Abstract
Improved development of remote sensing approaches to deliver timely and accurate measurements for environmental monitoring, particularly with respect to marine and estuarine environments is a priority. We describe a machine learning, cloud processing protocol for simultaneous mapping seagrass meadows in waters of variable quality across Moreton Bay, Australia. This method was adapted from a protocol developed for mapping coral reef areas. Georeferenced spot check field-survey data were obtained across Moreton Bay, covering areas of differing water quality, and categorized into either substrate or ≥25% seagrass cover. These point data with coincident Landsat 8 OLI satellite imagery (30 m resolution; pulled directly from Google Earth Engine’s public archive) and a bathymetric layer (30 m resolution) were incorporated to train a random forest classifier. The semiautomated machine learning algorithm was applied to map seagrass in shallow areas of variable water quality simultaneously, and a bay-wide map was created for Moreton Bay. The output benthic habitat map representing seagrass presence/absence was accurate (63%) as determined by validation with an independent data set.
1. Introduction
Seagrass meadows are typically found growing in the soft sediment of shallow waters that are often intertidal, but where enough light penetrates the water column to enable photosynthesis [1]. Here they act to stabilize the coastline by reducing disturbance of the substrate during storm events and tides [2]. These secret gardens provide not only food and nursery grounds for a diverse array of organisms from mollusks to fish to dugongs, but, collectively with mangroves and saltmarshes, seagrasses are part of the ‘coastal wetland blue carbon’, sequestering large amounts of carbon in their biomass [3]. Thus, seagrasses are of tremendous ecological and economical value. However, seagrass is constantly under threat of anthropological destruction through the effect of pollution (in the form of run off) and physical abrasion (boating, dredging) [4,5]. In addition, seagrass meadows are threatened by extreme environmental events, and collectively, are being lost at an estimated global rate of ~7% every year [6], the exact figure largely unknown due to variability in the determination of aerial extent [7]. As such, the ability to measure and monitor seagrass over local to global extents in optically complex and variable coastal waters is paramount.
Remote sensing has become a familiar tool for measurement, mapping and monitoring seagrass meadow properties and has included the application of high (<5 m pixels) to moderate spatial (5–30 m pixels) resolution satellite imagery [8], drone surveys [9] and the use of automated underwater vehicles (AUVs; [10]) across the different spatial scales (local [11] to global [12,13]). Seagrasses occur in inter- and subtidal areas where there are substantial resuspended particles, which reduce the ability to see the bottom and accurately detect and map seagrasses [14]. Poor water quality or deep water reduce or negate the ability to penetrate the water column, obscuring the meadows from view [15,16,17]. Additionally, seagrasses frequently occur as patches of low-density cover [18], and there are substantial species-specific differences with respect to size, color and shape [19] all of which contribute to the difficulty of defining seagrass meadows and their constituents via remote sensing [15,20,21]. In addition to these challenges, effective monitoring requires the application of a consistent methodology that enables comparison between successive mapping efforts [22].
Seagrass habitats have often been mapped using approaches determined by the ability to differentiate seagrass presence due to differences in water clarity. Moreton Bay, Queensland, Australia provides the ideal representation of some of the challenges mentioned. Here seagrass meadows survive in a variable water quality regime (Figure 1a), and historically, the Eastern Banks and Western Bay areas have been mapped independently, utilizing different mapping methods. Clear water seagrass meadows on the Eastern Banks have been mapped utilizing a remote sensing approach that incorporated satellite imagery and photoquadrat field survey data [23]. However, as decreasing water quality limits the ability for the satellite to “see” the bottom and subsequently the capability to utilize an automated mapping method to map the benthos, the Western Bay area has been mapped previously via manual digitization of satellite imagery guided by field data, expert knowledge and bathymetry. Through mosaicking of the two areas, a baywide map has been created for 2004, and also for 2011 using a similar protocol [22,24]. Mapping of poor-quality waters relies heavily upon investigator interpretation which is manually laborious and is highly subjective making subsequent comparable maps near impossible. The ability to map seagrass in waters that are both clear and of diminished quality simultaneously would enable regular, routine map creation for ongoing observation of seagrass dynamics. Additionally, to compare maps throughout time requires the application of a consistent methodology which minimizes mapping discrepancy and error.

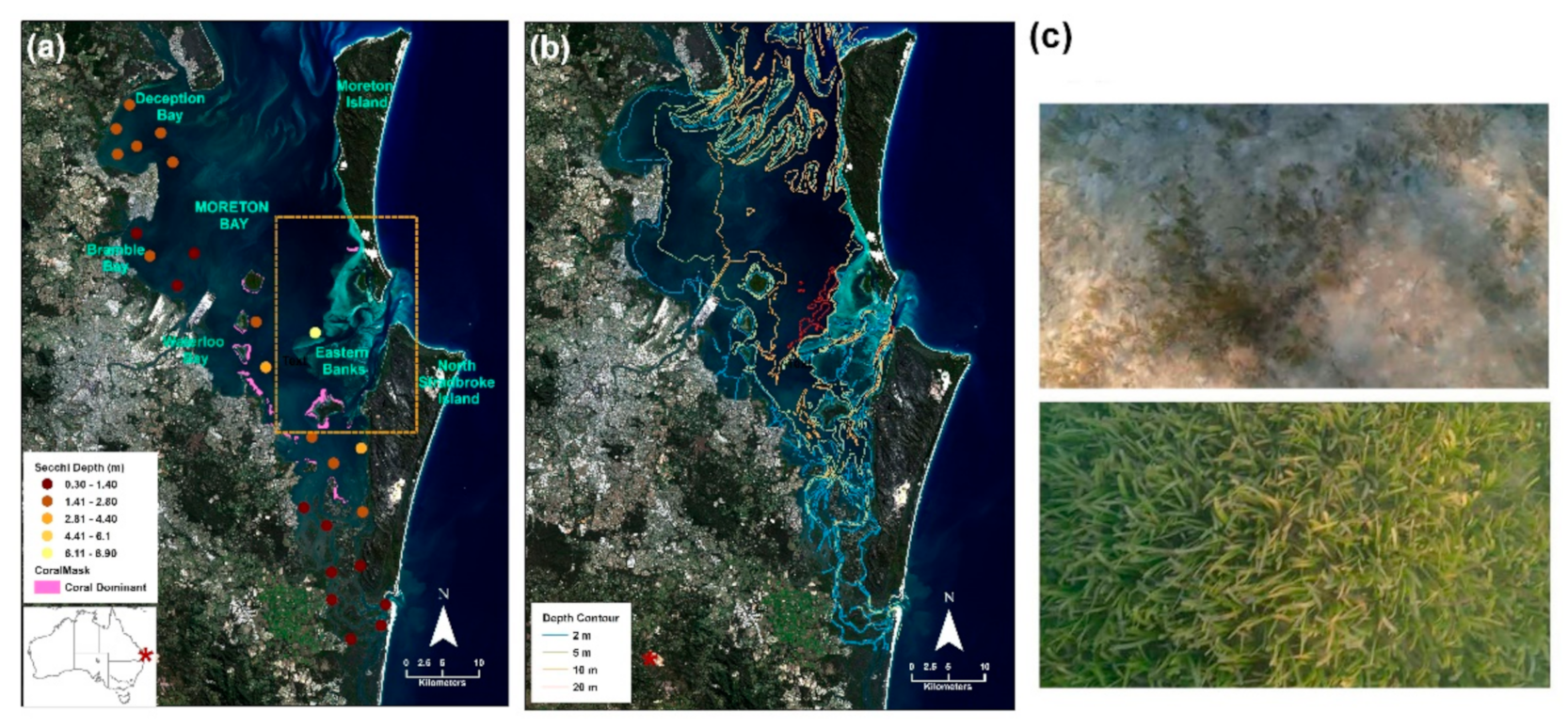
Figure 1.
The Moreton Bay Marine Park in Queensland, Australia (a) extends from Bribie Island in the north to the southern passage and is bordered by the mainland and Moreton and North Stradbroke Islands. Seagrass meadows can be found in the clear waters of the Eastern Banks (orange square) and in the less clear intertidal waters of the western bay along the coastline. Observed Secchi depth readings (May 2015) within the extent of the area of Moreton Bay mapped in this study indicate the variability in water clarity, with a higher number representing greater visibility [25]. Coral areas [26] that were masked from the seagrass habitat map are indicated in pink. (b) Depth contours used to delineate a depth cutoff for the input data load, and a depth cutoff for analysis and map production in this study. (c) Field survey photos (approx. 1 m2) of seagrass, Halophila spinulosa (top) and Cymodocea rotundata (bottom). Meadows in Moreton Bay consist of six species which occur in dense or sparse beds.
Here we describe an adaptation of a cloud-based processing methodology developed for the mapping of coral reef habitats [13], to map seagrass meadows in areas of optically clear waters, and poorer quality intertidal waters simultaneously. This method is consistent and incorporates georeferenced spot check data collected by citizen scientists, coincident satellite imagery, bathymetry, and a coral reef mask in a cloud processing environment to produce a uniform seagrass map for the whole of Moreton Bay. The method described is applicable for ongoing, repeat monitoring and time series analysis not only at the local scale of the whole of Moreton Bay, but to larger and potentially global extents.
2. Materials and Methods
2.1. Study Site
Moreton Bay (27°15′ S, 153°15′ E, 1582 km2, Figure 1a) is a relatively shallow and partially enclosed embayment. The maximum water depth within the Bay is approximately 30 m in the shipping channels, with the remainder shallower than ~12 m and a semi-diurnal tidal range of around 1.7 m (Figure 1b) [27]. The Moreton Bay Marine Park constitutes approximately 3400 km2 of coastal waters framed by the mainland to the west and extending to the east beyond Moreton and North Stradbroke Islands, to Bribie Island in the north with the Southern Passage the southernmost boundary (Figure 1a). Moreton Bay is home for wetland areas encompassing mangroves, seagrass (species include Halophila ovalis, H. decipiens, H. spinulosa, Halodule uninervis, Zostera muelleri, Cymodocea rotundata and Syringodium isoetifolium [28], Figure 1c) and salt marshes, as well as coral reef areas distributed throughout the region [26]. Across both the full extent of the western coastline and the Eastern Banks (Figure 1a, orange square) seagrass meadows exist in intertidal and subtidal waters, however the former is lower water quality and the latter clear. Secchi data for May 2015 indicate the difference in water clarity between the Western coastline and the Eastern Banks in the area mapped (Figure 1a; [25]). Seagrass meadows are present throughout the Bay persisting in waters to approximately 10 m in depth and at varying percentages of cover and species composition (Figure 1c).
2.2. Field Data
Spot check field data were collected by Science Under Sail volunteers and was distributed throughout the entire Bay, spanning variable water qualities (Figure 2a). The data were collected according to the Environmental Health Monitoring Program (EHMP) [29]. The field data were distributed such that only a single point fell within a 30 × 30 m Landsat pixel. The data were georeferenced and recorded to the level of seagrass species, percentage cover and substrate type. However, for the purposes of creating a baywide map, the data were collapsed to seagrass presence/absence (two classes: substrate, ≥25% seagrass cover). A subset of the data totaling 1076 points from January to October of 2015 was used and the points were randomly assigned to training (75%) or calibration (25%) data sets via the machine learning algorithm (Section 2.3 Mapping Method).
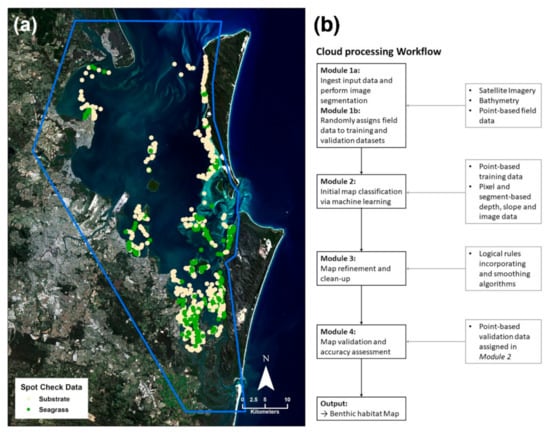
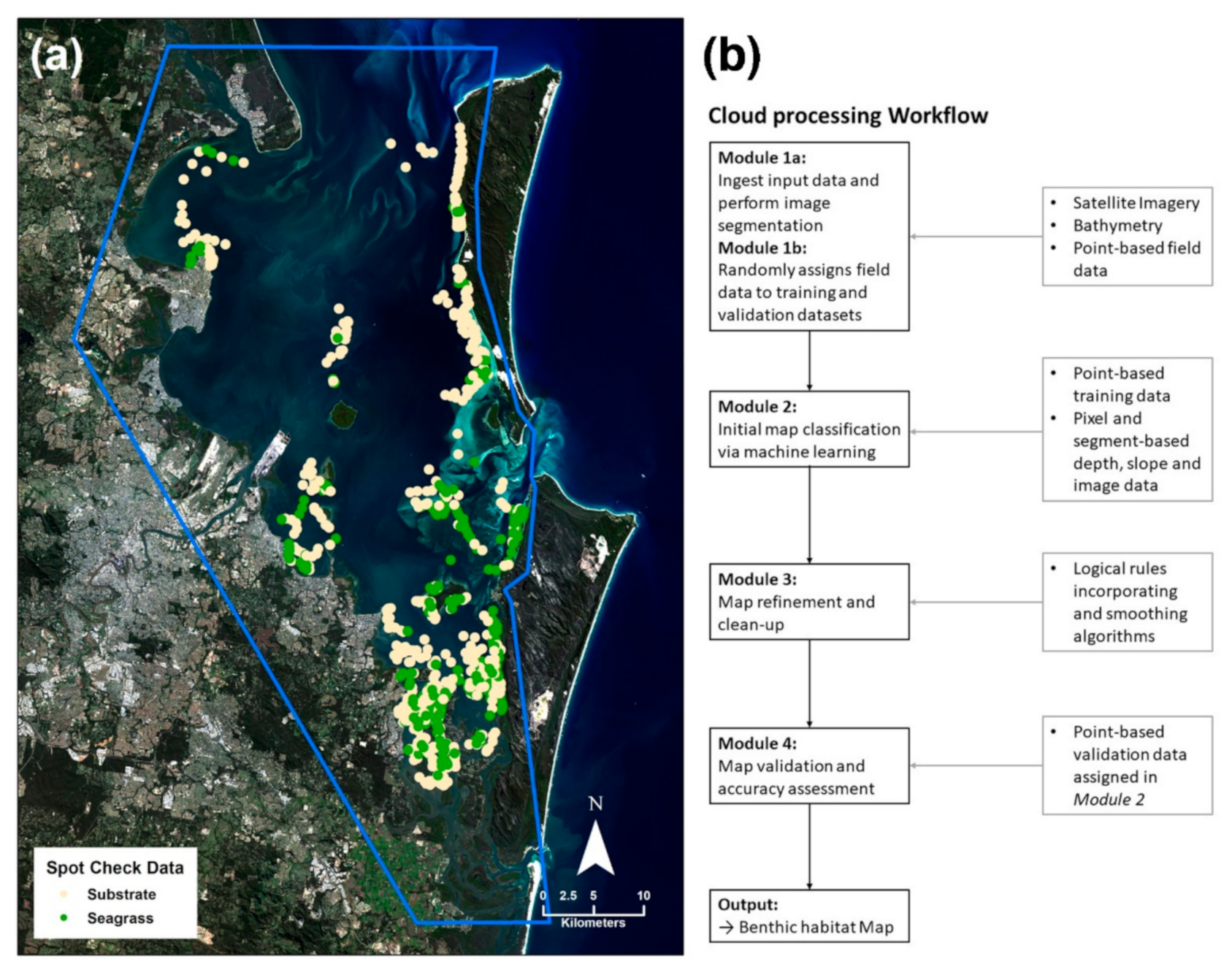
Figure 2.
Cloud processing method for seagrass mapping. (a) Imported satellite imagery (Landsat 8 OLI, 4 June 2015) and the distribution of field data with the extent of the mapping area indicated in blue (analysis was restricted to areas ≤5 m in depth). (b) Flowchart representing the workflow as applicable to seagrass mapping in the Google Earth Engine (modified from [13]).
2.3. Mapping Method
A cloud-based processing methodology developed for mapping coral reef habitat [13] was adapted in this study for seagrass environments (workflow shown in Figure 2b; adaptations are indicated in italicized text).
Bathymetry (30 m resolution; [30]), a coral reef mask [26], and spot check field data (georeferenced, distributed across the study area and assigned one of two classes: substrate or ≥25% seagrass cover), were ingested into the Google Earth Engine (GEE) cloud processing environment [31] from which Landsat 8 Operational Land Imager (OLI) imagery (4 June 2015) was imported directly through GEE from the United States Geological Survey (USGS; surface reflectance, Tier 1; ee. ‘LANDSAT/LC08/C01/T1_SR/LC08_089079_20150604′). This imagery is geometrically and radiometrically corrected and further corrected to surface reflectance using the Land Surface Reflectance Code (LaSRC) [32].
Input data sets were ingested into GEE (Figure 2b, Module 1a) and the area to be mapped was segmented into pixel-based objects, consisting of relatively homogeneous pixels based on color and texture [33]. This was done via ‘simple non-iterative clustering’ [34] using the blue, green and red bands of the satellite imagery, with the bathymetry layer, and slope as a derivative of depth. A depth limit for analysis was set at 5 m (Figure 1a). The mean value for each input layer was extracted for each pixel within an object.
Module 1b of the workflow randomly assigned the point-based input field data, which was relatively evenly distributed across the study area, to either training or validation data. The training data, representing known occurrences of bottom type, was used to sample the data created in Module 1a, and train a random forest algorithm (Figure 2b, Module 2) [35]. The random forest classifier was trained with 50 trees per class, a minimum leaf population of one and the square root of the total number of covariates as the number chosen at each node split. The random forest prediction was then used to predict the class membership (seagrass or substrate) of each individual pixel across the full extent of the mapping area. In this instance, only slope and depth auxiliary data was utilized. Mapping depth was limited to 5 m, as this represents the depth range of most seagrasses in Moreton Bay [28].
The seagrass map was refined via smoothing using a kernel size of three (Figure 2b, Module 3), a coral reef mask [26] was applied to remove known areas of coral reef from the output, while a manual mask was used to remove inland waterways. Finally, the output map was validated, and the map accuracy determined (Figure 2b, Module 4).
2.4. Validation
The accuracy of the output map was determined using the validation data not used for the mapping process (25% of field data). A traditional error matrix that provides overall accuracy, user and producer errors [36] was created, in addition to a 95% confidence based on the user accuracies for each class.
3. Results
Using the workflow presented in Figure 2b for the GEE geospatial processing service, a seagrass map for the whole of Moreton Bay was created (Figure 3a). Seagrass has been mapped in the less clear waters along the coastline, indicating seagrass presence in the northern Deception Bay and in Bramble Bay (Figure 3a). Additionally, seagrass is present in the clearer waters of the Eastern Banks.
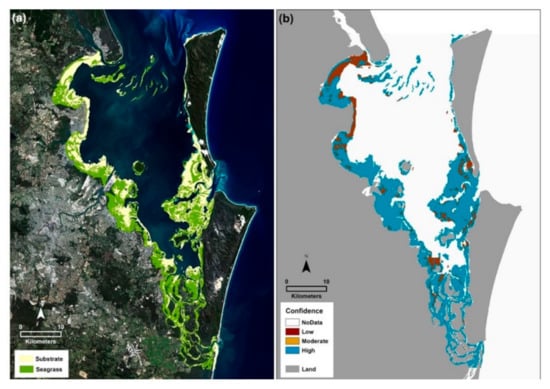
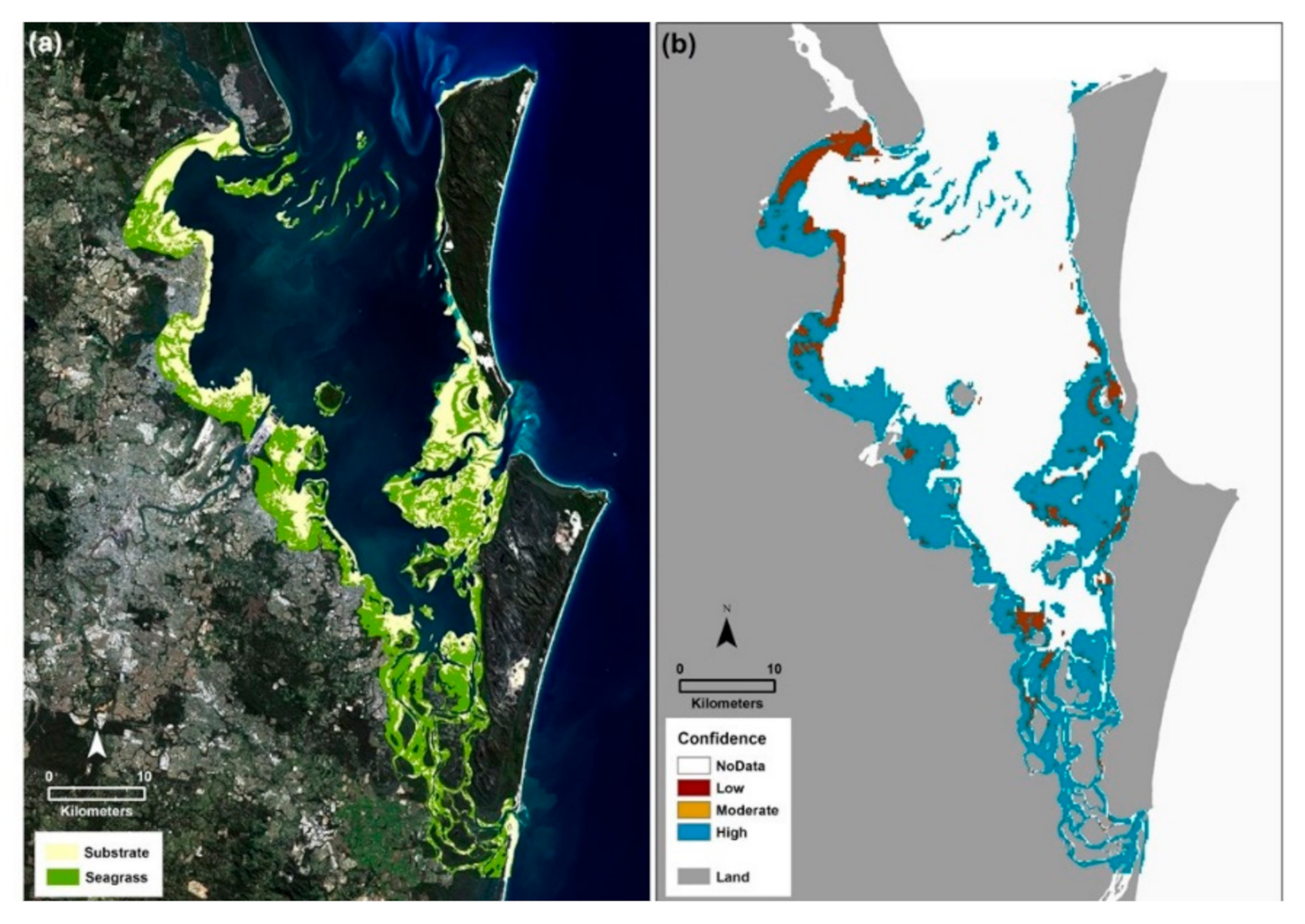
Figure 3.
Moreton Bay seagrass 2015. (a) Seagrass presence/absence map for the Moreton Bay Marine Park. Both the poorer quality intertidal waters of the Western Bay and the optically clear waters of the Eastern Banks were mapped consecutively using a workflow for the Google Earth Engine. Data input included Landsat 8 OLI satellite imagery (4 June 2015), citizen science spot check field data (2015), a coral reef mask [26] and a bathymetry layer [30]. (b) A map of 95% confidence based on the user accuracies for each class.
From a traditional error matrix [36], the overall map accuracy was calculated as 63% with user and producer accuracies for substrate of 54% and seagrass of 73%. A map of user confidence was generated and indicates that high confidence is seen for most areas in the bay, across all water quality levels and substrate types (Figure 3b). The lowest confidence occurs for mapped areas of Deception Bay in the north, the entrance to the Southern Passage, and the channel to the west of Moreton Island, at the top of the Eastern Banks.
4. Discussion
Use of GEE to integrate satellite and field data sets with ecological knowledge to map seagrass across Moreton Bay’s less clear intertidal waters and the clearer waters of the Eastern Banks, provides an accurate and complete coverage for the whole bay.
The field data used in this study had been collected for an ecological study, and therefore the field campaign had not been specifically designed for a mapping study. However, this dataset was the best available and provided a dominant class of cover (seagrass or substrate) determined from the information recorded at the time of observation. Each of the field points were spatially distinct to provide the classifier with a single field point representation for any pixel. Field data collection occurred throughout 2015 between the austral summer (January), through winter, and October. The satellite imagery, however, was obtained in our coolest and driest month, June 2015. Where seagrass is shown as present in the output map, seagrass cover may range from low (25%) to high (>80%). These seagrass meadows undergo seasonal change [37]. Although the final seagrass map represents a single point in time, there may be seasonal and shorter-term variation in the extent of seagrass in the Bay.
Results presented here demonstrate that georeferenced field data distributed across areas of varying water quality [38,39], enabled establishment of a machine learning classifier that could concomitantly map across differing water clarities. In fact, this method has produced a map product with high user confidence, particularly in seagrass areas (Figure 3b). Interestingly, the lowest confidence was observed for areas that were substrate dominant. This may arise from the vastly differing colors that contribute to the pool of training signatures for this class (dark mud to bright sand) or because of variations in water clarity. Decreased water clarity arises from particulate matter such as resuspended sediment, phytoplankton and dissolved inorganic molecules [40]. Average Secchi disc transparency depths measured in Moreton Bay waters in 2015 were 1.5 m for Deception Bay, 1.5 m for Bramble Bay and 5.5 m for the Eastern Banks ([41]; see Figure 1 for locations), a 3.6-fold difference from East to West.
It must be considered that in the mapped area there will be regions where one cannot see the bottom due to decreased water clarity. There are currently no automated approaches to remove these areas, and although manual delineation could be used to remove areas of less clear water, this is subjective. We ensured that the field data used represented a range of tidal stages and were in locations where water quality ranged from very clear to lower clarity [38,39]. A random forest classifier as used in this study, is well suited to this scenario. In contrast to a parametric classifier for example that may average spectral signatures across training samples, a random forest classifier uses each individual training point to inform the classification based on the specific tidal stage and water quality captured in the imagery [35]. This accounts for the clarity, or lack thereof, of the water at the time of observation, enabling assignment of a bottom class to all pixels in the mapping area—clear and less clear. Nevertheless, there is still likely to be some error associated with training points that fall in very low-quality water and future users may wish to consider dealing with this more systematically, perhaps by adding a low visibility water class.
Incorporation of publicly accessible Landsat or Sentinel 2a/b imagery to the process which can be directly imported through the GEE, provides greater choice over the imagery scenes used due to their frequent repeat cycles (Landsat satellites 5–8, 16 days; Sentinel constellation, 2–5 days). Thus, it would be possible to choose scenes where there is not only low cloud cover, but low tide or clearer waters, further accentuating the capability to map differing water clarities with high accuracy and reliability. The method described here may also be applied to input imagery of higher spatial resolution (such as Planet Dove (3 m) or World-View-2 imagery (3 m)) that can be acquired on-demand. To this end, in combination with seagrass field survey datasets being available and accessible in a range of areas globally, time series (years) or seasonal analyses can be performed to critically evaluate seagrass ecosystem dynamics at spatial and temporal resolutions appropriate to the scientific question or monitoring and management objectives.
There are many examples of combinations of field and image data used for mapping seagrass in clear and poor-quality waters [42,43,44,45]. Additionally, airborne hyperspectral imagery in combination with complex inversion models, has been used for mapping of seagrass to the species level [46] or eelgrass [47] in poorer-quality waters. In contrast, our method has been applied to mapping seagrass in areas of differing water quality simultaneously. The methodology described here provides a simple solution to the complex problem of mapping benthic constituents in less clear water, a method that has been demonstrated scalable worldwide [13]. Previous mapping of the Moreton Bay area has required intensive manual digitization and interpretation of satellite imagery, which is comparatively time consuming, and subjective, even when guided by field data [48]. The automated approach presented substantially reduces investigator error, incorporates water depth, and enables robust and rapid assimilation of the data for generation of reliable output map products. Additionally, the method can be applied on a global scale and enables incorporation of citizen science data, empowering local organizations and reducing costs.
Importantly, any capability to assess seagrass habitats accurately, quantitatively, and spatially over time requires a method that is robust, consistent, and repeatable [8] so that map outputs are comparable over time. Due to its inherent simplicity and applicability across a range of water clarities, the method described here potentially provides the capability for repeat, time series mapping of seagrass meadow extent and change analysis, supporting monitoring and management of these areas.
To further improve our capability of mapping and monitoring seagrass properties (species composition and percentage cover) or the presence of the blue-green algae Lyngbya majuscula, our focus will be on improving field data collection and analysis methods. Research will examine the automated annotation of benthic photoquadrats as input for the classifier and the optimal distribution of field data to enable focused surveys that yield the most representative map for monitoring. Additionally, we will explore the use of the higher-spatial-resolution Sentinel-2 imagery—with its more frequent coverage, this would enable the possibility of examining pixel stacks to create an optimal mosaic for annual or seasonal changes.
Author Contributions
E.M.K.: conceptualization 55%, methodology 50%, software 10%, analysis 80%, data curation 20%, validation 90%, investigation 100%, writing—original draft preparation 70%, writing—review and editing 80%, project administration 90%, funding acquisition 90%; C.R.: conceptualization 15%, methodology 15%, software 90%, writing—original draft preparation 20%, writing—review and editing 15%, project administration 10%, funding acquisition 10%; J.U.: analysis 10%, resources 80%, data curation 40%; S.B.: analysis 10%, resources 20%, data curation 40%; M.L.: conceptualization 20%, methodology 25%, validation 10%, writing—review and editing 5%; S.P.: conceptualization 10%, methodology 10%, writing—original draft preparation 10%. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a Goodman Foundation Research Grant, and Paul Allen Philanthropies and Vulcan Inc. as part of the Allen Coral Atlas.
Data Availability Statement
Earth engine code is available from https://doi.org./10.5281/zenodo.3714181, whilst a live version is available at github.com/CoralMapping/gee-mapping-source. The reef mask is available at Pangaea.de (https://doi.org/10.1594/PANGAEA.892402). Thirty-meter resolution bathymetry for the Great Barrier Reef (incorporating Moreton Bay) is available at https://ecat.ga.gov.au/geonetwork/srv/eng/catalog.search#/metadata/115066 (accessed on 10 June 2021). Water quality data and EHMP data are available at https://seqcatchments.maps.arcgis.com/apps/webappviewer3d/index.html?id=4c9c69cf54d34f499c478ae97b82e44c (accessed on 3 November 2021). The 2015 Moreton Bay seagrass map is available from Pangaea.de (https://doi.org/10.1594/PANGAEA.937501).
Acknowledgments
Thank you to our infield staff and students—Carolina Castro, Alexandra Ordonez, Chantel Say, Maria Kottermair, Lucas Langlois and Mitchell Lyons—and to the Moreton Bay research Station for their support during our stay.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ralph, P.J.; Durako, M.J.; Enríquez, S.; Collier, C.J.; Doblin, M.A. Impact of light limitation on seagrasses. J. Exp. Mar. Biol. Ecol. 2007, 350, 176–193. [Google Scholar] [CrossRef]
- Ondiviela, B.; Losada, I.J.; Lara, J.; Maza, M.; Galván, C.; Bouma, T.; van Belzen, J. The role of seagrasses in coastal protection in a changing climate. Coast. Eng. 2014, 87, 158–168. [Google Scholar] [CrossRef]
- Mcleod, E.; Chmura, G.L.; Bouillon, S.; Salm, R.; Bjork, M.; Duarte, C.M.; Lovelock, C.E.; Schlesinger, W.H.; Silliman, B.R. A blueprint for blue carbon: Toward an improved understanding of the role of vegetated coastal habitats in sequestering CO2. Front. Ecol. Environ. 2011, 9, 552–560. [Google Scholar] [CrossRef] [Green Version]
- Short, F.T.; Polidoro, B.; Livingstone, S.R.; Carpenter, K.E.; Bandeira, S.; Bujang, J.S.; Calumpong, H.P.; Carruthers, T.J.B.; Coles, R.G.; Dennison, W.C.; et al. Extinction risk assessment of the world’s seagrass species. Biol. Conserv. 2011, 144, 1961–1971. [Google Scholar] [CrossRef]
- Duarte, C.M. The future of seagrass meadows. Environ. Conserv. 2002, 29, 192–206. [Google Scholar] [CrossRef] [Green Version]
- Waycott, M.; Duarte, C.M.; Carruthers, T.J.B.; Orth, R.J.; Dennison, W.C.; Olyarnik, S.; Calladine, A.; Fourqurean, J.W.; Heck, K.L.; Hughes, A.R.; et al. Accelerating loss of seagrasses across the globe threatens coastal ecosystems. Proc. Natl. Acad. Sci. USA 2009, 106, 12377–12381. [Google Scholar] [CrossRef] [Green Version]
- Tan, Y.M.; Dalby, O.; Kendrick, G.A.; Statton, J.; Sinclair, E.A.; Fraser, M.W.; Macreadie, P.I.; Gillies, C.L.; Coleman, R.A.; Waycott, M.; et al. Seagrass Restoration Is Possible: Insights and Lessons from Australia and New Zealand. Front. Mar. Sci. 2020, 7, 617. [Google Scholar] [CrossRef]
- Phinn, S.; Roelfsema, C.; Kovacs, E.; Canto, R.; Lyons, M.; Saunders, M.; Maxwell, P. Mapping, monitoring and modelling seagrass using remote sensing techniques. In Seagrasses of Australia; Springer: Cham, Switzerland, 2018; pp. 445–487. [Google Scholar]
- Joyce, K.E.; Duce, S.; Leahy, S.M.; Leon, J.; Maier, S.W. Principles and practice of acquiring drone-based image data in marine environments. Mar. Freshw. Res. 2019, 70, 952–963. [Google Scholar] [CrossRef]
- Bonin-Font, F.; Burguera, A.; Lisani, J.L. Visual Discrimination and Large Area Mapping of Posidonia Oceanica Using a Lightweight AUV. IEEE Access 2017, 5, 24479–24494. [Google Scholar] [CrossRef]
- Wabnitz, C.C.; Andréfouët, S.; Torres-Pulliza, D.; Muller-Karger, F.E.; Kramer, P.A. Regional-scale seagrass habitat mapping in the Wider Caribbean region using Landsat sensors: Applications to conservation and ecology. Remote Sens. Environ. 2008, 112, 3455–3467. [Google Scholar] [CrossRef]
- Traganos, D.; Aggarwal, B.; Poursanidis, D.; Topouzelis, K.; Chrysoulakis, N.; Reinartz, P. Towards Global-Scale Seagrass Mapping and Monitoring Using Sentinel-2 on Google Earth Engine: The Case Study of the Aegean and Ionian Seas. Remote Sens. 2018, 10, 1227. [Google Scholar] [CrossRef] [Green Version]
- Lyons, M.B.; Roelfsema, C.M.; Kennedy, E.V.; Kovacs, E.M.; Borrego-Acevedo, R.; Markey, K.; Roe, M.; Yuwono, D.M.; Harris, D.L.; Phinn, S.R.; et al. Mapping the world’s coral reefs using a global multiscale earth observation framework. Remote Sens. Ecol. Conserv. 2020, 6, 557–568. [Google Scholar] [CrossRef] [Green Version]
- Leonard, M.; Lina Mtwana, N.; Benjamin Lawrence, J.; Leanne, C.C.-U.; Chris, M.R.; Richard, U. The global distribution of seagrass meadows. Environ. Res. Lett. 2020, 15, 074041. [Google Scholar]
- Phinn, S.; Roelfsema, C.; Dekker, A.; Brando, V.; Anstee, J. Mapping seagrass species, cover and biomass in shallow waters: An assessment of satellite multi-spectral and airborne hyper-spectral imaging systems in Moreton Bay (Australia). Remote Sens. Environ. 2008, 112, 3413–3425. [Google Scholar] [CrossRef]
- Phinn, S.R.; Dekker, A.G.; Brando, V.E.; Roelfsema, C.M. Mapping Water Quality and Substrate Cover in Optically Complex Coastal and Reef Waters: An Integrated Approach. Mar. Pollut. Bull. 2005, 51, 459–469. [Google Scholar] [CrossRef]
- Green, E.P.; Mumby, P.J.; Edwards, A.J.; Clark, C.D. A Review of Remote Sensing for the Assessment and Management of Tropical Coastal Resources. Coast. Manag. 1996, 24, 1–40. [Google Scholar] [CrossRef]
- Alsaffar, Z.; Pearman, J.K.; Cúrdia, J.; Ellis, J.; Calleja, M.L.; Ruiz-Compean, P.; Roth, F.; Villalobos, R.; Jones, B.H.; Morán, X.A.G.; et al. The role of seagrass vegetation and local environmental conditions in shaping benthic bacterial and macroinvertebrate communities in a tropical coastal lagoon. Sci. Rep. 2020, 10, 13550. [Google Scholar] [CrossRef]
- Kuo, J.; Den Hartog, C. Seagrass Taxonomy and Identification Key. In Global Seagrass Research Methods; Short, F.C.R., Short, C., Eds.; Elsevier Science B.V.: Amsterdam, The Netherlands, 2001; pp. 31–58. [Google Scholar]
- Kovacs, E.; Roelfsema, C.; Lyons, M.; Zhao, S.; Phinn, S. Seagrass habitat mapping: How do Landsat 8 OLI, Sentinel-2, ZY-3A, and Worldview-3 perform? Remote Sens. Lett. 2018, 9, 686–695. [Google Scholar] [CrossRef]
- Knudby, A.; Nordlund, L. Remote sensing of seagrasses in a patchy multi-species environment. Int. J. Remote Sens. 2011, 32, 2227–2244. [Google Scholar] [CrossRef]
- Roelfsema, C.; Kovacs, E.M.; Saunders, M.I.; Phinn, S.; Lyons, M.; Maxwell, P. Challenges of remote sensing for quantifying changes in large complex seagrass environments. Estuar. Coast. Shelf Sci. 2013, 133, 161–171. [Google Scholar] [CrossRef]
- Roelfsema, C.M.; Lyons, M.; Kovacs, E.M.; Maxwell, P.; Saunders, M.I.; Samper-Villarreal, J.; Phinn, S.R. Multi-temporal mapping of seagrass cover, species and biomass: A semi-automated object based image analysis approach. Remote Sens. Environ. 2014, 150, 172–187. [Google Scholar] [CrossRef]
- Roelfsema, C.; Phinn, S.; Udy, N.; Maxwell, P. An integrated field and remote sensing approach for mapping seagrass cover, Moreton Bay, Australia. J. Spat. Sci. 2009, 54, 45–62. [Google Scholar] [CrossRef]
- HLW. Ecosystem Health Monitoring Program; Dataset, Healthy Land and Water: Brisbane, Australia, 2015. [Google Scholar]
- Roelfsema, C.; Loder, J.; Host, R.; Kovacs, E. Benthic Inventory of Reefal Areas of Inshore Moreton Bay, Queensland, Australia; Reef Check Australia: Brisbane, Australia, 2018. [Google Scholar]
- Dennison, W.C.; Carruthers, T.J.B.; Thomas, J.E.; Glibert, P.M. A comparison of issues and management approaches in Moreton Bay, Australia and Chesapeake Bay, USA. In Developments in Ecosystems, Volume 1: Wetlands Ecosystems in Asia: Function and Management; Wong, M.H., Ed.; Elsevier: Amsterdam, The Netherlands, 2004; p. 451. [Google Scholar]
- Maxwell, P.; Connolly, R.; Roelfsema, C.; Burfeind, D.; Udy, J.; O’Brien, K.; Saunders, M.; Barnes, R.; Olds, A.D.; Hendersen, C.J. Seagrasses of Moreton Bay Quandamooka: Diversity, ecology and resilience. In Moreton Bay Quandamooka & Catchment: Past, Present, and Future; Moreton Bay Foundation Ltd.: Brisbane, Australia, 2019; pp. 279–298. [Google Scholar]
- EHMP. Ecosystem Health Monitoring Program (EHMP) 2008–09; Annual Technical Report; Healthy Land and Water: Brisbane, Austria, 2010. [Google Scholar]
- Beaman, R.J. High-Resolution Depth Model for the Great Barrier Ree-30 m; Commonwealth of Australia (Geoscience Australia): Canberra, Australia, 2017. [Google Scholar] [CrossRef]
- Gorelick, N.; Hancher, M.; Dixon, M.; Ilyushchenko, S.; Thau, D.; Moore, R. Google Earth Engine: Planetary-scale geospatial analysis for everyone. Remote Sens. Environ. 2017, 202, 18–27. [Google Scholar] [CrossRef]
- USGS. Landsat 8–9 Calibration and Validation (Cal/Val) Algorithm Description Document (ADD); United States Geological Society: Sioux Falls, SD, USA, 2021. [Google Scholar]
- Blaschke, T. Object based image analysis for remote sensing. J. Photogramm. Remote Sens. 2010, 65, 2–16. [Google Scholar] [CrossRef] [Green Version]
- Achanta, R.; Susstrunk, S. Superpixels and Polygons using Simple Non-Iterative Clustering. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 4895–4904. [Google Scholar] [CrossRef] [Green Version]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning; Springer: New York, NY, USA, 2013; Volume 112. [Google Scholar]
- Congalton, R.G.; Green, K. Assessing the accuracy of remotely sensed data: Principles and practices. In Mapping Science, 2nd ed.; CRC Press: Boca Rotan, FL, USA, 2008; p. 200. [Google Scholar]
- Kilminster, K.; McMahon, K.; Waycott, M.; Kendrick, G.A.; Scanes, P.; McKenzie, L.; O’Brien, K.R.; Lyons, M.; Ferguson, A.; Maxwell, P.; et al. Unravelling complexity in seagrass systems for management: Australia as a microcosm. Sci. Total Environ. 2015, 534, 97–109. [Google Scholar] [CrossRef]
- Roelfsema, C.M.; Phinn, R.S. Validation. In Coral Reef Remote Sensing: A Guide for Multi-Level Sensing Mapping and Assessment; Goodman, J., Purkis, S., Phinn, S.R., Eds.; Elsiver: Dordrecht, Germany, 2013. [Google Scholar]
- Roelfsema, C.M.; Lyons, M.; Murray, N.; Kovacs, E.M.; Kennedy, E.; Markey, K.; Borrego-Acevedo, R.; Alvarez, A.O.; Say, C.; Tudman, P.; et al. Workflow for the Generation of Expert-Derived Training and Validation Data: A View to Global Scale Habitat Mapping. Front. Mar. Sci. 2021, 8, 643381. [Google Scholar] [CrossRef]
- Effler, S.W. Secchi Disk Transparency and Turbidity. J. Environ. Eng. Asce 1988, 114, 1436–1447. [Google Scholar] [CrossRef]
- HLW. Trends over Time; Healthy Land and Water: Brisbane, Australia, 2021. [Google Scholar]
- Dekker, A.G.; Brando, V.E.; Anstee, J.M. Retrospective seagrass change detection in a shallow coastal tidal Australian lake. Remote Sens. Environ. 2005, 97, 415–433. [Google Scholar] [CrossRef]
- Hossain, M.S.; Bujang, J.S.; Zakaria, M.H.; Hashim, M. The application of remote sensing to seagrass ecosystems: An overview and future research prospects. Int. J. Remote Sens. 2014, 36, 61–114. [Google Scholar] [CrossRef]
- Koedsin, W.; Intararuang, W.; Ritchie, R.J.; Huete, A. An Integrated Field and Remote Sensing Method for Mapping Seagrass Species, Cover, and Biomass in Southern Thailand. Remote Sens. 2016, 8, 292. [Google Scholar] [CrossRef] [Green Version]
- Nahirnick, N.K.; Reshitnyk, L.; Campbell, M.; Heesiing-Lewis, M.; Costa, M.; Yakimishyn, J.; Lee, L. Mapping with Confidence; delineating seagrass habitats using Unoccupied Aerial Systems (UAS). Remote Sens. Ecol. Conserv. 2019, 5, 121–135. [Google Scholar] [CrossRef]
- Hedley, J.D.; Russell, B.J.; Randolph, K.; Perez-Castro, M.A.; Vasquez-Elizondo, R.M.; Enriquez, S.; Dierssen, H.M. Remote Sensing of Seagrass Leaf Area Index and Species: The Capability of a Model Inversion Method Assessed by Sensitivity Analysis and Hyperspectral Data of Florida Bay. Front. Mar. Sci. 2017, 4, 362. [Google Scholar] [CrossRef] [Green Version]
- Dierssen, H.M.; Bostrom, K.J.; Chlus, A.; Hammerstrom, K.; Thompson, D.R.; Lee, Z. Pushing the Limits of Seagrass Remote Sensing in the Turbid Waters of Elkhorn Slough, California. Remote Sens. 2019, 11, 1664. [Google Scholar] [CrossRef] [Green Version]
- McKenzie, L. Guidelines for the Rapid Assessment and Mapping of Tropical Seagrass Habitats State of Queensland; State of Queensland, Department of Primary Industries: Brisbane, Australia, 2003; p. 46. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).