A Voxel-Based Individual Tree Stem Detection Method Using Airborne LiDAR in Mature Northeastern U.S. Forests
Abstract
:1. Introduction
1.1. Evolution of Remote Sensing for Next-Gen Forest Inventories
1.2. Application and Challenges of Individual Tree Detection in Hardwood Forests
1.3. Inconsistency in ITD Evaluation Metrics and Methods
1.4. Study Objectives
2. Materials and Methods
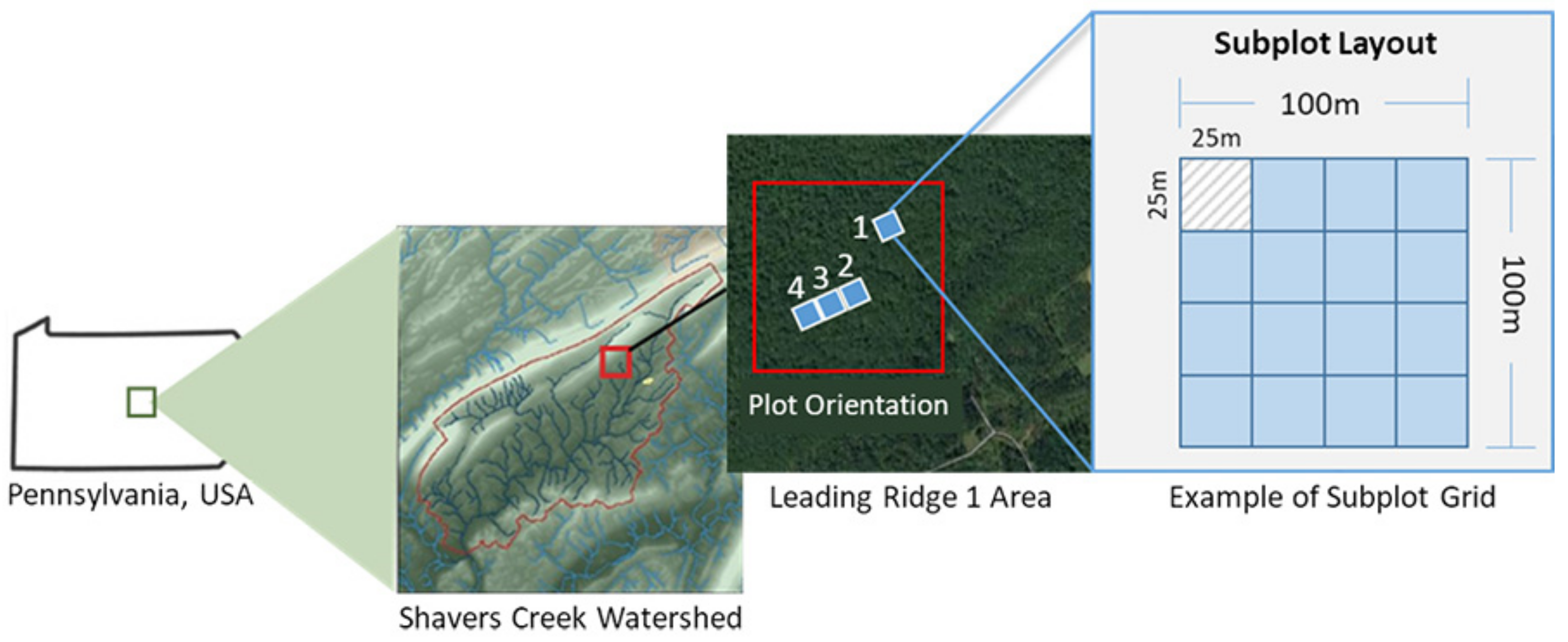
2.1. Study Area
2.2. Reference Tree Data
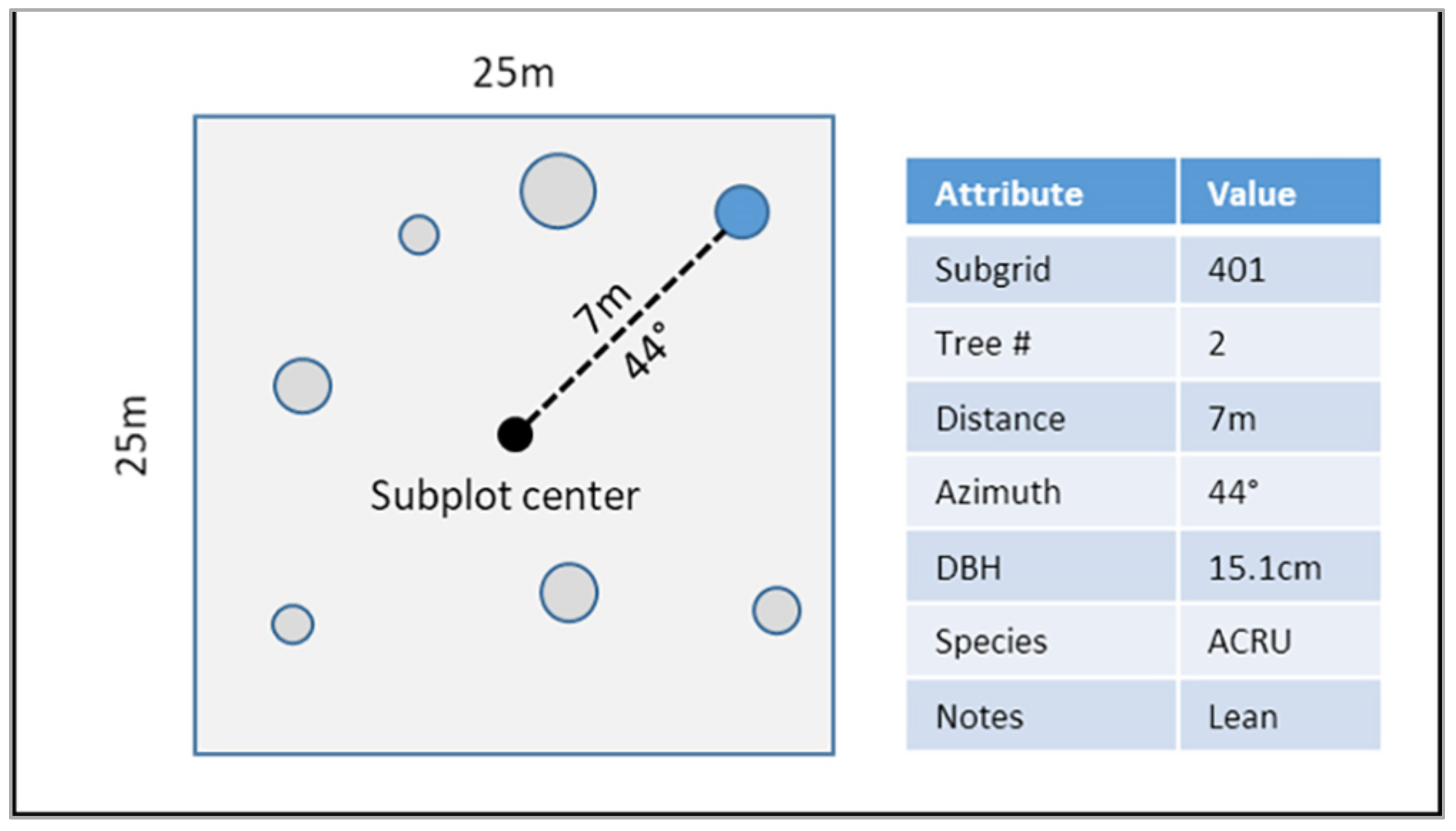
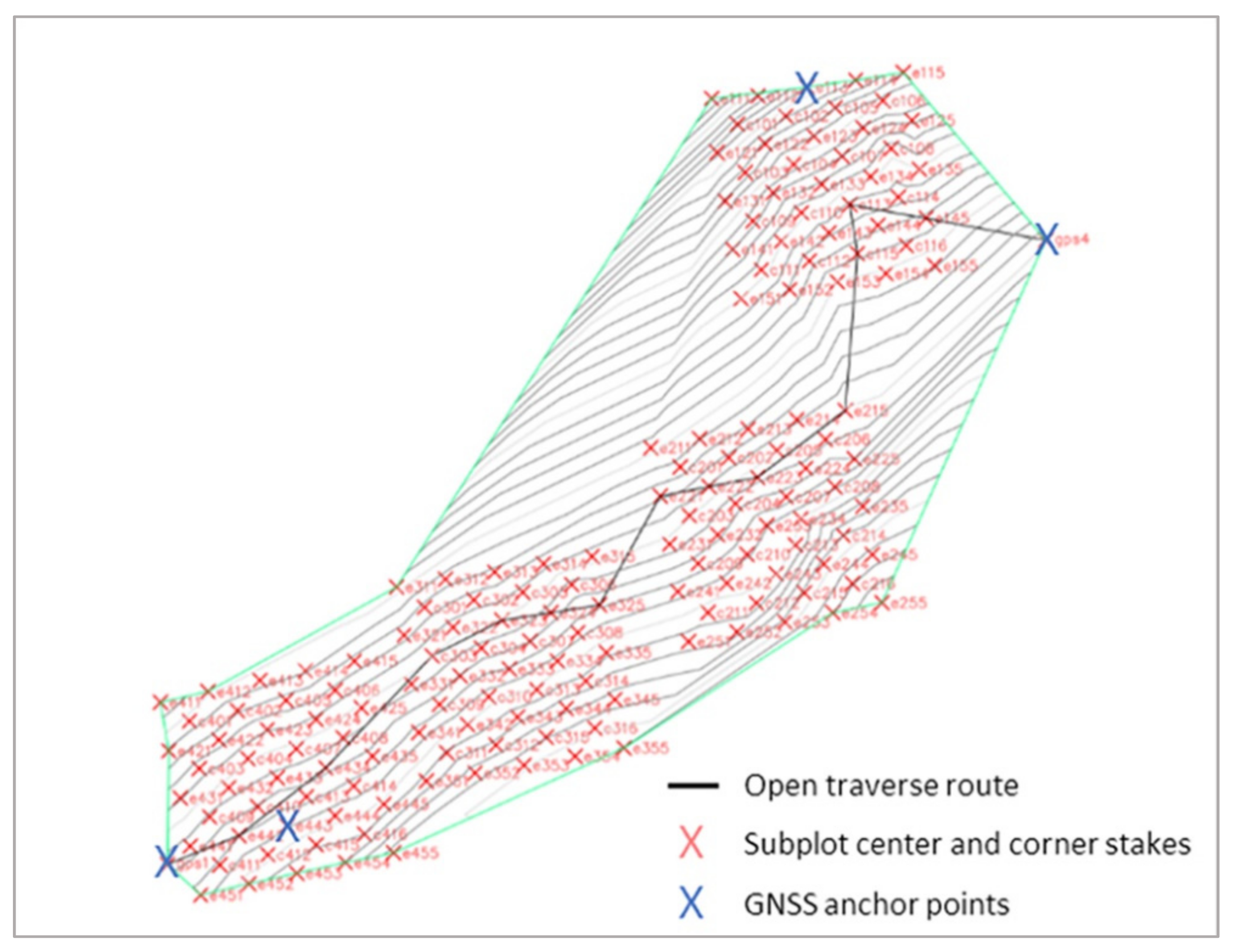
2.2.1. Surveying Ground Plots
2.2.2. Linking the Local Coordinate System to Geographic Coordinates via GPS
2.3. Airborne Laser Scanning (ALS) Data
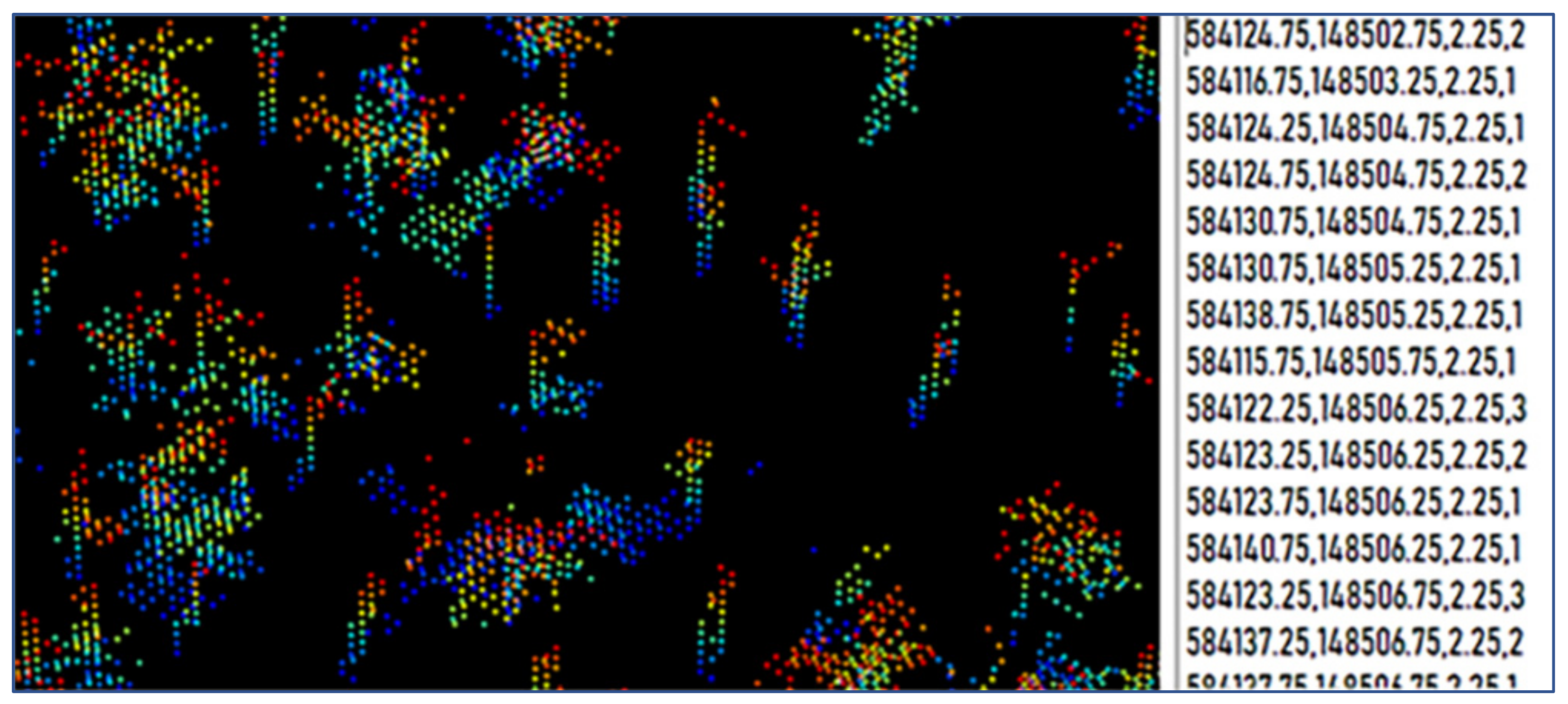
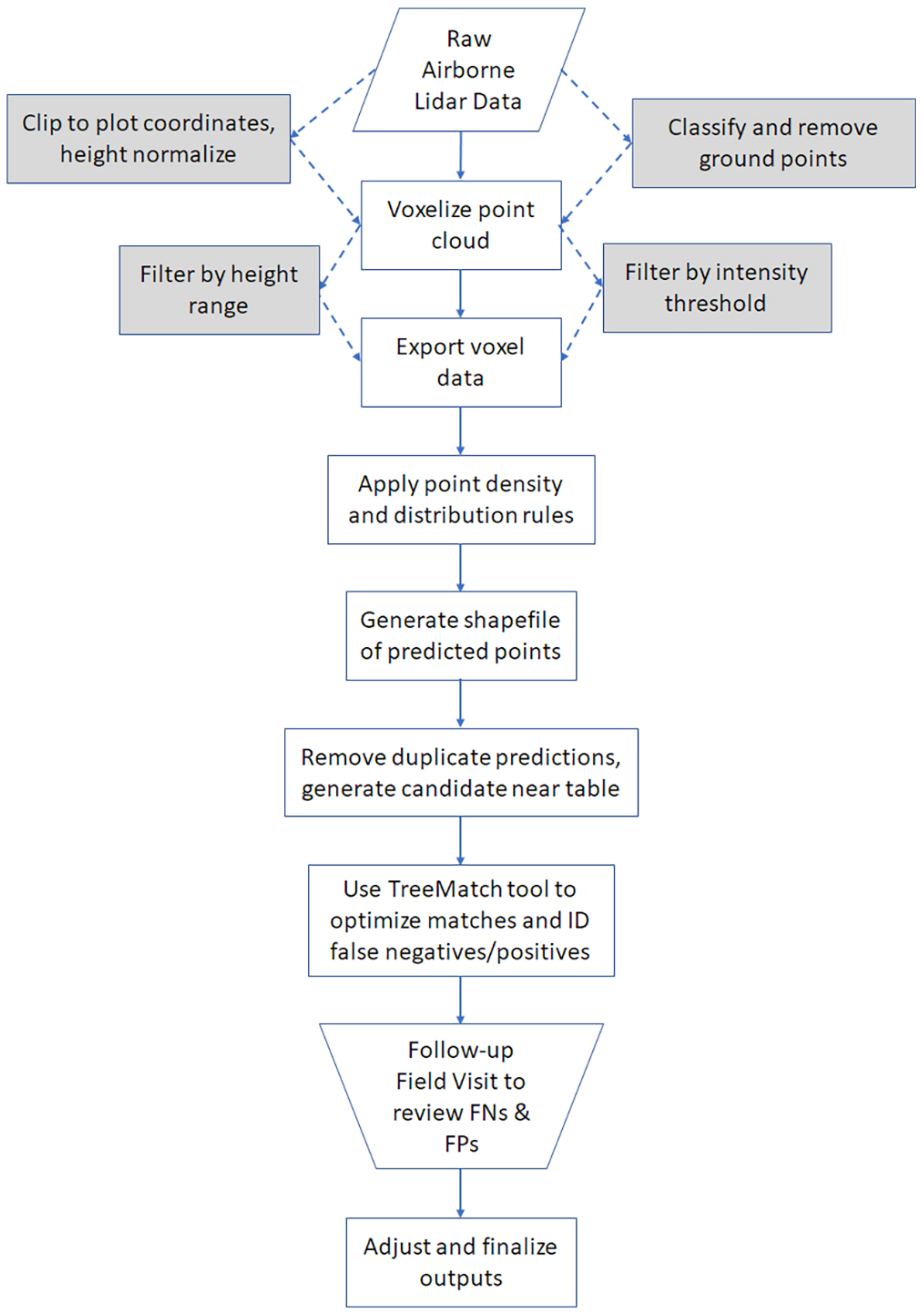
LiDAR Data Preprocessing
2.4. Tree Detection
2.4.1. Background
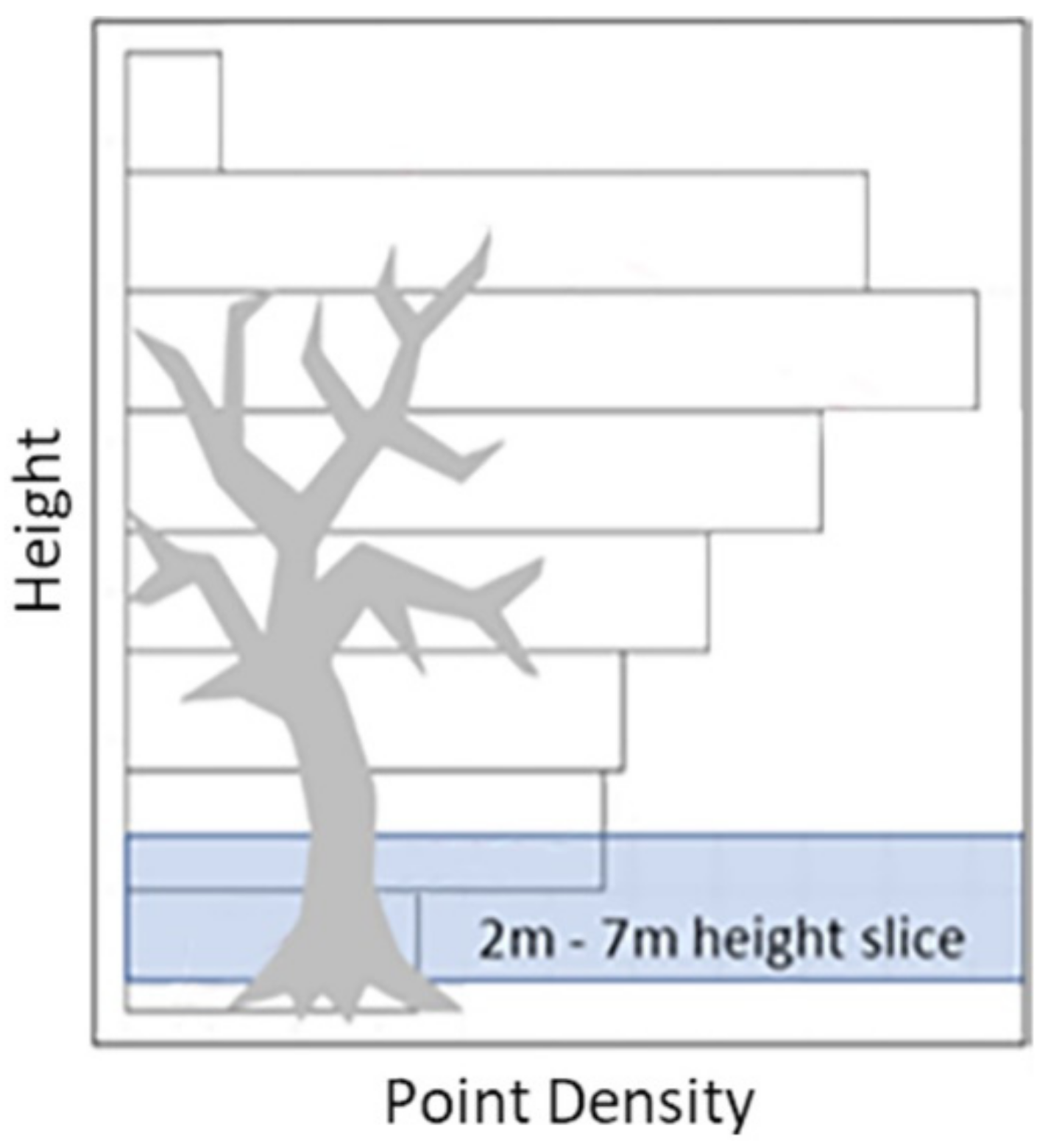
2.4.2. Vertical Segmentation of the Point Cloud
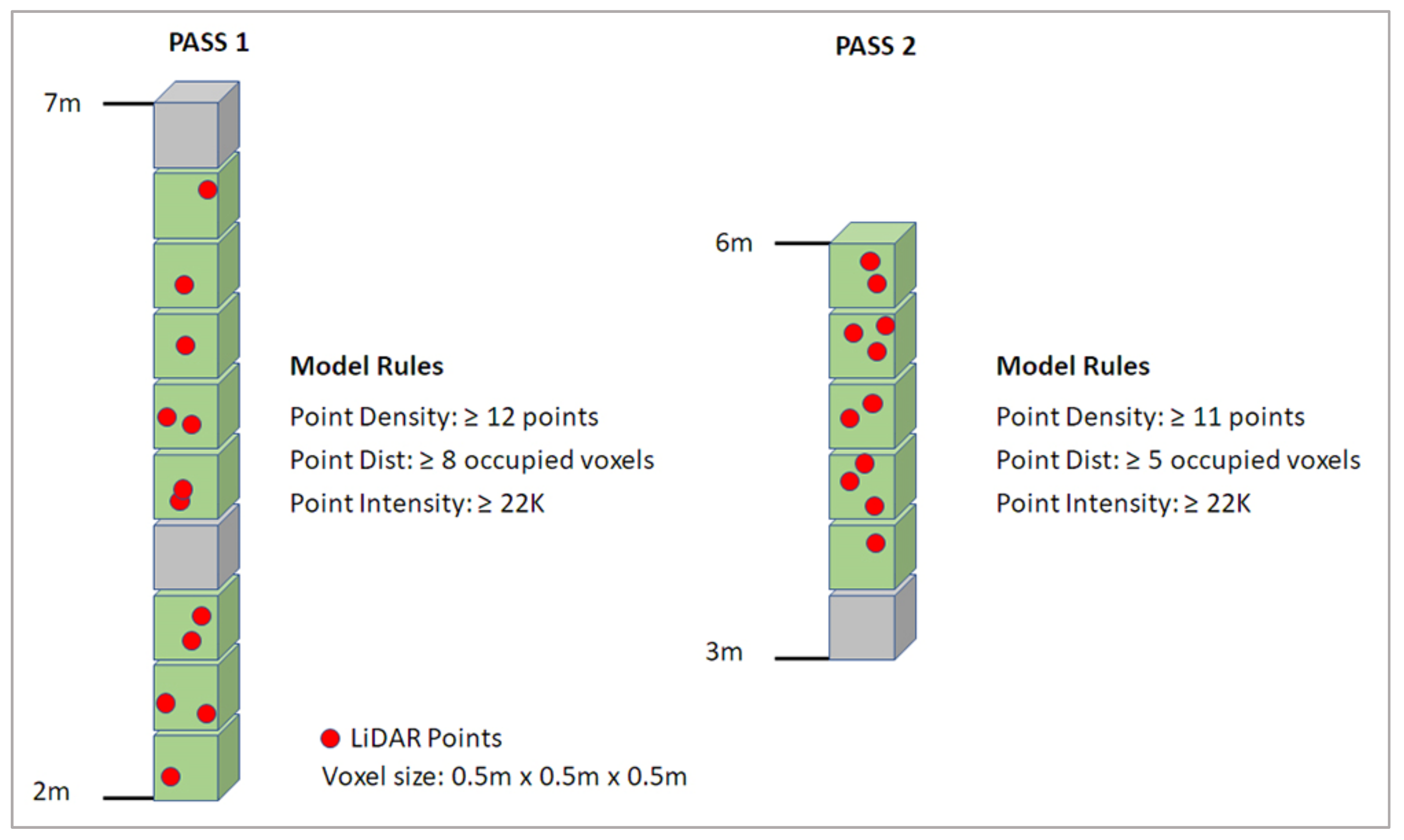
2.4.3. Voxel Column Analysis
2.4.4. Merging Duplicate Predictions
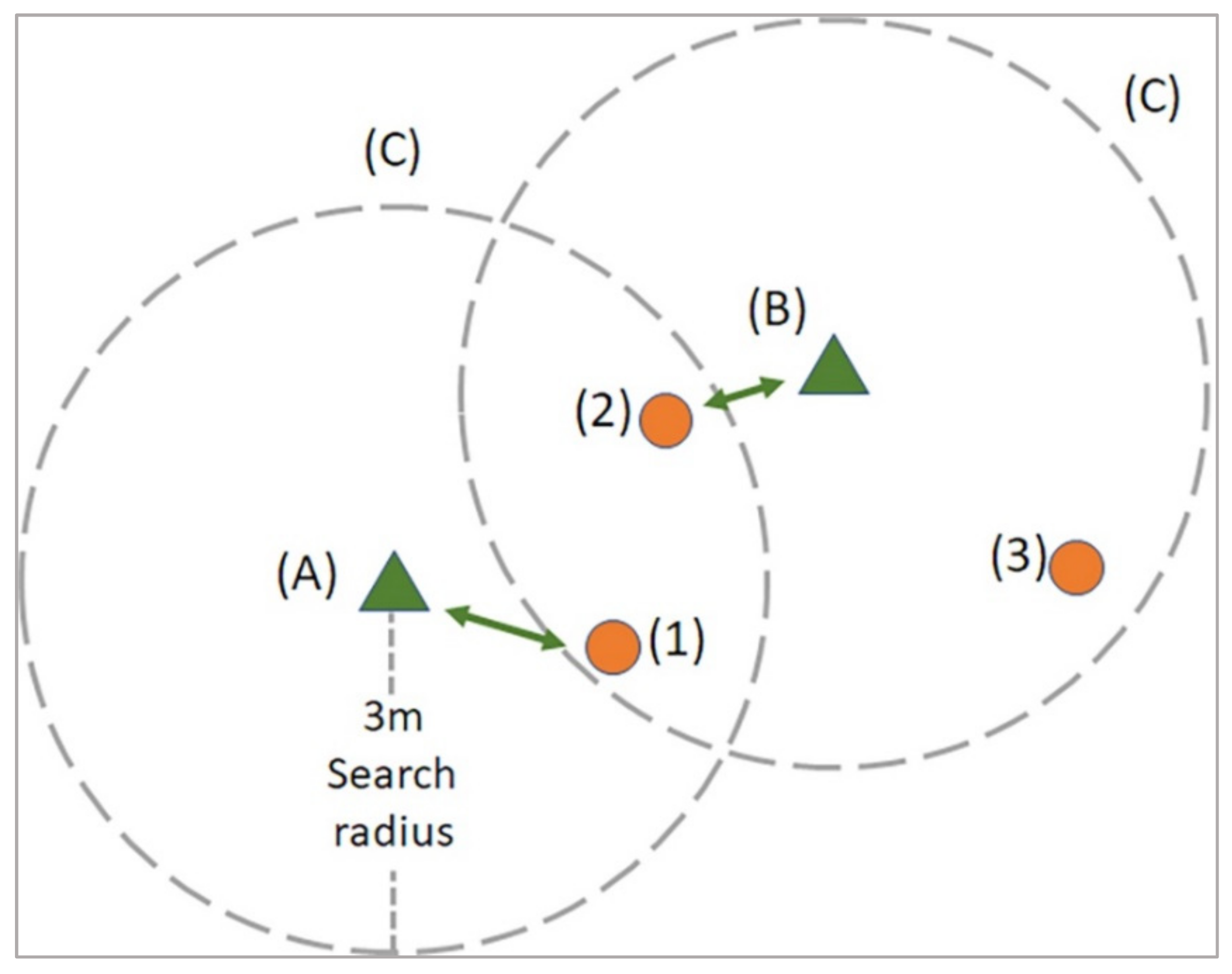
2.4.5. Tree-Matching and Evaluation Process
2.4.6. TreeMatch Data Postprocessing
3. Results
3.1. Overall Tree Detection Performance
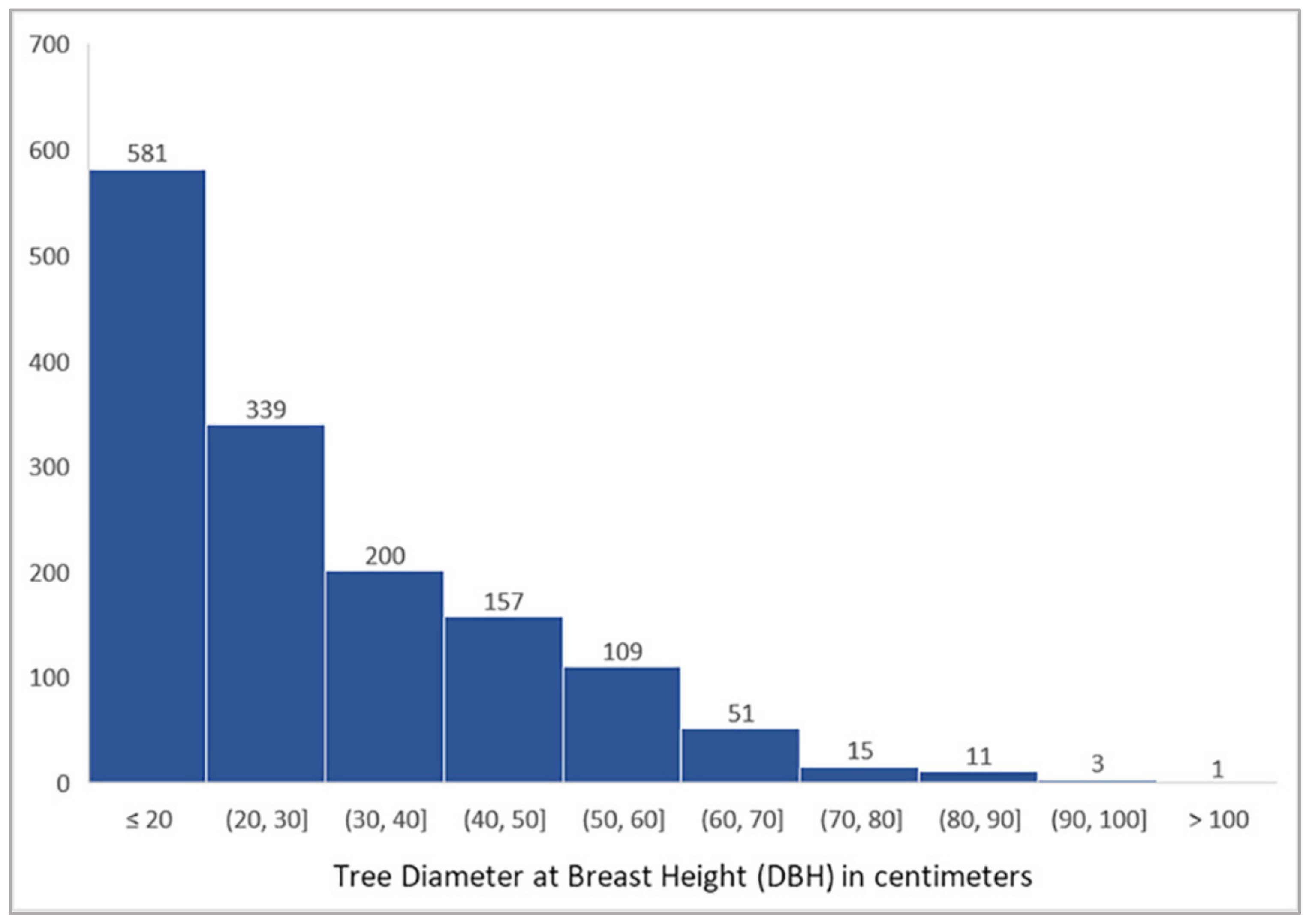
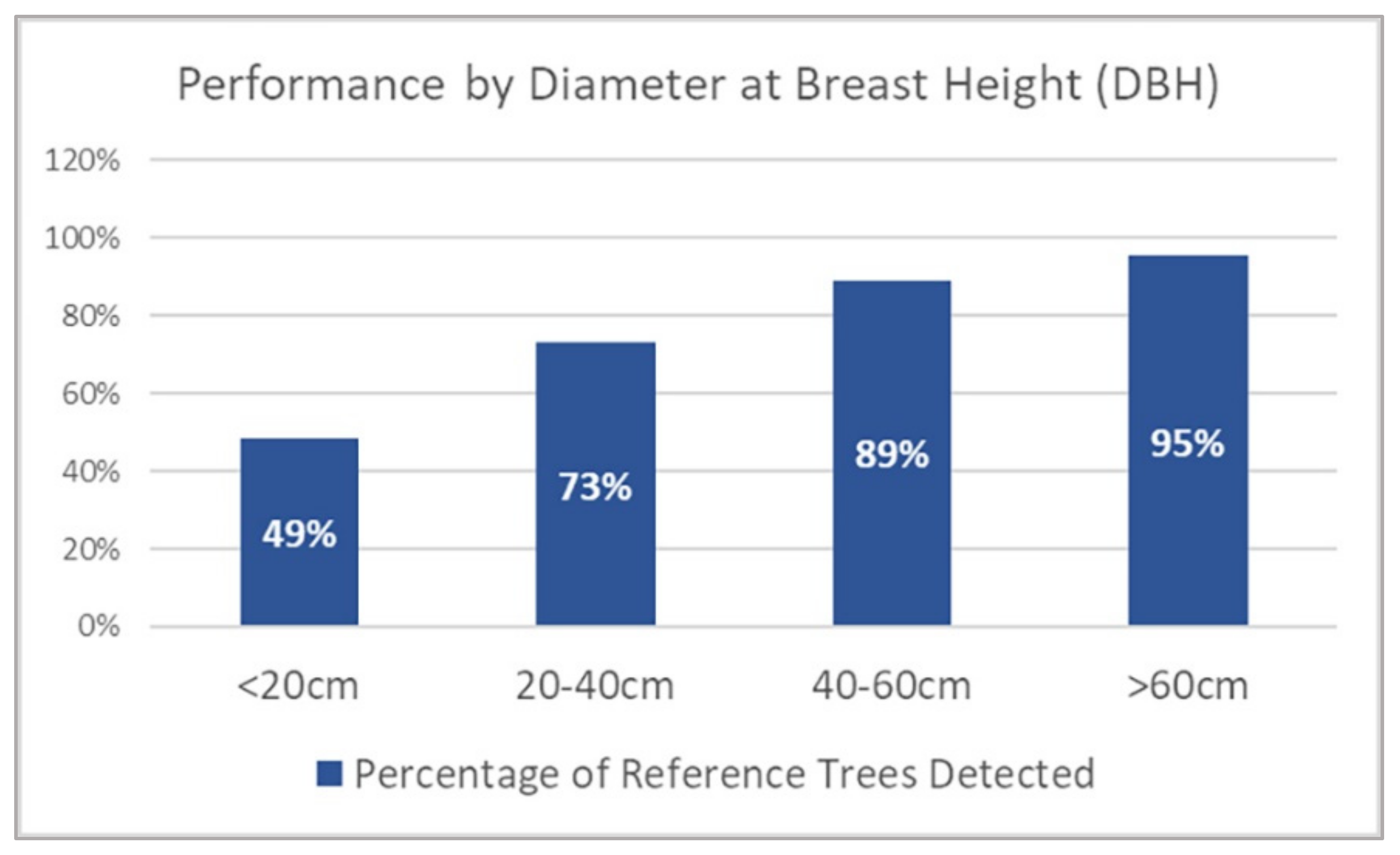
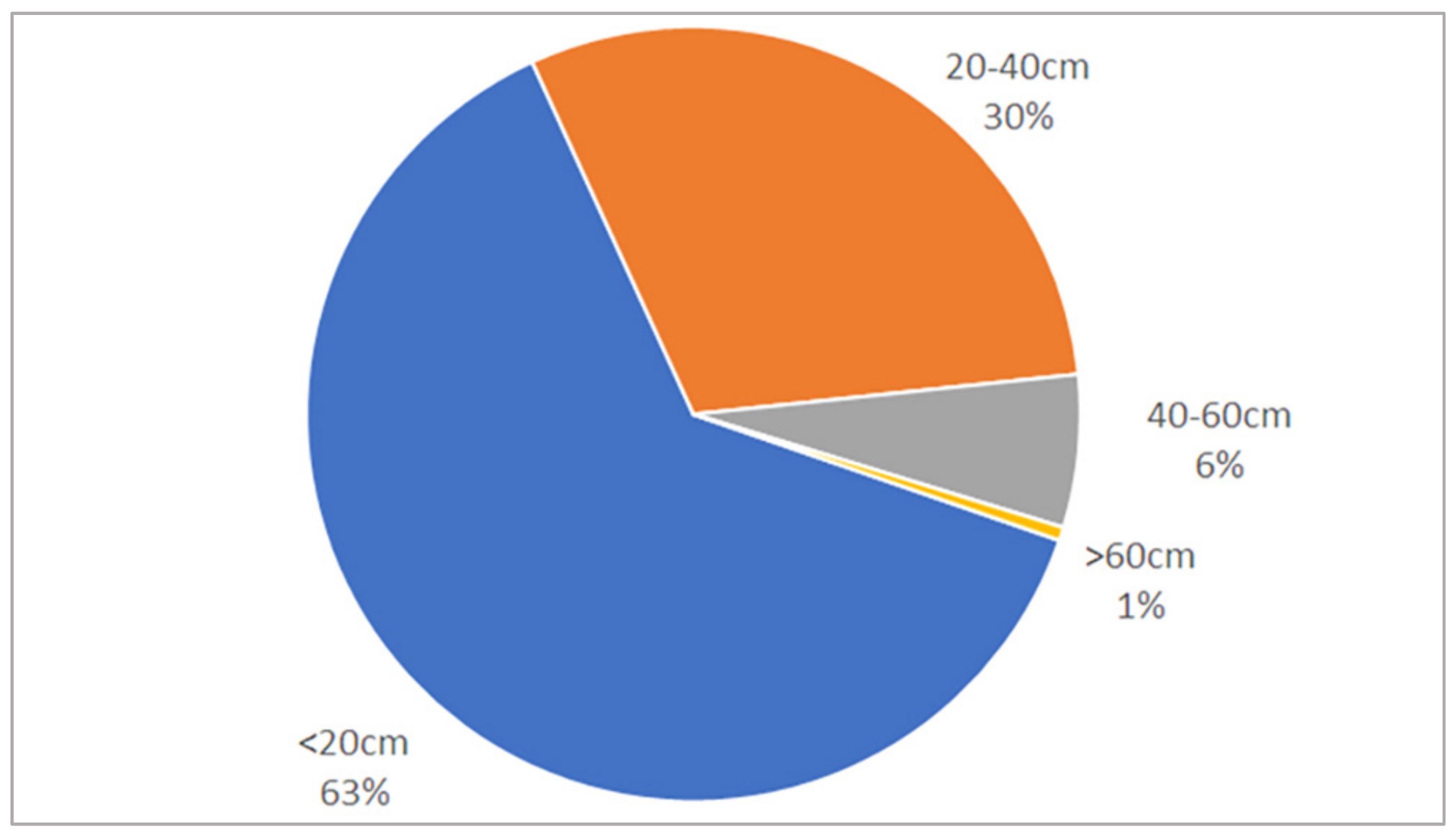
3.2. Tree Detection Performance by DBH Range
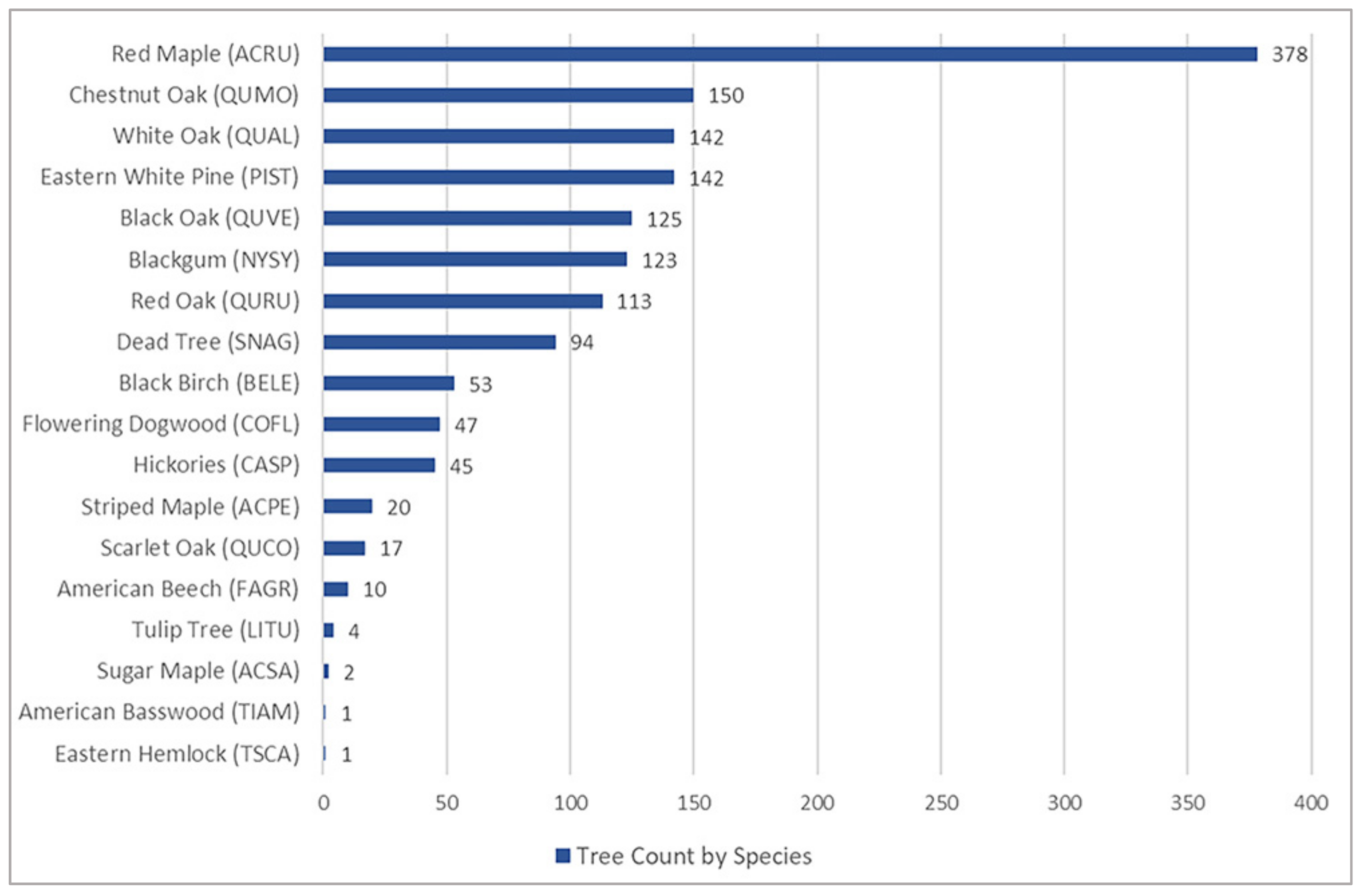
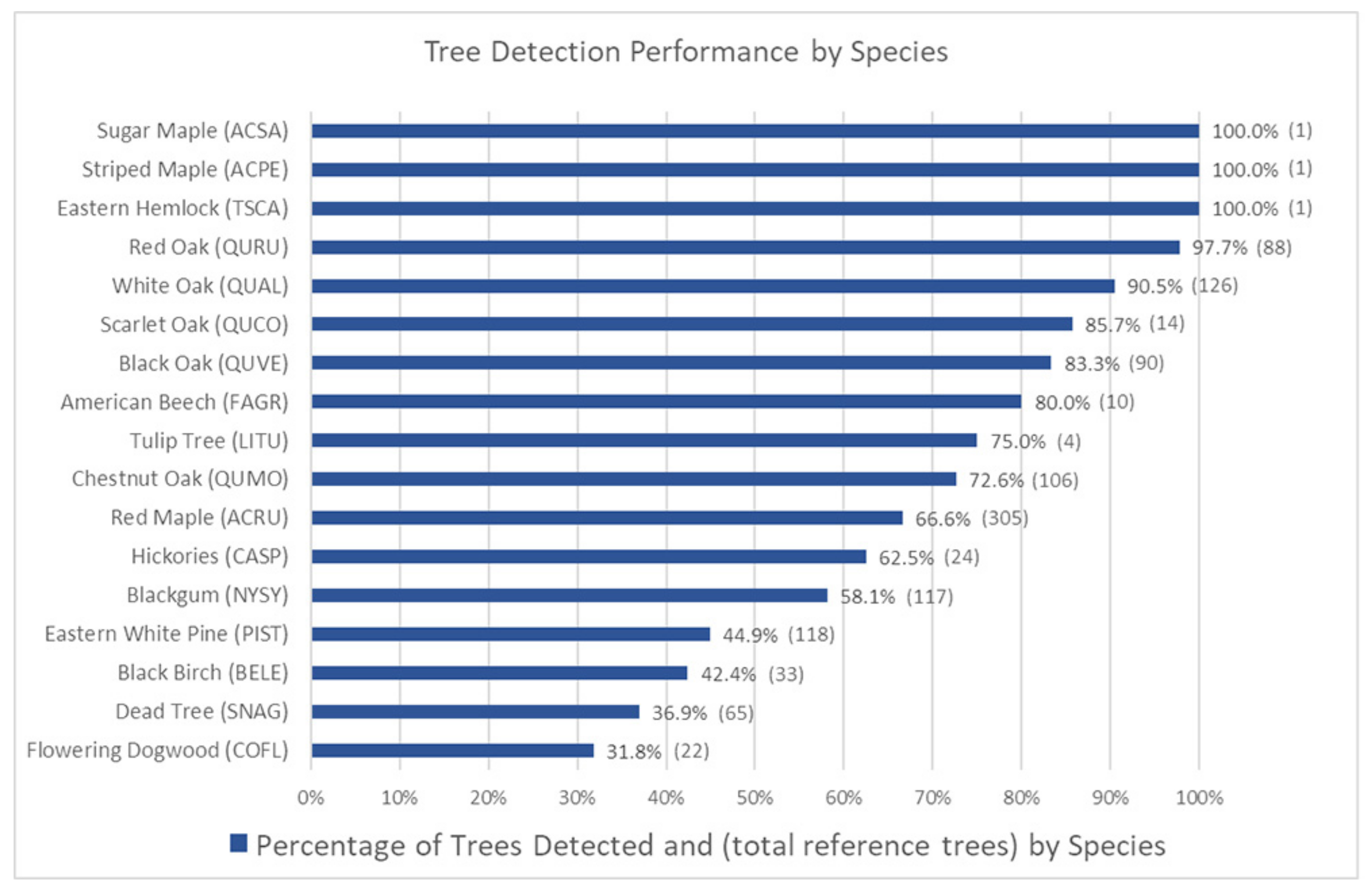
3.3. Tree Detection Performance by Species
3.4. False Positives
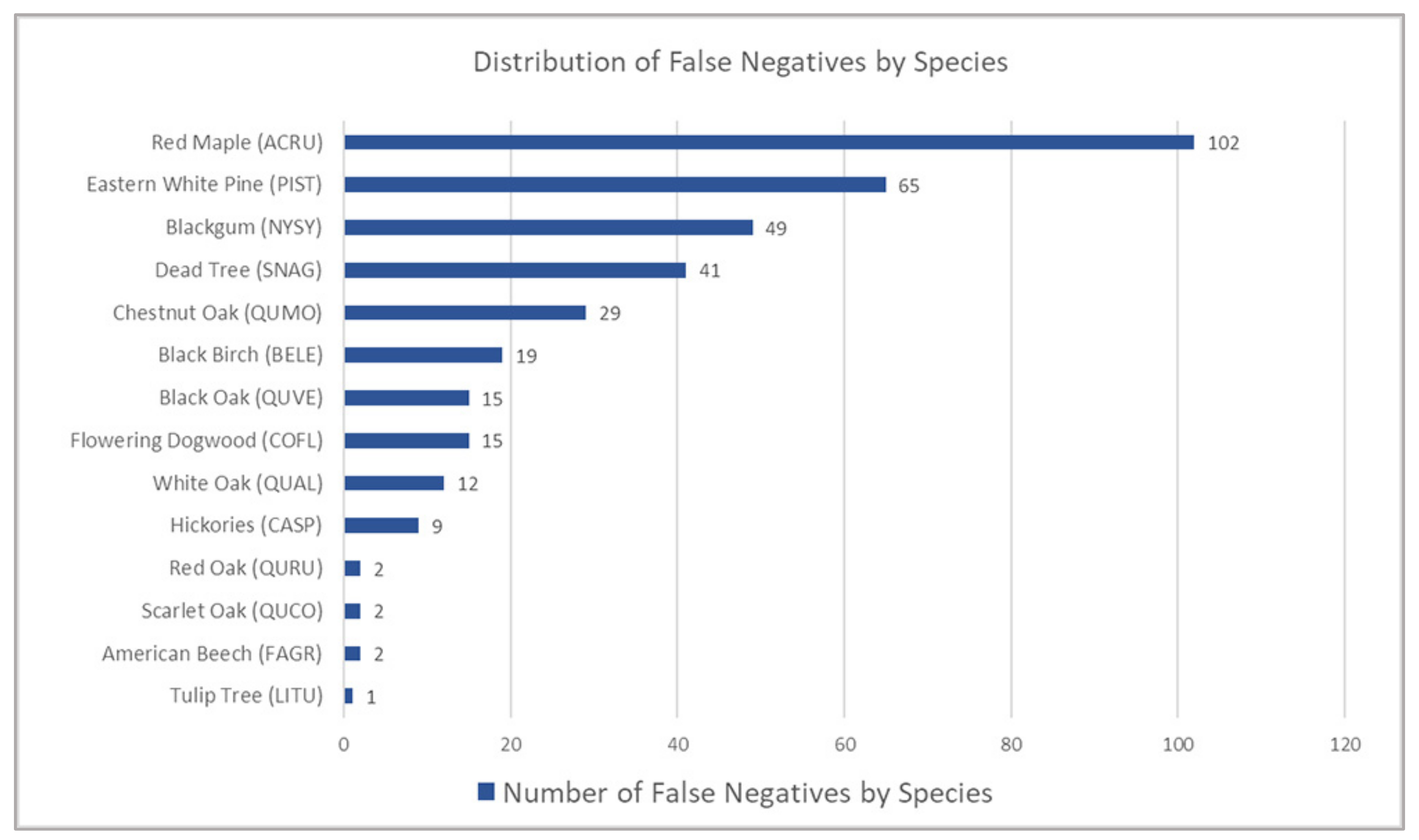
3.5. False Negatives
3.6. Spatial Accuracy of Detected Trees
4. Discussion
4.1. Context and Interpretation of Results
4.2. Contributions toward Standardization of ITD Evaluation
4.3. Opportunities for Future Research
4.4. Recommendations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, Y.; Kershaw, J.A.; Hsu, Y.H.; Yang, T.R. Carbon estimation using sampling to correct lidar-assisted enhanced forest inventory estimates. For. Chron. 2020, 96, 9–19. [Google Scholar] [CrossRef]
- Huang, W.; Dolan, K.; Swatantran, A.; Johnson, K.; Tang, H.; O’Neil-Dunne, J.; Dubayah, R.; Hurtt, G. High-resolution mapping of aboveground biomass for forest carbon monitoring system in the Tri-State region of Maryland, Pennsylvania and Delaware, USA. Environ. Res. Lett. 2019, 14, 095002. [Google Scholar] [CrossRef] [Green Version]
- Evans, D.L.; Roberts, S.D.; Parker, R.C. LiDAR-A new tool for forest measurements? For. Chron. 2006, 82, 211–218. [Google Scholar] [CrossRef]
- White, J.C.; Coops, N.C.; Wulder, M.A.; Vastaranta, M.; Hilker, T.; Tompalski, P. Remote sensing technologies for enhancing forest inventories: A review. Can. J. Remote Sens. 2016, 42, 619–641. [Google Scholar] [CrossRef] [Green Version]
- Kaartinen, H.; Hyyppä, J.; Yu, X.; Vastaranta, M.; Hyyppä, H.; Kukko, A.; Holopainen, M.; Heipke, C.; Hirschmugl, M.; Morsdorf, F.; et al. An international comparison of individual tree detection and extraction using airborne laser scanning. Remote Sens. 2012, 4, 950–974. [Google Scholar] [CrossRef] [Green Version]
- White, J.C.; Wulder, M.A.; Varhola, A.; Vastaranta, M.; Coops, N.C.; Cook, B.D.; Pitt, D.; Woods, M. A best practices guide for generating forest inventory attributes from airborne laser scanning data using an area-based approach. For. Chron. 2013, 89, 722–723. [Google Scholar] [CrossRef] [Green Version]
- White, J.C.; Tompalski, P.; Vastaranta, M.; Wulder, M.A.; Saarinen, N.; Stepper, C.; Coops, N.C. A Model Development and Application Guide for Generating an Enhanced Forest Inventory Using Airborne Laser Scanning Data and an Area-Based Approach; Information Report FI-X-018; Canadian Forest Service, Canadian Wood Fibre Centre: Victoria, BC, Canada, 2017. [Google Scholar]
- Yu, X.; Hyyppä, J.; Vastaranta, M.; Holopainen, M.; Viitala, R. Predicting individual tree attributes from airborne laser point clouds based on the random forests technique. ISPRS J. Photogramm. Remote Sens. 2011, 66, 28–37. [Google Scholar] [CrossRef]
- Pascual, A. Using tree detection based on airborne laser scanning to improve forest inventory considering edge effects and the co-registration factor. Remote Sens. 2019, 11, 2675. [Google Scholar] [CrossRef] [Green Version]
- Solberg, S.; Naesset, E.; Bollandsas, O.M. Single tree segmentation using airborne laser scanner data in a structurally heterogeneous spruce forest. Photogramm. Eng. Remote Sens. 2006, 72, 1369–1378. [Google Scholar] [CrossRef]
- Zhen, Z.; Quackenbush, L.J.; Zhang, L. Trends in automatic individual tree crown detection and delineation-evolution of LiDAR data. Remote Sens. 2016, 8, 333. [Google Scholar] [CrossRef] [Green Version]
- Hyyppä, J.; Hyyppä, H.; Leckie, D.; Gougeon, F.; Yu, X.; Maltamo, M. Review of methods of small-footprint airborne laser scanning for extracting forest inventory data in boreal forests. Int. J. Remote Sens. 2008, 29, 1339–1366. [Google Scholar] [CrossRef]
- Duncanson, L.I.; Cook, B.D.; Hurtt, G.C.; Dubayah, R.O. An efficient, multi-layered crown delineation algorithm for mapping individual tree structure across multiple ecosystems. Remote Sens. Environ. 2014, 154, 378–386. [Google Scholar] [CrossRef]
- Parkan, M.; Tuia, D. Individual tree segmentation in deciduous forests using geodesic voting. Int. Geosci. Remote Sens. Symp. 2015, 2015, 637–640. [Google Scholar] [CrossRef]
- Falkowski, M.J.; Smith, A.M.S.; Hudak, A.T.; Gessler, P.E.; Vierling, L.A.; Crookston, N.L. Automated estimation of individual conifer tree height and crown diameter via two-dimensional spatial wavelet analysis of lidar data. Can. J. Remote Sens. 2006, 32, 153–161. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, M. Estimation of tree heights and stand volume using an airborne lidar system. Remote Sens. Environ. 1996, 56, 1–7. [Google Scholar] [CrossRef]
- Straub, C.; Koch, B. Estimating single tree stem volume of Pinus sylvestris using airborne laser scanner and multispectral line scanner data. Remote Sens. 2011, 3, 929–944. [Google Scholar] [CrossRef] [Green Version]
- Kandare, K.; Dalponte, M.; Gianelle, D.; Chan, J.C.W. A new procedure for identifying single trees in understory layer using discrete LiDAR data. In Proceedings of the International Geoscience and Remote Sensing Symposium (IGARSS), Quebec City, QC, Canada, 13–18 July 2014; pp. 1357–1360. [Google Scholar]
- Koch, B.; Heyder, U.; Welnacker, H. Detection of individual tree crowns in airborne lidar data. Photogramm. Eng. Remote Sensing 2006, 72, 357–363. [Google Scholar] [CrossRef] [Green Version]
- Vauhkonen, J.; Ene, L.; Gupta, S.; Heinzel, J.; Holmgren, J.; Pitkänen, J.; Solberg, S.; Wang, Y.; Weinacker, H.; Hauglin, K.M.; et al. Comparative testing of single-tree detection algorithms under different types of forest. Forestry 2012, 85, 27–40. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Hyyppä, J.; Holopainen, M.; Vastaranta, M. Comparison of area-based and individual tree-based methods for predicting plot-level forest attributes. Remote Sens. 2010, 2, 1481–1495. [Google Scholar] [CrossRef] [Green Version]
- Ferraz, A.; Bretar, F.; Jacquemoud, S.; Gonçalves, G.; Pereira, L.; Tomé, M.; Soares, P. 3-D mapping of a multi-layered Mediterranean forest using ALS data. Remote Sens. Environ. 2012, 121, 210–223. [Google Scholar] [CrossRef]
- Lu, X.; Guo, Q.; Li, W.; Flanagan, J. A bottom-up approach to segment individual deciduous trees using leaf-off lidar point cloud data. ISPRS J. Photogramm. Remote Sens. 2014, 94, 1–12. [Google Scholar] [CrossRef]
- Pitkänen, J.; Maltamo, M.; Hyyppä, J.; Yu, X. Adaptive methods for individual tree detection on airborne laser based canopy height model. Int. Arch. 2004, 36, 187–191. [Google Scholar]
- Wang, Y.; Hyyppa, J.; Liang, X.; Kaartinen, H.; Yu, X.; Lindberg, E.; Holmgren, J.; Qin, Y.; Mallet, C.; Ferraz, A.; et al. International Benchmarking of the Individual Tree Detection Methods for Modeling 3-D Canopy Structure for Silviculture and Forest Ecology Using Airborne Laser Scanning. IEEE Trans. Geosci. Remote Sens. 2016, 54, 5011–5027. [Google Scholar] [CrossRef] [Green Version]
- Eysn, L.; Hollaus, M.; Lindberg, E.; Berger, F.; Monnet, J.M.; Dalponte, M.; Kobal, M.; Pellegrini, M.; Lingua, E.; Mongus, D.; et al. A benchmark of lidar-based single tree detection methods using heterogeneous forest data from the alpine space. Forests 2015, 6, 1721–1747. [Google Scholar] [CrossRef] [Green Version]
- Sačkov, I.; Kulla, L.; Bucha, T. A comparison of two tree detection methods for estimation of forest stand and ecological variables from airborne LiDAR data in central european forests. Remote Sens. 2019, 11, 1431. [Google Scholar] [CrossRef] [Green Version]
- Aubry-Kientz, M.; Dutrieux, R.; Ferraz, A.; Saatchi, S.; Hamraz, H.; Williams, J.; Coomes, D.; Piboule, A.; Vincent, G. A comparative assessment of the performance of individual tree crowns delineation algorithms from ALS data in tropical forests. Remote Sens. 2019, 11, 1086. [Google Scholar] [CrossRef] [Green Version]
- Ayrey, E.; Fraver, S.; Kershaw, J.A.; Kenefic, L.S.; Hayes, D.; Weiskittel, A.R.; Roth, B.E. Layer Stacking: A novel algorithm for individual forest tree segmentation from LiDAR point clouds. Can. J. Remote Sens. 2017, 43, 16–27. [Google Scholar] [CrossRef]
- Vega, C.; Hamrouni, A.; El Mokhtari, A.; Morel, M.; Bock, J.; Renaud, J.P.; Bouvier, M.; Durrieue, S. PTrees: A point-based approach to forest tree extractionfrom lidar data. Int. J. Appl. Earth Obs. Geoinf. 2014, 33, 98–108. [Google Scholar] [CrossRef]
- Chen, W.; Hu, X.; Chen, W.; Hong, Y.; Yang, M. Airborne LiDAR remote sensing for individual tree forest inventory using trunk detection-aided mean shift clustering techniques. Remote Sens. 2018, 10, 1078. [Google Scholar] [CrossRef] [Green Version]
- Silva, C.A.; Hudak, A.T.; Vierling, L.A.; Loudermilk, E.L.; O’Brien, J.J.; Hiers, J.K.; Jack, S.B.; Gonzalez-Benecke, C.; Lee, H.; Falkowski, M.J.; et al. Imputation of individual longleaf pine (Pinus palustris Mill.) tree attributes from field and LiDAR data. Can. J. Remote Sens. 2016, 42, 554–573. [Google Scholar] [CrossRef]
- Wang, Y.; Weinacker, H.; Koch, B. A Lidar point cloud based procedure for vertical canopy structure analysis and 3D single tree modelling in forest. Sensors 2008, 8, 3938–3951. [Google Scholar] [CrossRef] [Green Version]
- Lamprecht, S.; Stoffels, J.; Dotzler, S.; Haß, E.; Udelhoven, T. aTrunk-an ALS-based trunk detection algorithm. Remote Sens. 2015, 7, 9975–9997. [Google Scholar] [CrossRef] [Green Version]
- ESRI. ArcGIS Pro. Version 2.9.0 [Computer Software]. 2020. Available online: https://www.esri.com/en-us/arcgis/products/arcgis-pro/ (accessed on 18 April 2021).
- Roussel, J.; Auty, D. lidR: Airborne LiDAR Data Manipulations and Visualization for Forestry Applications. Version 3.0.3 [Computer Software]. 2020. Available online: https://github.com/Jean-Romain/lidR (accessed on 18 April 2021).
- R Core Team. R: A Language and Environment for Statistical Computing. Version 4.0 [Computer Software]. 2020. Available online: https://www.R-project.org/ (accessed on 18 April 2021).
- Reitberger, J.; Schnörr, C.; Krzystek, P.; Stilla, U. 3D segmentation of single trees exploiting full waveform LIDAR data. ISPRS J. Photogramm. Remote Sens. 2009, 64, 561–574. [Google Scholar] [CrossRef]
- Isenburg, M. LAStools—Efficient Tools for LiDAR Processing. Version 200619 [Computer Software]. 2020. Available online: http://rapidlasso.com/LAStools (accessed on 18 April 2021).
- Li, W.; Guo, Q.; Jakubowski, M.K.; Kelly, M. A new method for segmenting individual trees from the lidar point cloud. Photogramm. Eng. Remote Sens. 2012, 78, 75–84. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Weinacker, H.; Koch, B. Comparative analysis of clustering-based approaches for 3-D single tree detection using airborne fullwave LIDAR data. Remote Sens. 2010, 2, 968–989. [Google Scholar] [CrossRef] [Green Version]
- Tiede, D.; Hochleitner, G.; Blaschke, T. A full GIS-based workflow for tree identification and tree crown delineation using laser scanning. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. C Vienna Austria 2005, XXXVI, 9–14. [Google Scholar]
- ESRI. What Is Lidar Intensity Data? Available online: https://desktop.arcgis.com/en/arcmap/10.3/manage-data/las-dataset/what-is-intensity-data-.htm (accessed on 18 April 2021).
- Gurboi Optimization, LLC. Gurobi Optimizer. Version [Computer Software]. 2021. Available online: https://www.gurobi.com (accessed on 18 April 2021).
- Popescu, S.C.; Zhao, K. A voxel-based lidar method for estimating crown base height for deciduous and pine trees. Remote Sens. Environ. 2008, 112, 767–781. [Google Scholar] [CrossRef]
- Smits, I.; Prieditis, G.; Dagis, S.; Dubrovskis, D. Individual tree identification using different LIDAR and optical imagery data processing methods. Biosyst. Inf. Technol. 2012, 1, 19–24. [Google Scholar] [CrossRef]
- Jeronimo, S.M.A.; Kane, V.R.; Churchill, D.J.; McGaughey, R.J.; Franklin, J.F. Applying LiDAR individual tree detection to management of structurally diverse forest landscapes. J. For. 2018, 116, 336–346. [Google Scholar] [CrossRef] [Green Version]








| Attribute | Value |
|---|---|
| Flight date | 22 March 2020 |
| Condition | Leaf-off |
| Sensor | Riegl VQ1560i |
| Flying height | 950 m |
| Flight speed | 150 kts |
| Field of view | 60° |
| Scan rate | 422 Hz |
| Pulse rate | 2000 kHz |
| Average point spacing | 0.18 m |
| Average point density | 31.2 pts/m2 |
| Attribute | Value |
|---|---|
| Test Subplots | 48 |
| Total Trees | 1125 |
| Trees Detected (TPs) | 762 |
| False Negatives (FN) | 363 |
| False Positives (FPs) | 38 |
| False Positive Rate | 4.75% |
| Overall Detection Rate (DR) | 67.73% |
| Minimum DR (across 48 subplots) | 47.62% |
| Maximum DR (across 48 subplots) | 85.00% |
| Mean DR (across 48 subplots) | 68.00% |
| Median DR (across 48 subplots) | 68.21% |
| Standard Deviation for DR (across 48 subplots) | 9.33% |
| Attribute | Value |
|---|---|
| Total Matches | 742 * |
| Minimum | 0.020 m |
| Maximum | 2.999 m |
| Mean | 0.590 m |
| Median | 0.460 m |
| Standard Deviation | 0.504 m |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hershey, J.L.; McDill, M.E.; Miller, D.A.; Holderman, B.; Michael, J.H. A Voxel-Based Individual Tree Stem Detection Method Using Airborne LiDAR in Mature Northeastern U.S. Forests. Remote Sens. 2022, 14, 806. https://doi.org/10.3390/rs14030806
Hershey JL, McDill ME, Miller DA, Holderman B, Michael JH. A Voxel-Based Individual Tree Stem Detection Method Using Airborne LiDAR in Mature Northeastern U.S. Forests. Remote Sensing. 2022; 14(3):806. https://doi.org/10.3390/rs14030806
Chicago/Turabian StyleHershey, Jeff L., Marc E. McDill, Douglas A. Miller, Brennan Holderman, and Judd H. Michael. 2022. "A Voxel-Based Individual Tree Stem Detection Method Using Airborne LiDAR in Mature Northeastern U.S. Forests" Remote Sensing 14, no. 3: 806. https://doi.org/10.3390/rs14030806
APA StyleHershey, J. L., McDill, M. E., Miller, D. A., Holderman, B., & Michael, J. H. (2022). A Voxel-Based Individual Tree Stem Detection Method Using Airborne LiDAR in Mature Northeastern U.S. Forests. Remote Sensing, 14(3), 806. https://doi.org/10.3390/rs14030806







