Assessing the Dynamics of Plant Species Invasion in Eastern-Mediterranean Coastal Dunes Using Cellular Automata Modeling and Satellite Time-Series Analyses
Abstract
1. Introduction
2. Materials and Methods
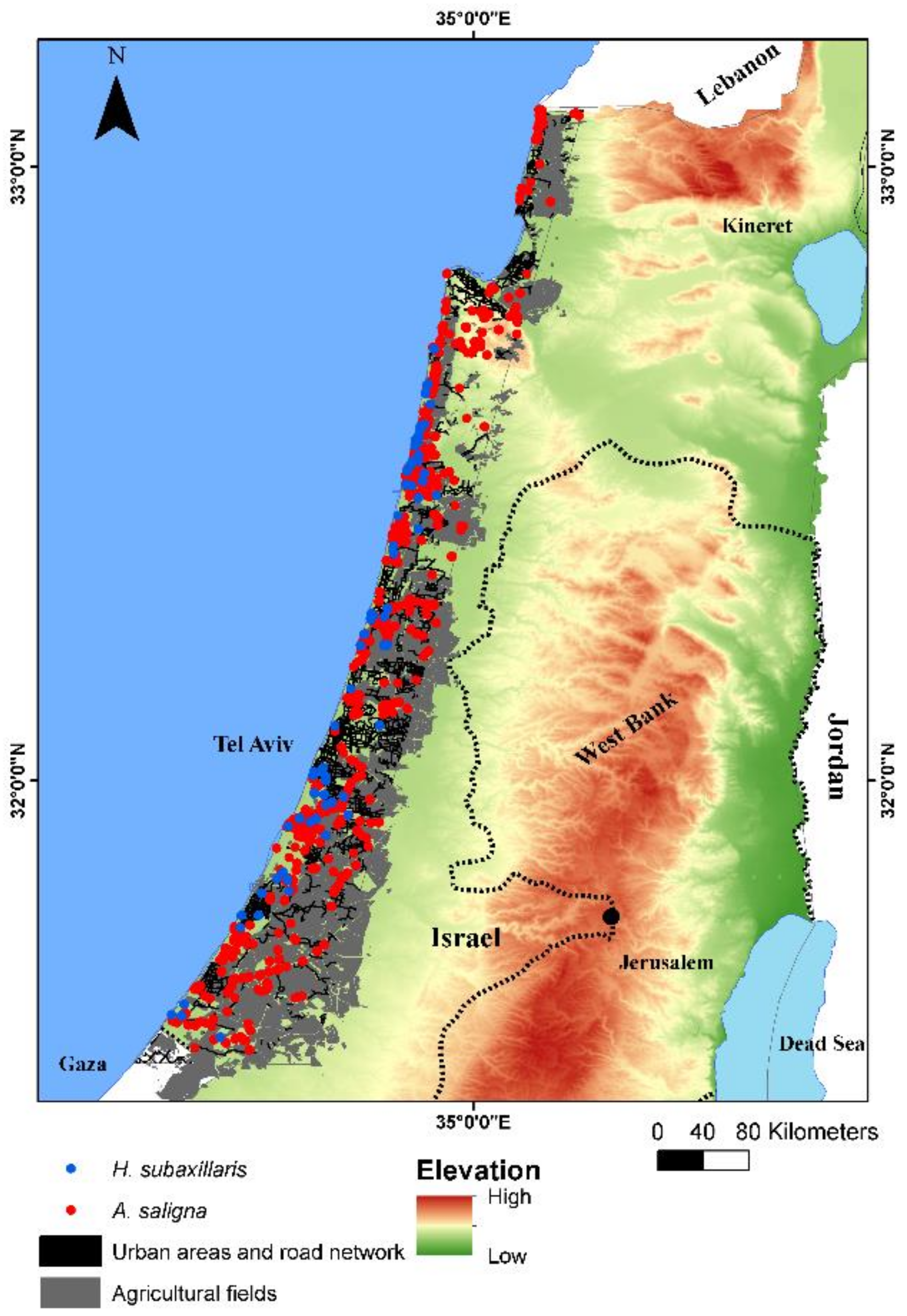
2.1. Research Area and Climate Trends
2.2. The Focal Plant Species
2.3. Spatial Data from Local Archive and Field Campaign
2.4. Spatial Data from Remote Sensing
2.5. Remote Sensing Classification
2.6. Modeling Expansion and Future Projections of the Plant Distributions
2.7. Accuracy Assessment of Remote Sensing Classification and the Plant Expansion Model
2.8. Landscape Studies
3. Results
3.1. Remote Sensing Classification of A. saligna and H. subaxillaris
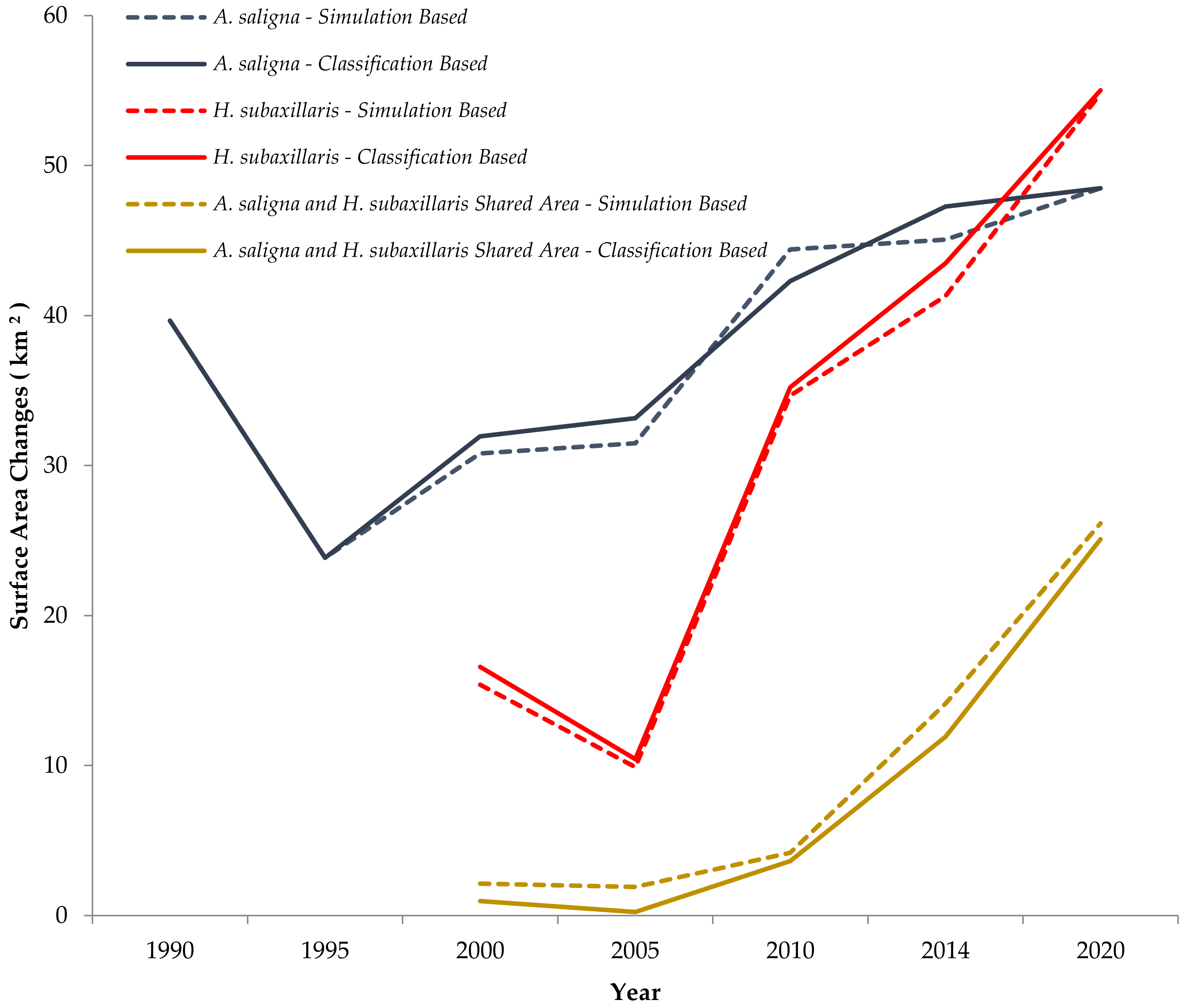
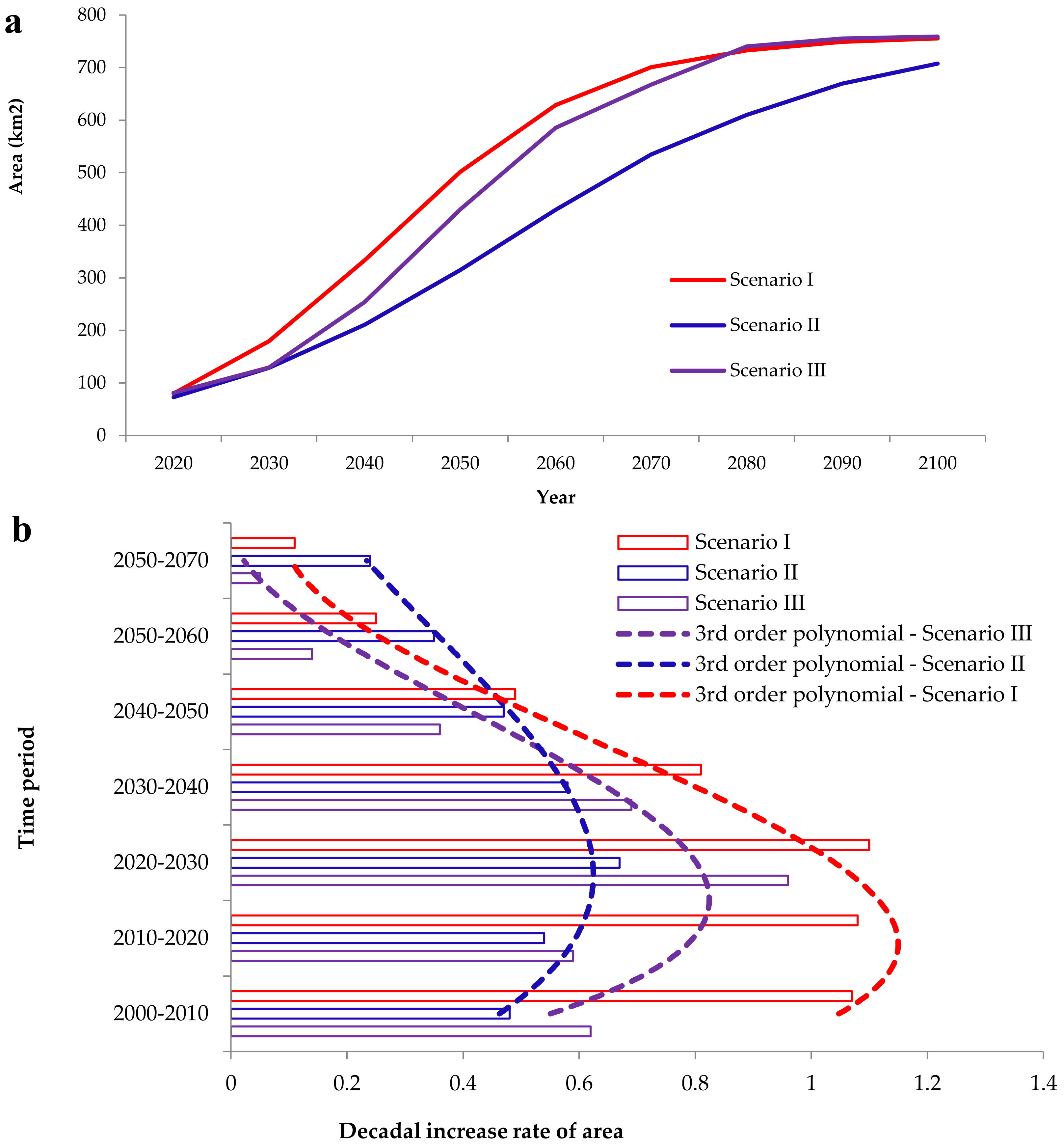
3.2. Simulation of Focal Species Expansion from 1990 to 2020 and Modeling Future Cover for the Period 2020–2100
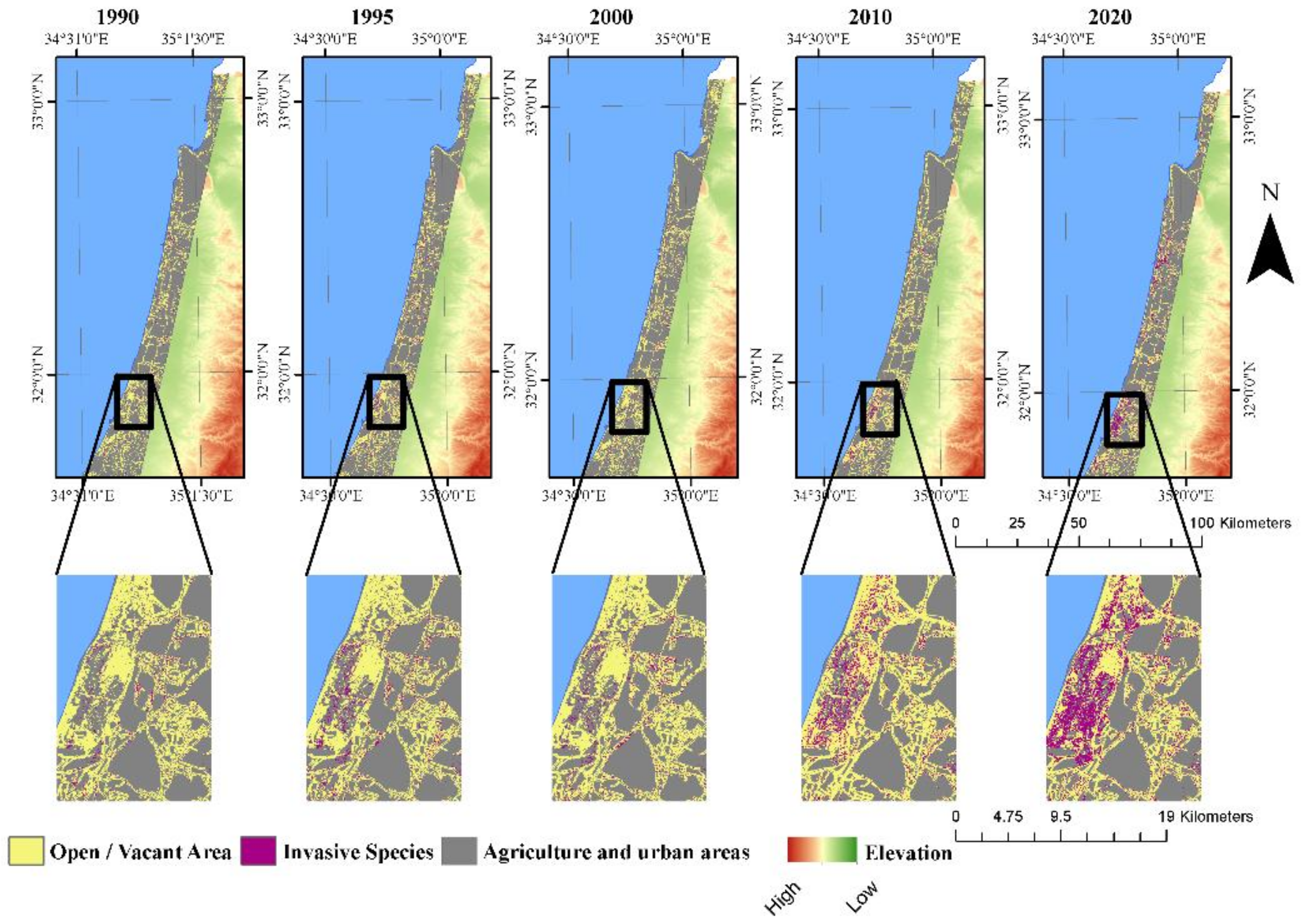
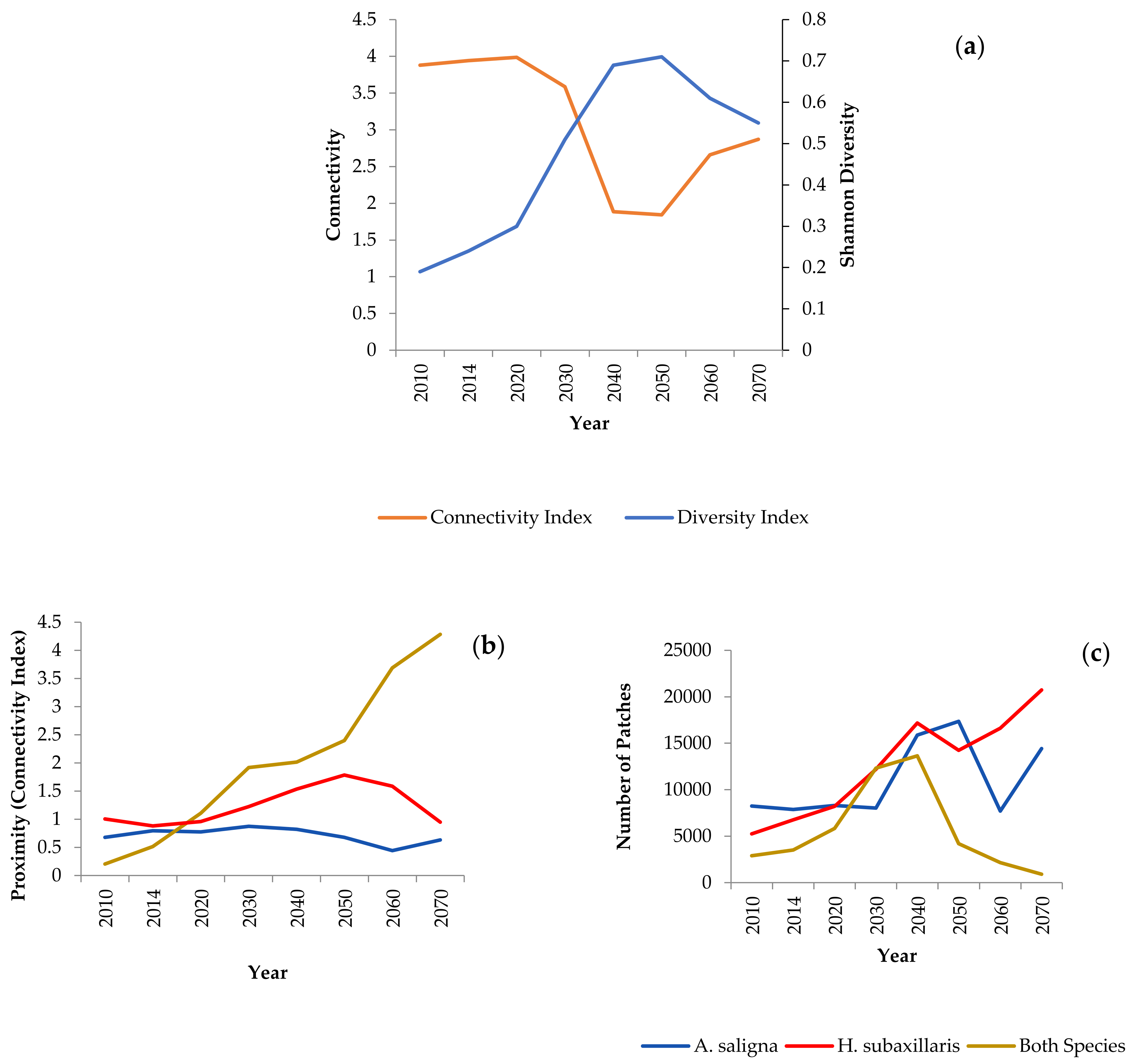
3.3. Landscape Dynamics
4. Discussion
4.1. Land Cover Changes of A. saligna and H. subaxillaris and Their Expansion Rate from 2000 to 2020
4.2. Future Projections of A. saligna and H. subaxillaris Expansion
4.3. Potential Effects of A. saligna and H. subaxillaris on the Landscape Matrix
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Year | A. saligna | H. subaxillaris |
|---|---|---|
| 1990–2000 | 47 | N/A |
| 2000–2005 | 56 | 67 |
| 2005–2010 | 110 | 156 |
| 2010–2015 | 159 | 256 |
| 2018–2020 | 357 | 393 |
| Image ID | Sensor | Date | Level |
|---|---|---|---|
| LT05_L2SP_175037_19901101_20200915_02_T1 | TM 5 | 1 November 1990 | L2 |
| LT05_L2SP_175038_19901101_20200915_02_T1 | TM 5 | 1 November 1990 | L2 |
| LT05_L2SP_174037_19900315_20200916_02_T1 | TM 5 | 15 March 1990 | L2 |
| LT05_L2SP_174038_19900315_20200916_02_T1 | TM 5 | 15 March 1990 | L2 |
| LT05_L2SP_175038_19951115_20200912_02_T1 | TM 5 | 15 November 1995 | L2 |
| LT05_L2SP_175037_19951115_20200912_02_T1 | TM 5 | 15 November 1995 | L2 |
| LT05_L2SP_175038_19950405_20200912_02_T1 | TM 5 | 5 April 1995 | L2 |
| LT05_L2SP_175037_19950405_20200912_02_T1 | TM 5 | 5 April 1995 | L2 |
| LT05_L2SP_175037_20001112_20200906_02_T1 | TM 5 | 12 November 2000 | L2 |
| LT05_L2SP_175038_20001112_20200906_02_T1 | TM 5 | 12 November 2000 | L2 |
| LT05_L2SP_174037_20000326_20200907_02_T1 | TM 5 | 26 March 2000 | L2 |
| LT05_L2SP_174038_20000326_20200907_02_T1 | TM 5 | 26 March 2000 | L2 |
| LT05_L2SP_175038_20050331_20200902_02_T1 | TM 5 | 31 March 2005 | L2 |
| LT05_L2SP_175037_20050331_20200902_02_T1 | TM 5 | 31 March 2005 | L2 |
| LT05_L2SP_174038_20061005_20200831_02_T1 | TM 5 | 5 October 2006 | L2 |
| LT05_L2SP_174037_20061005_20200831_02_T1 | TM 5 | 5 October 2006 | L2 |
| LT05_L2SP_175038_20090411_20200828_02_T1 | TM 5 | 11 April 2009 | L2 |
| LT05_L2SP_175037_20090411_20200828_02_T1 | TM 5 | 11 April 2009 | L2 |
| LT05_L2SP_175038_20101108_20200823_02_T1 | TM 5 | 18 November 2010 | L2 |
| LT05_L2SP_175037_20101108_20200823_02_T1 | TM 5 | 18 November 2010 | L2 |
| LC08_L2SP_174037_20140418_20200911_02_T1 | OLI 8 | 18 April 2014 | L2 |
| LC08_L2SP_174037_20141027_20200910_02_T1 | OLI 8 | 27 October 2014 | L2 |
| LC08_L2SP_174038_20140418_20200911_02_T1 | OLI 8 | 18 April 2014 | L2 |
| LC08_L2SP_174038_20141027_20200910_02_T1 | OLI 8 | 27 October 2014 | L2 |
| LC08_L2SP_175038_20200308_20200822_02_T1 | OLI 8 | 8 March 2020 | L2 |
| LC08_L2SP_175037_20200308_20200822_02_T1 | OLI 8 | 8 March 2020 | L2 |
| LC08_L2SP_175037_20201018_20201105_02_T1 | OLI 8 | 18 October 2020 | L2 |
| LC08_L2SP_175038_20201018_20201105_02_T1 | OLI 8 | 18 October 2020 | L2 |
| H. subaxillaris | A. saligna | |||||
|---|---|---|---|---|---|---|
| Year | Overall Accuracy | Kappa Coefficient | Number of Samples | Overall Accuracy | Kappa Coefficient | Number of Samples |
| 2010 | 79% | 0.54 | 280 | 83% | 0.62 | 360 |
| 2011 | 80% | 0.57 | 280 | 79% | 0.5 | 354 |
| 2012 | 81% | 0.45 | 240 | 79% | 0.49 | 326 |
| 2013 | 84% | 0.53 | 216 | 84% | 0.43 | 302 |
| 2014 | 88% | 0.45 | 201 | 82% | 0.47 | 306 |
| 2015 | 84% | 0.58 | 150 | 82% | 0.51 | 293 |
| 2016 | 85% | 0.59 | 148 | 82% | 0.47 | 305 |
| 2017 | 74% | 0.48 | 211 | 82% | 0.54 | 316 |
| 2018 | 81% | 0.54 | 176 | 85% | 0.6 | 326 |
| 2019 | 80% | 0.55 | 184 | 80% | 0.55 | 341 |
| 2020 | 84% | 0.61 | 259 | 83% | 0.63 | 369 |
| Model Training | |||
|---|---|---|---|
| A. saligna 1990–2015 | Arima | HOLT | TBATS |
| RMSE | 1.33 | 1.55 | 2.67 |
| MAPE | 1.85 | 2.69 | 5.98 |
| AIC | 68.8 | 92.4 | 109.2 |
| H. subaxillaris (2000–2015) | Arima | HOLT | TBATS |
| RMSE | 2.32 | 2.32 | 3.5 |
| MAPE | 3.79 | 4.01 | 12.86 |
| AIC | 58.1 | 70.55 | 76 |
| Model Validation (2015–2020) | |||
| A. saligna | ARIMA | HOLT | TBATS |
| RMSE | 0.22 | 0.47 | 15.8 |
| MAPE | 0.22 | 0.59 | 20.99 |
| H. subaxillaris | ARIMA | HOLT | TBATS |
| RMSE | 0.42 | 0.46 | 17.7 |
| MAPE | 0.47 | 0.49 | 21.52 |
References
- Hejda, M.; Pyšek, P.; Jarošík, V. Impact of Invasive Plants on the Species Richness, Diversity and Composition of Invaded Communities. J. Ecol. 2009, 97, 393–403. [Google Scholar] [CrossRef]
- Wardle, D.A.; Peltzer, D.A. Impacts of Invasive Biota in Forest Ecosystems in an Aboveground–Belowground Context. Biol. Invasions 2017, 19, 3301–3316. [Google Scholar] [CrossRef]
- Langmaier, M.; Lapin, K. A Systematic Review of the Impact of Invasive Alien Plants on Forest Regeneration in European Temperate Forests. Front. Plant Sci. 2020, 11, 1349. [Google Scholar] [CrossRef] [PubMed]
- Kumar Rai, P.; Singh, J.S. Invasive Alien Plant Species: Their Impact on Environment, Ecosystem Services and Human Health. Ecol. Indic. 2020, 111, 106020. [Google Scholar] [CrossRef] [PubMed]
- Kutiel, P.B.; Cohen, O.; Shoshany, M. Invasion Rate of the Alien Species Acacia Saligna within Coastal Sand Dune Habitats in Israel. Isr. J. Plant Sci. 2004, 52, 115–124. [Google Scholar] [CrossRef]
- Chechina, M.; Hamann, A. Choosing Species for Reforestation in Diverse Forest Communities: Social Preference versus Ecological Suitability. Ecosphere 2015, 6, 1–13. [Google Scholar] [CrossRef]
- Makowski, C.; Finkl, C.W. Impacts of Invasive Species on Coastal Environments: Coast in Crisis; Springer Nature: Berlin, Germany, 2018; ISBN 9783319913827. [Google Scholar]
- Brown, A.C.; McLachlan, A. Sandy Shore Ecosystems and the Threats Facing Them: Some Predictions for the Year 2025. Environ. Conserv. 2002, 29, 62–77. [Google Scholar] [CrossRef]
- Mijović, A.; Popović, Z.; Vukov, T.; Smiljanić, M.; Matić, R.; Bojović, S. Sand Topography Influences the Distribution of Xerohalophytic Vegetation on a Southern Adriatic Beach in Montenegro. Plant Biosyst. 2012, 146, 664–673. [Google Scholar] [CrossRef]
- Farris, E.; Pisanu, S.; Ceccherelli, G.; Filigheddu, R. Human Trampling Effects on Mediterranean Coastal Dune Plants. Plant Biosystems 2013, 147, 1043–1051. [Google Scholar] [CrossRef]
- Kutiel, P. Conservation and Management of the Mediterranean Coastal Sand Dunes in Israel. J. Coast. Conserv. 2001, 2, 183–192. [Google Scholar] [CrossRef]
- Avis, A.M. A Review of Coastal Dune Stabilization in the Cape Province of South Africa. Landsc. Urban Plan. 1989, 18, 55–68. [Google Scholar] [CrossRef]
- Kettenring, K.M.; Adams, C.R. Lessons Learned from Invasive Plant Control Experiments: A Systematic Review and Meta-Analysis. J. Appl. Ecol. 2011, 48, 970–979. [Google Scholar] [CrossRef]
- Richardson, D.M.; Rejmánek, M. Trees and Shrubs as Invasive Alien Species—A Global Review. Divers. Distrib. 2011, 17, 788–809. [Google Scholar] [CrossRef]
- Dafni, A.; Heller, D. Invasions of Adventive Plants in Israel. In Biological Invasions in Europe and the Mediterranean Basin; Springer: Dodrecht, Germany, 1990; pp. 135–160. [Google Scholar] [CrossRef]
- Weidlich, E.W.A.; Flórido, F.G.; Sorrini, T.B.; Brancalion, P.H.S. Controlling Invasive Plant Species in Ecological Restoration: A Global Review. J. Appl. Ecol. 2020, 57, 1806–1817. [Google Scholar] [CrossRef]
- Stone, C.M.; Witt, A.B.R.; Walsh, G.C.; Foster, W.A.; Murphy, S.T. Would the Control of Invasive Alien Plants Reduce Malaria Transmission? A Review. Parasites Vectors 2018, 11, 76. [Google Scholar] [CrossRef]
- Jones, B.A. Tree Shade, Temperature, and Human Health: Evidence from Invasive Species-Induced Deforestation. Ecol. Econ. 2019, 156, 12–23. [Google Scholar] [CrossRef]
- Reaser, J.K.; Meyerson, L.A.; Cronk, Q.; De Poorter, M.; Eldrege, L.G.; Green, E.; Kairo, M.; Latasi, P.; Mack, R.N.; Mauremootoo, J.; et al. Ecological and Socioeconomic Impacts of Invasive Alien Species in Island Ecosystems. Environ. Conserv. 2007, 34, 98–111. [Google Scholar] [CrossRef]
- Pyšek, P.; Richardson, D.M. Invasive Species, Environmental Change and Management, and Health. Annu. Rev. Environ. Resour. 2010, 35, 25–55. [Google Scholar] [CrossRef]
- Young, H.S.; Parker, I.M.; Gilbert, G.S.; Sofia Guerra, A.; Nunn, C.L. Introduced Species, Disease Ecology, and Biodiversity–Disease Relationships. Trends Ecol. Evol. 2017, 32, 41–54. [Google Scholar] [CrossRef]
- Jones, B.A.; McDermott, S.M. Health Impacts of Invasive Species Through an Altered Natural Environment: Assessing Air Pollution Sinks as a Causal Pathway. Environ. Resour. Econ. 2018, 71, 23–43. [Google Scholar] [CrossRef]
- Bartz, R.; Kowarik, I. Assessing the Environmental Impacts of Invasive Alien Plants: A Review of Assessment Approaches. NeoBiota 2019, 43, 69–99. [Google Scholar] [CrossRef]
- Shrestha, U.B.; Shrestha, B.B. Climate Change Amplifies Plant Invasion Hotspots in Nepal. Divers. Distrib. 2019, 25, 1599–1612. [Google Scholar] [CrossRef]
- Jesse, W.A.M.; Molleman, J.; Franken, O.; Lammers, M.; Berg, M.P.; Behm, J.E.; Helmus, M.R.; Ellers, J. Disentangling the Effects of Plant Species Invasion and Urban Development on Arthropod Community Composition. Glob. Change Biol. 2020, 26, 3294–3306. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, E.; Buccheri, M.; Martini, F.; Boscutti, F. Agricultural Land Use Curbs Exotic Invasion but Sustains Native Plant Diversity at Intermediate Levels. Sci. Rep. 2021, 11, 8385. [Google Scholar] [CrossRef]
- Har-Edom, O.L.; Sternberg, M. Invasive Species and Climate Change: Conyza Canadensis (L.) Cronquist as a Tool for Assessing the Invasibility of Natural Plant Communities along an Aridity Gradient. Biol. Invasions 2010, 12, 1953–1960. [Google Scholar] [CrossRef]
- Adams, V.M.; Petty, A.M.; Douglas, M.M.; Buckley, Y.M.; Ferdinands, K.B.; Okazaki, T.; Ko, D.W.; Setterfield, S.A. Distribution, Demography and Dispersal Model of Spatial Spread of Invasive Plant Populations with Limited Data. Methods Ecol. Evol. 2015, 6, 782–794. [Google Scholar] [CrossRef]
- Fournier, A.; Barbet-Massin, M.; Rome, Q.; Courchamp, F. Predicting Species Distribution Combining Multi-Scale Drivers. Glob. Ecol. Conserv. 2017, 12, 215–226. [Google Scholar] [CrossRef]
- Manzoor, S.A.; Griffiths, G.; Lukac, M. Species Distribution Model Transferability and Model Grain Size-Finer May Not Always Be Better. Sci. Rep. 2018, 8, 7168. [Google Scholar] [CrossRef]
- Lake, T.A.; Briscoe Runquist, R.D.; Moeller, D.A. Predicting Range Expansion of Invasive Species: Pitfalls and Best Practices for Obtaining Biologically Realistic Projections. Divers. Distrib. 2020, 26, 1767–1779. [Google Scholar] [CrossRef]
- von Neumann, J. Theory of Self-Reproducing Automata; University of Illionois Press: Champain, IL, USA, 1966. [Google Scholar]
- Jongejans, E.; Shea, K.; Skarpaas, O.; Kelly, D.; Sheppard, A.W.; Woodburn, T.L. Dispersal and Demography Contributions to Population Spread of Carduus Nutans in Its Native and Invaded Ranges. J. Ecol. 2008, 96, 687–697. [Google Scholar] [CrossRef]
- Berger, U.; Piou, C.; Schiffers, K.; Grimm, V. Competition among Plants: Concepts, Individual-Based Modelling Approaches, and a Proposal for a Future Research Strategy. Perspect. Plant Ecol. Evol. Syst. 2008, 9, 121–135. [Google Scholar] [CrossRef]
- Merow, C.; Lafleur, N.; Silander, J.A.; Wilson, A.M.; Rubega, M. Developing Dynamic Mechanistic Species Distribution Models: Predicting Bird-Mediated Spread of Invasive Plants across Northeastern North America. Am. Nat. 2011, 178, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Maeda, E.E.; de Almeida, C.M.; de Carvalho Ximenes, A.A.; Formaggio, A.R.; Shimabukuro, Y.E.; Pellikka, P. Dynamic Modeling of Forest Conversion: Simulation of Past and Future Scenarios of Rural Activities Expansion in the Fringes of the Xingu National Park, Brazilian Amazon. Int. J. Appl. Earth Obs. Geoinf. 2011, 13, 435–446. [Google Scholar] [CrossRef]
- Hewitt, R.; van Delden, H.; Escobar, F. Participatory Land Use Modelling, Pathways to an Integrated Approach. Environ. Model. Softw. 2014, 52, 149–165. [Google Scholar] [CrossRef]
- Lopez, S. Modeling Agricultural Change through Logistic Regression and Cellular Automata: A Case Study on Shifting Cultivation. J. Geogr. Inf. Syst. 2014, 6, 220–235. [Google Scholar] [CrossRef][Green Version]
- George, J.S.; Paul, S.K.; Dhawale, R. A Cellular-Automata Model for Assessing the Sensitivity of the Street Network to Natural Terrain. Ann. GIS 2021, 27, 261–272. [Google Scholar] [CrossRef]
- Sternberg, M.; Yu, S.L.; Bar, P. Soil Seed Banks, Habitat Heterogeneity, and Regeneration Strategies in a Mediterranean Coastal Sand Dune. Isr. J. Plant Sci. 2004, 52, 213–221. [Google Scholar] [CrossRef]
- Stanley, T.D.; Ross, E.M. Flora of South-Eastern Queensland; Queensland Department of Primary Industries: Brisbane, Australia, 1983; Volume 1. [Google Scholar]
- Sternberg, M. From America to the Holy Land: Disentangling Plant Traits of the Invasive Heterotheca subaxillaris (Lam.) Britton & Rusby. Plant Ecol. 2016, 217, 1307–1314. [Google Scholar] [CrossRef]
- Nativ, R.; Ephrath, J.E.; Berliner, P.R.; Saranga, Y. Drought Resistance and Water Use Efficiency in Acacia saligna. Aust. J. Bot. 1999, 47, 577–586. [Google Scholar] [CrossRef]
- Morimoto, M.; Cantrell, C.L.; Libous-Bailey, L.; Duke, S.O. Phytotoxicity of Constituents of Glandular Trichomes and the Leaf Surface of Camphorweed, Heterotheca Subaxillaris. Phytochemistry 2009, 70, 69–74. [Google Scholar] [CrossRef]
- El-Gendy, S.A.; Abd Elmoniem, E.M.; Al-Abdulla, M.M.; Eisa, S.S. Morphological and Physiological Responses of Acacia saligna (Labill.) to Water Stress. Aust. J. Basic Appl. Sci. 2012, 6, 90–97. [Google Scholar]
- Holben, B.N. Characteristics of Maximum-Value Composite Images from Temporal AVHRR Data. Int. J. Remote Sens. 1986, 7, 1417–1434. [Google Scholar] [CrossRef]
- Weier, J.; Herring, D. Measuring Vegetation (NDVI & EVI)—Normalized Difference Vegetation Index (NDVI); NASA Earth Observatory: Greenbelt, MD, USA, 2000. [Google Scholar]
- Fone, A.L. A Comparative Demographic Study of Annual and Perennial Hypochoeris (Asteraceae). J. Ecol. 1989, 77, 495–508. [Google Scholar] [CrossRef]
- Brodie, J.F.; Roland, C.A.; Stehn, S.E.; Smirnova, E. Variability in the Expansion of Trees and Shrubs in Boreal Alaska. Ecology 2019, 100, e02660. [Google Scholar] [CrossRef]
- Hagoort, M.; Geertman, S.; Ottens, H. Spatial Externalities, Neighbourhood Rules and CA Land-Use Modelling. Ann. Reg. Sci. 2008, 42, 39–56. [Google Scholar] [CrossRef]
- Liao, J.; Tang, L.; Shao, G.; Su, X.; Chen, D.; Xu, T. Incorporation of Extended Neighborhood Mechanisms and Its Impact on Urban Land-Use Cellular Automata Simulations. Environ. Model. Softw. 2016, 75, 163–175. [Google Scholar] [CrossRef]
- Congalton, R.G. A Review of Assessing the Accuracy of Classifications of Remotely Sensed Data. Remote Sens. Environ. 1991, 37, 35–46. [Google Scholar] [CrossRef]
- Dabija, A.; Kluczek, M.; Zagajewski, B.; Raczko, E.; Kycko, M.; Al-Sulttani, A.H.; Tardà, A.; Pineda, L.; Corbera, J. Comparison of Support Vector Machines and Random Forests for Corine Land Cover Mapping. Remote Sens. 2021, 13, 777. [Google Scholar] [CrossRef]
- Sheykhmousa, M.; Mahdianpari, M.; Ghanbari, H.; Mohammadimanesh, F.; Ghamisi, P.; Homayouni, S. Support Vector Machine Versus Random Forest for Remote Sensing Image Classification: A Meta-Analysis and Systematic Review. IEEE J. Sel. Top. Appl. Earth Observ. Remote Sens. 2020, 13, 6308–6325. [Google Scholar] [CrossRef]
- Huang, C.; Davis, L.S.; Townshend, J.R.G. An Assessment of Support Vector Machines for Land Cover Classification. Int. J. Remote Sens. 2002, 23, 725–749. [Google Scholar] [CrossRef]
- Li, C.; Wang, J.; Wang, L.; Hu, L.; Gong, P. Comparison of Classification Algorithms and Training Sample Sizes in Urban Land Classification with Landsat Thematic Mapper Imagery. Remote Sens. 2014, 6, 964–983. [Google Scholar] [CrossRef]
- Xie, Y.; Sha, Z.; Yu, M. Remote Sensing Imagery in Vegetation Mapping: A Review. J. Plant Ecol. 2008, 1, 9–23. [Google Scholar] [CrossRef]
- Xiao, H.; Su, F.; Fu, D.; Lyne, V.; Liu, G.; Pan, T.; Teng, J. Optimal and Robust Vegetation Mapping in Complex Environments Using Multiple Satellite Imagery: Application to Mangroves in Southeast Asia. Int. J. Appl. Earth Obs. Geoinf. 2021, 99, 102320. [Google Scholar] [CrossRef]
- National Research Council. Firewood Crops: Shrub and Tree Species for Energy Production. Available online: https://www.nap.edu/catalog/21317/firewood-crops-shrub-and-tree-species-for-energy-production (accessed on 11 January 2022).
- Zhu, K.; Woodall, C.W.; Clark, J.S. Failure to Migrate: Lack of Tree Range Expansion in Response to Climate Change. Glob. Change Biol. 2012, 18, 1042–1052. [Google Scholar] [CrossRef]
- Cohen, O.; Bar, P. The impact of Acacia saligna invasion on the indigenous vegetation in various coastal habitats in Israel and its implication for nature conservation. Isr. J. Plant Sci. 2017, 45, 1–11. [Google Scholar] [CrossRef]
- Baskin, J.M.; Baskin, C.C. Germination Dimorphism in Heterotheca subaxillaris Var. subaxillaris. Bull. Torrey Bot. Club 1976, 103, 201–206. [Google Scholar] [CrossRef]
- Lonard, R.I.; Judd, F.W.; Stalter, R. Biological Flora of Coastal Dunes and Wetlands: Heterotheca subaxillaris (J. de Lamarck) N. Britton & H. Rusby. J. Coast. Res. 2011, 27, 1052–1058. [Google Scholar] [CrossRef]
- Le Maitre, D.C.; Gaertner, M.; Marchante, E.; Ens, E.J.; Holmes, P.M.; Pauchard, A.; O’Farrell, P.J.; Rogers, A.M.; Blanchard, R.; Blignaut, J.; et al. Impacts of Invasive Australian Acacias: Implications for Management and Restoration. Divers. Distrib. 2011, 17, 1015–1029. [Google Scholar] [CrossRef]
- Lockwood, J.L.; Cassey, P.; Blackburn, T. The Role of Propagule Pressure in Explaining Species Invasions. Trends Ecol. Evol. 2005, 20, 223–228. [Google Scholar] [CrossRef]
- Oliver, T.; Roy, D.B.; Hill, J.K.; Brereton, T.; Thomas, C.D. Heterogeneous Landscapes Promote Population Stability. Ecol. Lett. 2010, 13, 473–484. [Google Scholar] [CrossRef]
- Eilts, J.A.; Mittelbach, G.G.; Reynolds, H.L.; Gross, K.L. Resource Heterogeneity, Soil Fertility, and Species Diversity: Effects of Clonal Species on Plant Communities. Am. Nat. 2011, 177, 574–588. [Google Scholar] [CrossRef] [PubMed]
- Hochman, A.; Kunin, P.; Alpert, P.; Harpaz, T.; Saaroni, H.; Rostkier-Edelstein, D. Weather regimes and analogues downscaling of seasonal precipitation: A case study over Israel. Int. J. Climatol. 2020, 40, 2062–2077. [Google Scholar] [CrossRef]

| A. saligna | H. subaxillaris | |||||||
|---|---|---|---|---|---|---|---|---|
| Year | OA (%) | PA/UA (%) | Kappa Coefficient | Number of Validation Samples | OA (%) | PA/UA (%) | Kappa Coefficient | Number of Validation Samples |
| 1990 | 77 | 86/63 | 0.68 | 30 | ||||
| 1995 | 89 | 97/74 | 0.69 | 30 | ||||
| 2000 | 91 | 98/87 | 0.7 | 30 | 85 | 95/74 | 0.65 | 25 |
| 2005 | 94 | 100/78 | 0.71 | 158 | 84 | 88/81 | 0.69 | 25 |
| 2010 | 90 | 98/83 | 0.77 | 360 | 86 | 94/78 | 0.70 | 280 |
| 2014 | 94 | 100/84 | 0.82 | 306 | 85 | 90/81 | 0.64 | 216 |
| 2020 | 87 | 98/77 | 0.75 | 369 | 85 | 84/89 | 0.68 | 259 |
| Year | Area Covered Exclusively by A. saligna (km2) | Area Covered Exclusively by H. subaxillaris (km2) | Mixed Land Cover Class (km2) | Vacant Area (%) | Vacant Area (km2) |
|---|---|---|---|---|---|
| 1990 | 39.7 | N/A | N/A | 94 | 730 |
| 1995 | 23.8 | N/A | N/A | 96 | 746 |
| 2000 | 31.2 | 17.0 | 0.97 | 92 | 720 |
| 2006 | 33.2 | 10.4 | 0.24 | 93 | 726 |
| 2010 | 42.3 | 35.2 | 3.64 | 88 | 689 |
| 2014 | 47.3 | 43.5 | 11.9 | 86 | 667 |
| 2020 | 48.7 | 55.0 | 25.9 | 82 | 640 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kozhoridze, G.; Dor, E.B.; Sternberg, M. Assessing the Dynamics of Plant Species Invasion in Eastern-Mediterranean Coastal Dunes Using Cellular Automata Modeling and Satellite Time-Series Analyses. Remote Sens. 2022, 14, 1014. https://doi.org/10.3390/rs14041014
Kozhoridze G, Dor EB, Sternberg M. Assessing the Dynamics of Plant Species Invasion in Eastern-Mediterranean Coastal Dunes Using Cellular Automata Modeling and Satellite Time-Series Analyses. Remote Sensing. 2022; 14(4):1014. https://doi.org/10.3390/rs14041014
Chicago/Turabian StyleKozhoridze, Giorgi, Eyal Ben Dor, and Marcelo Sternberg. 2022. "Assessing the Dynamics of Plant Species Invasion in Eastern-Mediterranean Coastal Dunes Using Cellular Automata Modeling and Satellite Time-Series Analyses" Remote Sensing 14, no. 4: 1014. https://doi.org/10.3390/rs14041014
APA StyleKozhoridze, G., Dor, E. B., & Sternberg, M. (2022). Assessing the Dynamics of Plant Species Invasion in Eastern-Mediterranean Coastal Dunes Using Cellular Automata Modeling and Satellite Time-Series Analyses. Remote Sensing, 14(4), 1014. https://doi.org/10.3390/rs14041014







