; Quintano, C. SENTINEL-2A Red-Edge Spectral Indices Suitability for Discriminating Burn Severity. Int. J. Appl. Earth Obs. Geoinf. 2016, 50, 170–175. [Google Scholar] [CrossRef]
- Gitelson, A.; Merzlyak, M.N. Quantitative Estimation of Chlorophyll-a Using Reflectance Spectra: Experiments with Autumn Chestnut and Maple Leaves. J. Photochem. Photobiol. B Biol. 1994, 22, 247–252. [Google Scholar] [CrossRef]
- Vescovo, L.; Wohlfahrt, G.; Balzarolo, M.; Pilloni, S.; Sottocornola, M.; Rodeghiero, M.; Gianelle, D. New Spectral Vegetation Indices Based on the Near-Infrared Shoulder Wavelengths for Remote Detection of Grassland Phytomass. Int. J. Remote Sens. 2012, 33, 2178–2195. [Google Scholar] [CrossRef]
- Kim, M.S.; Daughtry, C.S.T.; Chappelle, E.W.; McMurtrey, J.E.; Walthall, C.L. The Use of High Spectral Resolution Bands for Estimating Absorbed Photosynthetically Active Radiation (A Par). In Proceedings of the 6th Symposium on Physical Measurements and Signatures in Remote Sensing, Val d’Isère, France, 17–21 January 1994. [Google Scholar]
- Huete, A.R. A Soil-Adjusted Vegetation Index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Rondeaux, G.; Steven, M.; Baret, F. Optimization of Soil-Adjusted Vegetation Indices. Remote Sens. Environ. 1996, 55, 95–107. [Google Scholar] [CrossRef]
- Yeom, J.; Jung, J.; Chang, A.; Ashapure, A.; Maeda, M.; Maeda, A.; Landivar, J. Comparison of Vegetation Indices Derived from UAV Data for Differentiation of Tillage Effects in Agriculture. Remote Sens. 2019, 11, 1548. [Google Scholar] [CrossRef]
- Hernández-Clemente, R.; Navarro-Cerrillo, R.M.; Zarco-Tejada, P.J. Carotenoid Content Estimation in a Heterogeneous Conifer Forest Using Narrow-Band Indices and PROSPECT + DART Simulations. Remote Sens. Environ. 2012, 127, 32–46. [Google Scholar] [CrossRef]
- Demmig-Adams, B.; Adams, W.W. Photoprotection and Other Responses of Plants to High Light Stress. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1992, 43, 599–626. [Google Scholar] [CrossRef]
- Wu, C.; Niu, Z.; Tang, Q.; Huang, W. Estimating Chlorophyll Content from Hyperspectral Vegetation Indices: Modeling and Validation. Agric. For. Meteorol. 2008, 148, 1230–1241. [Google Scholar] [CrossRef]
- Roujean, J.L.; Breon, F.M. Estimating PAR Absorbed by Vegetation from Bidirectional Reflectance Measurements. Remote Sens. Environ. 1995, 51, 375–384. [Google Scholar] [CrossRef]
- ESRI. ArcGIS PRO: Essential Workflows; ESRI: Redlands, CA, USA, 2016. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Feurer, M.; Eggensperger, K.; Falkner, S.; Lindauer, M.; Hutter, F. Auto-Sklearn 2.0: The Next Generation. arXiv 2020, arXiv:2007.0407424. [Google Scholar]
- Hutter, F.; Hoos, H.H.; Leyton-Brown, K. Sequential Model-Based Optimization for General Algorithm Configuration. In Proceedings of the Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heidelberg, Germany, 2011; Volume 6683. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 24, 123–140. [Google Scholar] [CrossRef]
- Olson, R.S.; Urbanowicz, R.J.; Andrews, P.C.; Lavender, N.A.; Kidd, L.C.; Moore, J.H. Automating Biomedical Data Science through Tree-Based Pipeline Optimization. In Proceedings of the Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heidelberg, Germany, 2016; Volume 9597. [Google Scholar]
- Franceschi, L.; Frasconi, P.; Salzo, S.; Grazzi, R.; Pontil, M. Bilevel Programming for Hyperparameter Optimization and Meta-Learning. In International Conference on Machine Learning; PMLR; 2018; Volume 80, pp. 1568–1577. Available online: http://proceedings.mlr.press/v80/franceschi18a.html?ref=https://githubhelp.com (accessed on 26 January 2022).
- Feurer, M.; Springenberg, J.T.; Hutter, F. Initializing Bayesian Hyperparameter Optimization via Meta-Learning. In Proceedings of the National Conference on Artificial Intelligence, Austin, TX, USA, 25–30 January 2015; Volume 2. [Google Scholar]
- Yue, J.; Yang, G.; Li, C.; Li, Z.; Wang, Y.; Feng, H.; Xu, B. Estimation of Winter Wheat Above-Ground Biomass Using Unmanned Aerial Vehicle-Based Snapshot Hyperspectral Sensor and Crop Height Improved Models. Remote Sens. 2017, 9, 708. [Google Scholar] [CrossRef]
- Ono, J.P.; Castelo, S.; Lopez, R.; Bertini, E.; Freire, J.; Silva, C. PipelineProfiler: A Visual Analytics Tool for the Exploration of AutoML Pipelines. IEEE Trans. Vis. Comput. Graph. 2021, 27, 390–400. [Google Scholar] [CrossRef]
- Qi, Y.; Minka, T.P.; Picard, R.W.; Ghahramani, Z. Predictive Automatic Relevance Determination by Expectation Propagation. In Proceedings of the Twenty-First International Conference on Machine Learning, ICML 2004, Banff, AB, Canada, 4–8 July 2004. [Google Scholar]
- Seeger, M. Gaussian Processes for Machine Learning. Int. J. Neural Syst. 2004, 14, 69–106. [Google Scholar] [CrossRef]
- Van Diedenhoven, B.; Hasekamp, O.P.; Aben, I. Surface Pressure Retrieval from SCIAMACHY Measurements in the O2A Band: Validation of the Measurements and Sensitivity on Aerosols. Atmos. Chem. Phys. Discuss. 2005, 5, 2109–2120. [Google Scholar] [CrossRef]
- Riris, H.; Rodriguez, M.; Mao, J.; Allan, G.; Abshire, J. Airborne Demonstration of Atmospheric Oxygen Optical Depth Measurements with an Integrated Path Differential Absorption Lidar. Opt. Express 2017, 25, 29307–29327. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Gritz, Y.; Merzlyak, M.N. Relationships between Leaf Chlorophyll Content and Spectral Reflectance and Algorithms for Non-Destructive Chlorophyll Assessment in Higher Plant Leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Darvishzadeh, R.; Skidmore, A.; Atzberger, C.; van Wieren, S. Estimation of Vegetation LAI from Hyperspectral Reflectance Data: Effects of Soil Type and Plant Architecture. Int. J. Appl. Earth Obs. Geoinf. 2008, 10, 358–373. [Google Scholar] [CrossRef]
- Martínez-Usó, A.; Pla, F.; Sotoca, J.M.; García-Sevilla, P. Clustering-Based Hyperspectral Band Selection Using Information Measures. Proc. IEEE Trans. Geosci. Remote Sens. 2007, 45, 4158–4171. [Google Scholar]
- Thenkabail, P.S.; Smith, R.B.; de Pauw, E. Hyperspectral Vegetation Indices and Their Relationships with Agricultural Crop Characteristics. Remote Sens. Environ. 2000, 71, 158–182. [Google Scholar] [CrossRef]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Yang, M.D.; Tseng, H.H.; Hsu, Y.C.; Tsai, H.P. Semantic Segmentation Using Deep Learning with Vegetation Indices for Rice Lodging Identification in Multi-Date UAV Visible Images. Remote Sens. 2020, 12, 633. [Google Scholar] [CrossRef]
- Yang, M.D.; Boubin, J.G.; Tsai, H.P.; Tseng, H.H.; Hsu, Y.C.; Stewart, C.C. Adaptive Autonomous UAV Scouting for Rice Lodging Assessment Using Edge Computing with Deep Learning EDANet. Comput. Electron. Agric. 2020, 179, 683–696. [Google Scholar] [CrossRef]
- Luan, J.; Zhang, C.; Xu, B.; Xue, Y.; Ren, Y. The Predictive Performances of Random Forest Models with Limited Sample Size and Different Species Traits. Fish. Res. 2020, 227, 105534. [Google Scholar] [CrossRef]
- Feurer, M.; Hutter, F. Towards Further Automation in AutoML. In Proceedings of the ICML 2018 AutoML Workshop, Bangkok, Thailand, 25–28 October 2018. [Google Scholar]
- Ma, L.; Li, M.; Ma, X.; Cheng, L.; Du, P.; Liu, Y. A Review of Supervised Object-Based Land-Cover Image Classification. ISPRS J. Photogramm. Remote Sens. 2017, 130, 277–293. [Google Scholar] [CrossRef]
- Neal, R. Bayesian Learning for Neural Networks. In Lecture Notes in Statistics; Springer: New York, NY, USA, 1996; Volume 1. [Google Scholar]
- Mackay, D.J.C. Probable Networks and Plausible Predictions—A Review of Practical Bayesian Methods for Supervised Neural Networks. Netw. Comput. Neural Syst. 1995, 6, 469. [Google Scholar] [CrossRef]


| Vegetation Index | Description | Equation | Reference |
|---|---|---|---|
| NDVI | Normalized Difference Vegetation Index | (R800 − R680)/(R800 + R680) | [73] |
| NDVI2 | Normalized Difference Vegetation Index 2 | (R750−R705)/(R750 + R705) | [75] |
| OSAVI | Optimized Soil Adjusted Vegetation Index | (1 + 0.16) × (R800 − R670)/(R800 + R670 + 0.16) | [79] |
| OSAVI2 | Optimized Soil Adjusted Vegetation Index 2 | (1 + 0.16) × (R750 − R705)/(R750 + R705 + 0.16) | [83] |
| RDVI | Renormalized Difference Vegetation Index | (R800 − R670)/√(R800 + R670) | [84] |
| SR | Simple Ratio | R515/R550 | [81] |
| SAVI | Soil-Adjusted Vegetation Index | (1 + L 1) × (R800 − R670)/(R800 + R670 + L) | [78] |
| TCARI | Transformed Chlorophyll Absorption Reflectance Index | ((R700 − R670) − 0.2 × (R700 − R550) × (R700/R670) | [6] |
| Parameter Name | Range Value | Description |
|---|---|---|
| time_left_for_this_task | 30 s | The time restriction for seeking suitable models. |
| per_run_time_limit | 10 s | The maximum amount of time a single call to the ML model could perform. |
| ensemble_size | 50 (default) | Several models were added to the ensemble from Ensemble libraries. |
| ensemble_nbest | 50 (default) | The amount of best models for building an ensemble model. |
| resampling_strategy | CV; folds = 3 | (CV = cross-validation); to deal with overfitting |
| seed | 47 | Used to seed SMAC. |
| training/testing split | (0.5; 0.5) | Data partitioning way |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, K.-Y.; Sampaio de Lima, R.; Burnside, N.G.; Vahtmäe, E.; Kutser, T.; Sepp, K.; Cabral Pinheiro, V.H.; Yang, M.-D.; Vain, A.; Sepp, K. Toward Automated Machine Learning-Based Hyperspectral Image Analysis in Crop Yield and Biomass Estimation. Remote Sens. 2022, 14, 1114. https://doi.org/10.3390/rs14051114
Li K-Y, Sampaio de Lima R, Burnside NG, Vahtmäe E, Kutser T, Sepp K, Cabral Pinheiro VH, Yang M-D, Vain A, Sepp K. Toward Automated Machine Learning-Based Hyperspectral Image Analysis in Crop Yield and Biomass Estimation. Remote Sensing. 2022; 14(5):1114. https://doi.org/10.3390/rs14051114
Chicago/Turabian StyleLi, Kai-Yun, Raul Sampaio de Lima, Niall G. Burnside, Ele Vahtmäe, Tiit Kutser, Karli Sepp, Victor Henrique Cabral Pinheiro, Ming-Der Yang, Ants Vain, and Kalev Sepp. 2022. "Toward Automated Machine Learning-Based Hyperspectral Image Analysis in Crop Yield and Biomass Estimation" Remote Sensing 14, no. 5: 1114. https://doi.org/10.3390/rs14051114
APA StyleLi, K.-Y., Sampaio de Lima, R., Burnside, N. G., Vahtmäe, E., Kutser, T., Sepp, K., Cabral Pinheiro, V. H., Yang, M.-D., Vain, A., & Sepp, K. (2022). Toward Automated Machine Learning-Based Hyperspectral Image Analysis in Crop Yield and Biomass Estimation. Remote Sensing, 14(5), 1114. https://doi.org/10.3390/rs14051114








