Comparing Gaofen-5, Ground, and Huanjing-1A Spectra for the Monitoring of Soil Salinity with the BP Neural Network Improved by Particle Swarm Optimization
Abstract
1. Introduction
2. Study Area and Method
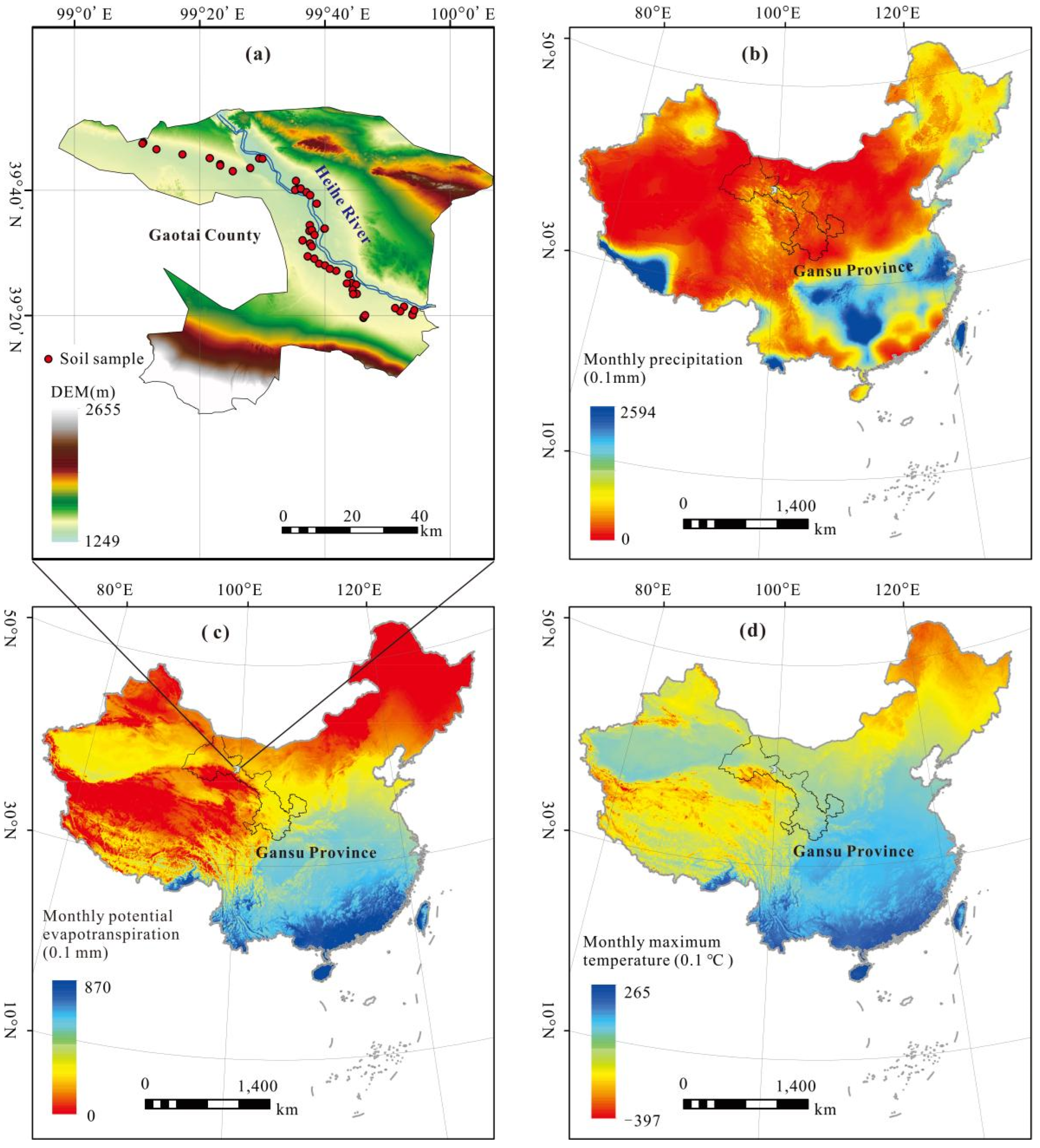
2.1. Study Area
2.2. Field Sampling and Spectra Process
2.2.1. Field Sampling
2.2.2. Laboratory Spectra Process Using Analytical Spectral Devices (ASD, USA)
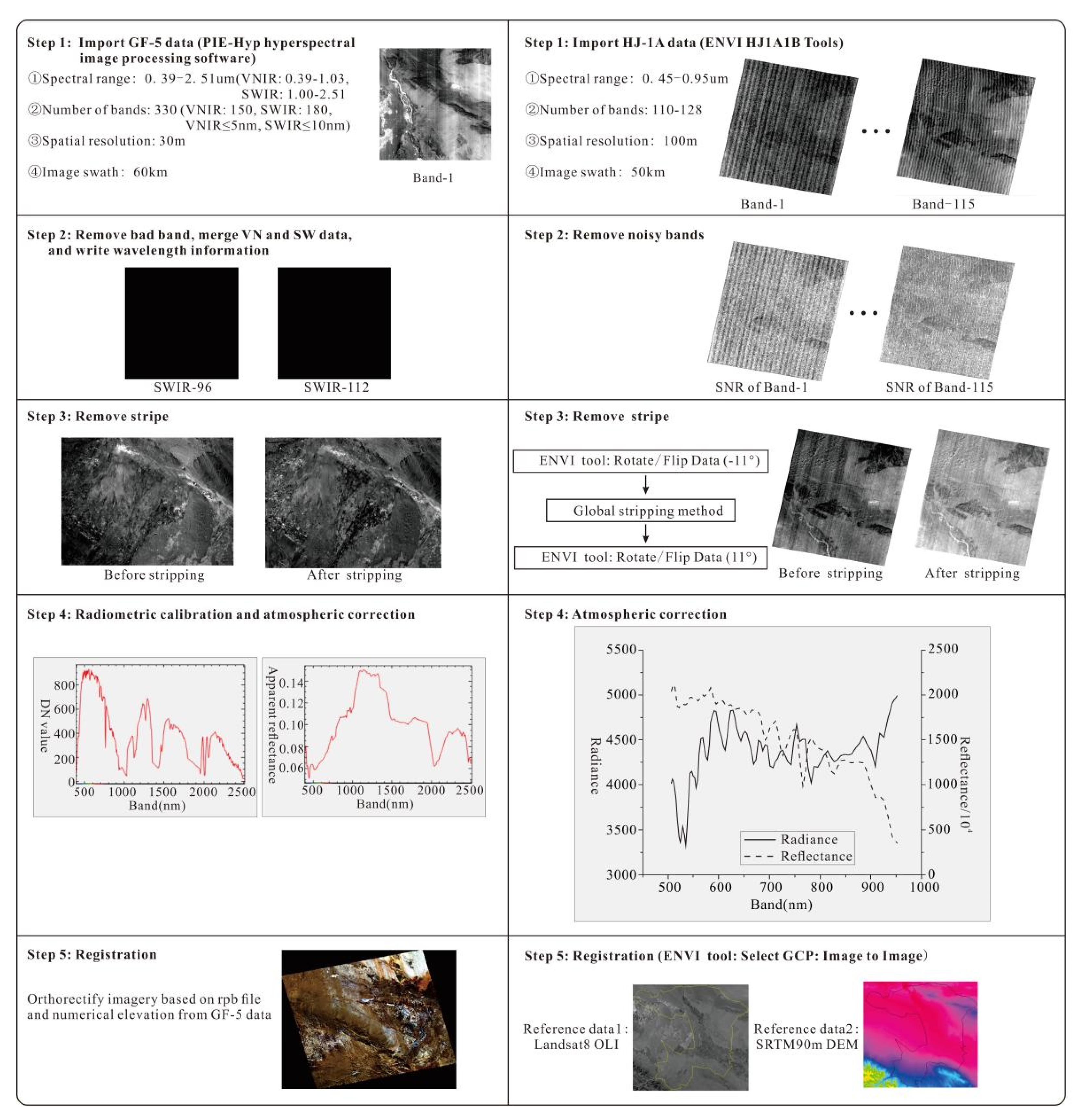
2.2.3. GF-5 and HJ-1A Hyperspectral Imagery Process
2.3. Model Establishment and Verification
3. Results
3.1. Descriptive Statistics of EC
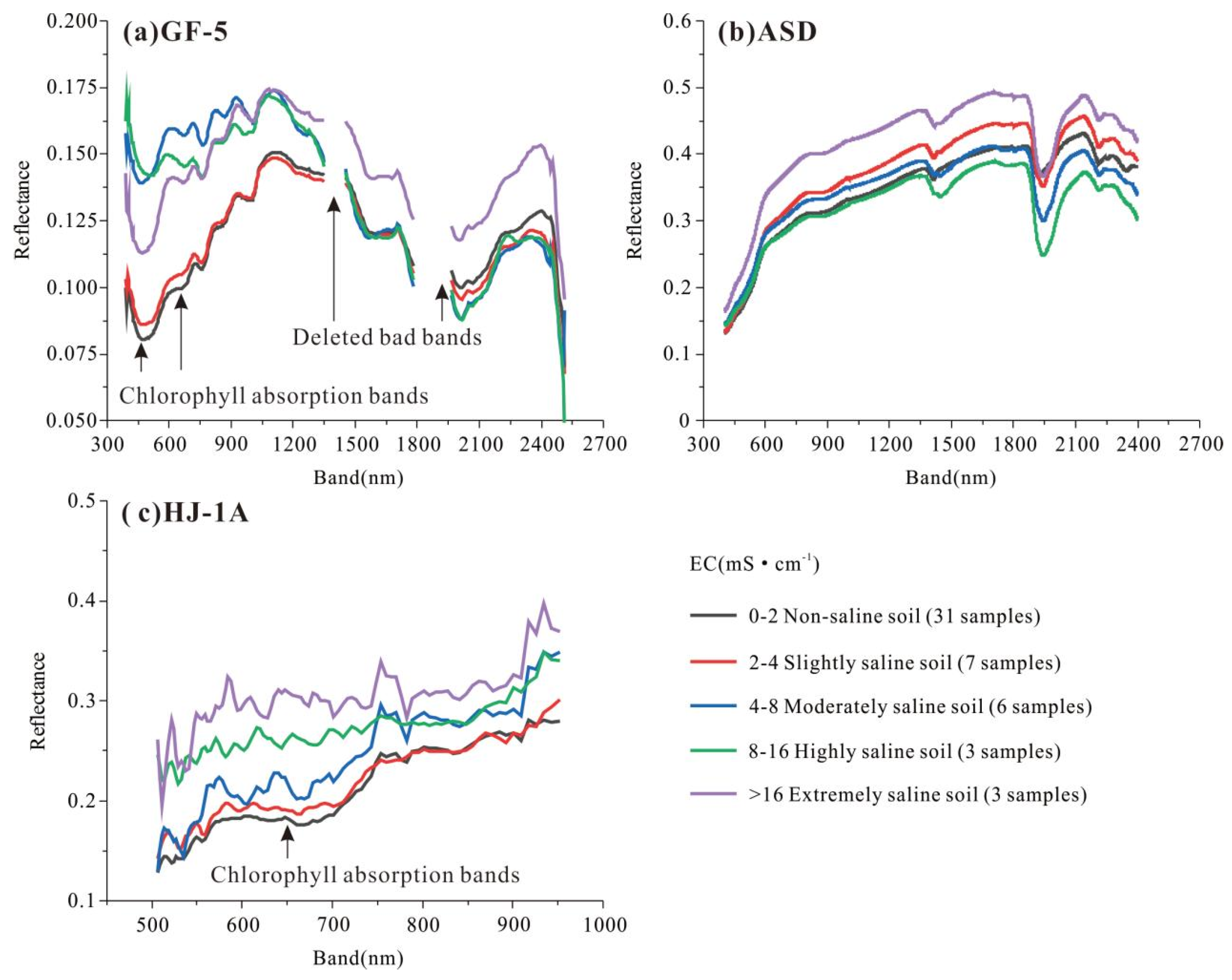
3.2. Hyperspectral Curve of Soil Samples
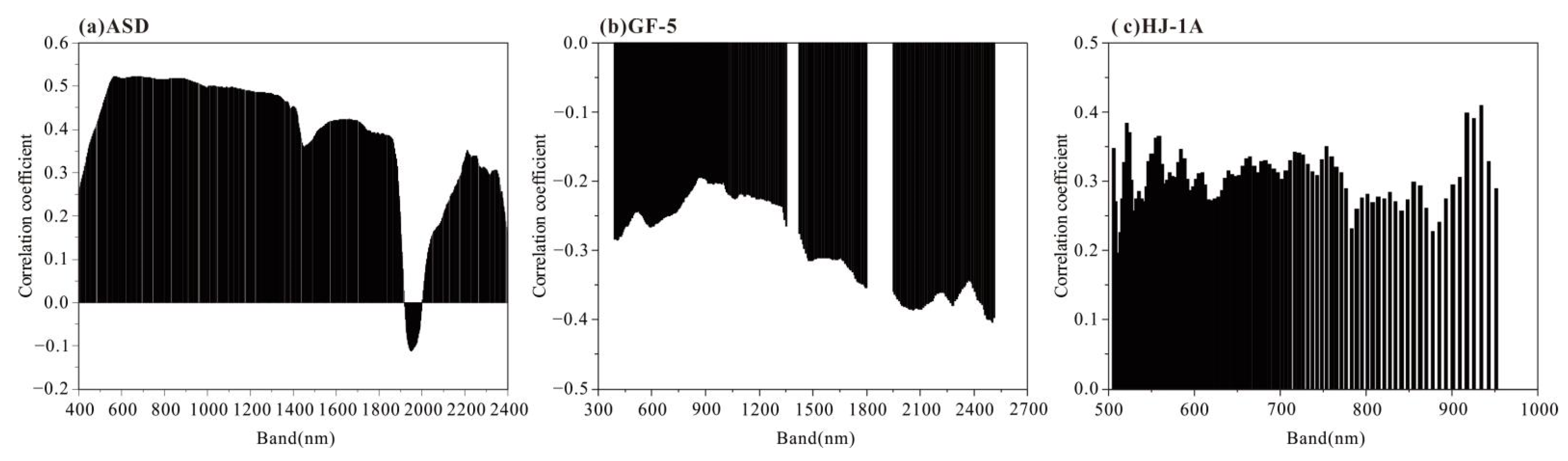
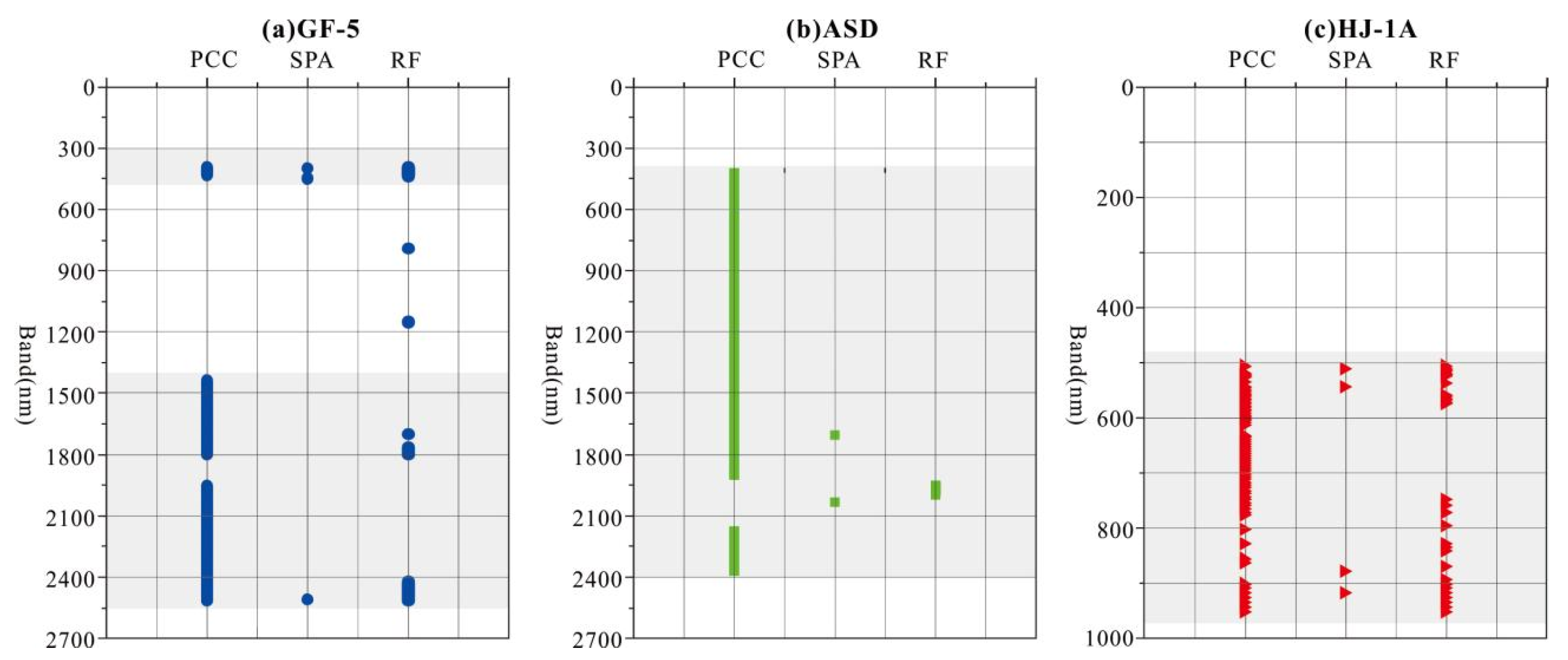
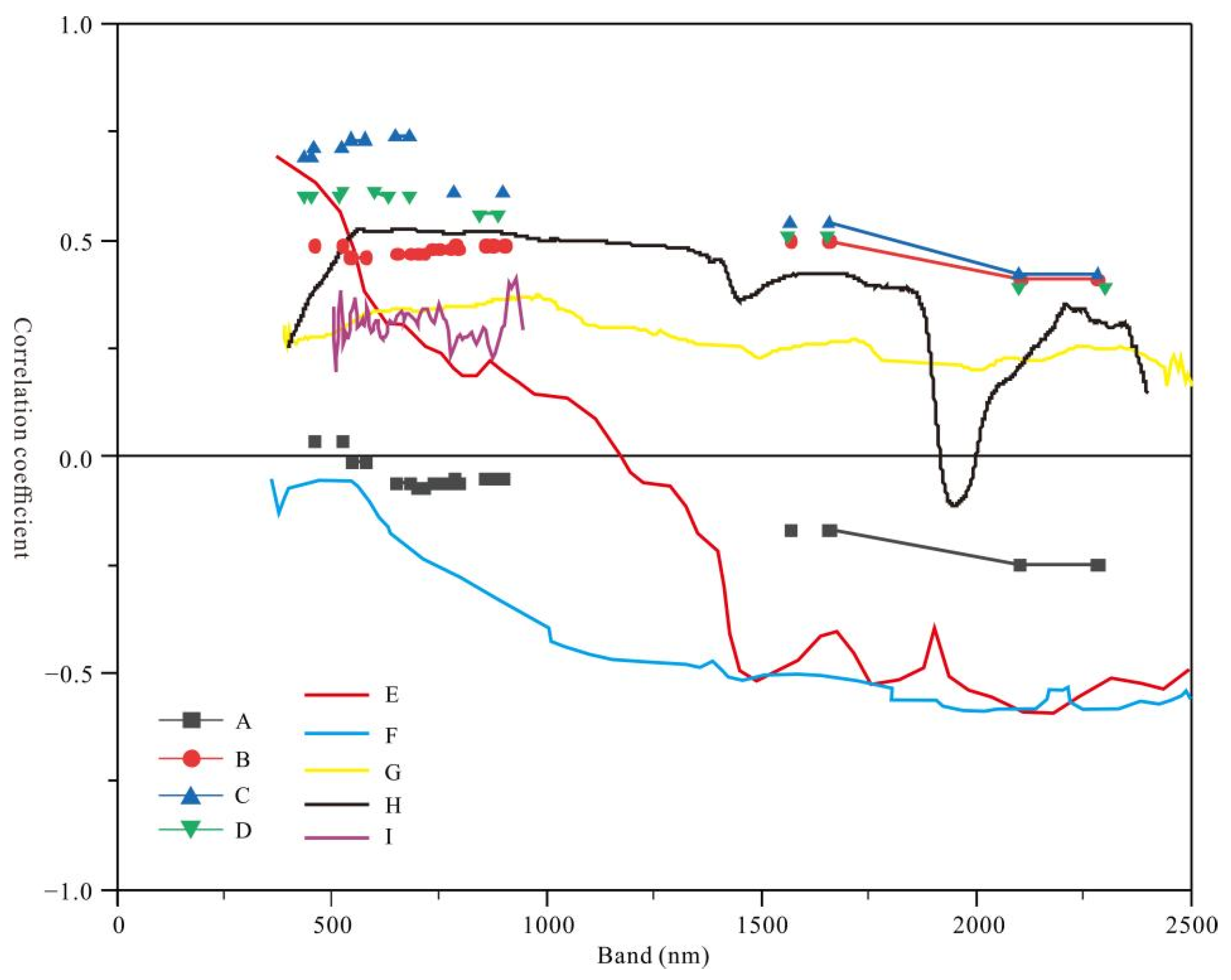
3.3. The Results of Four Band Screening Methods
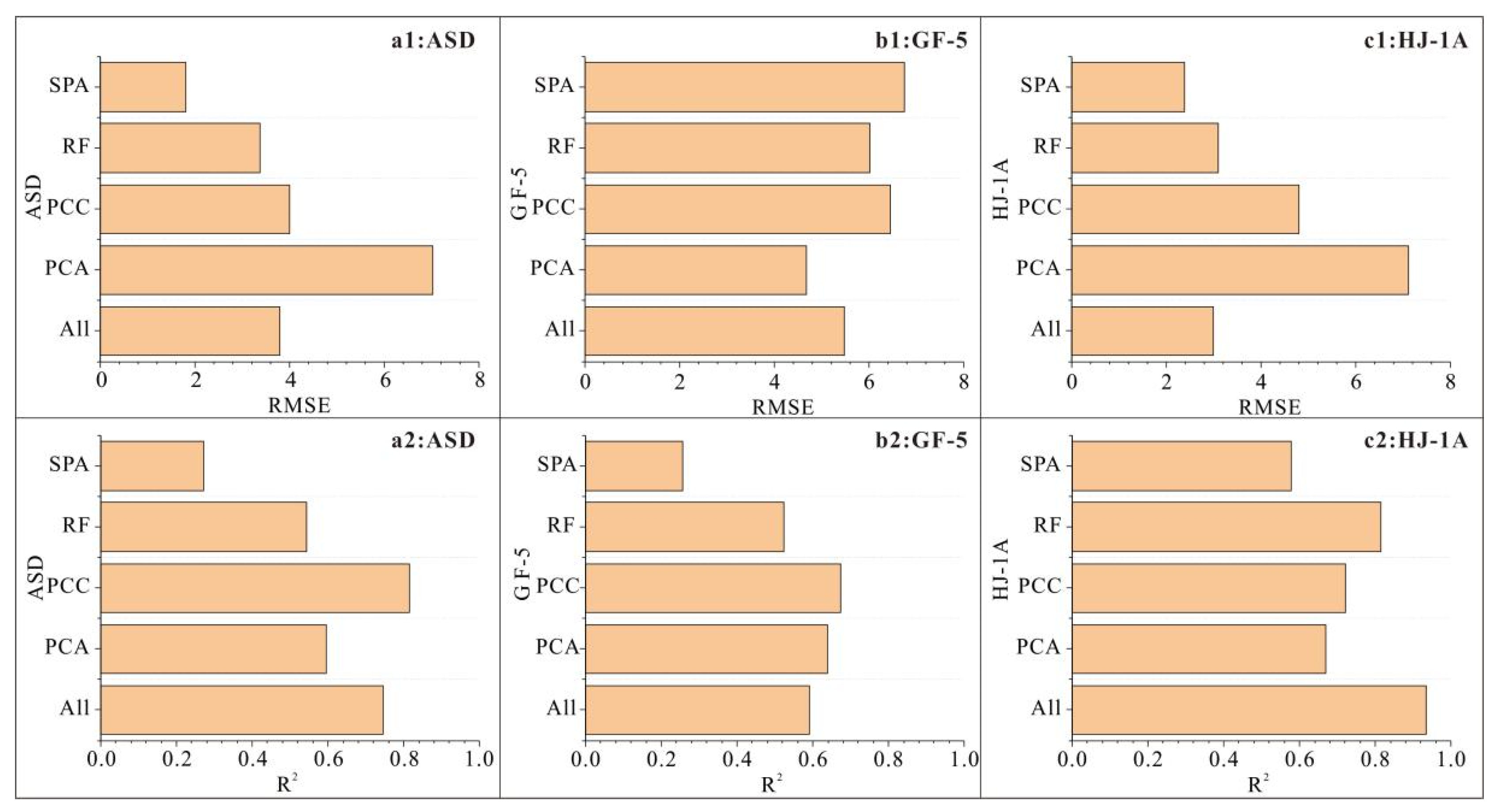
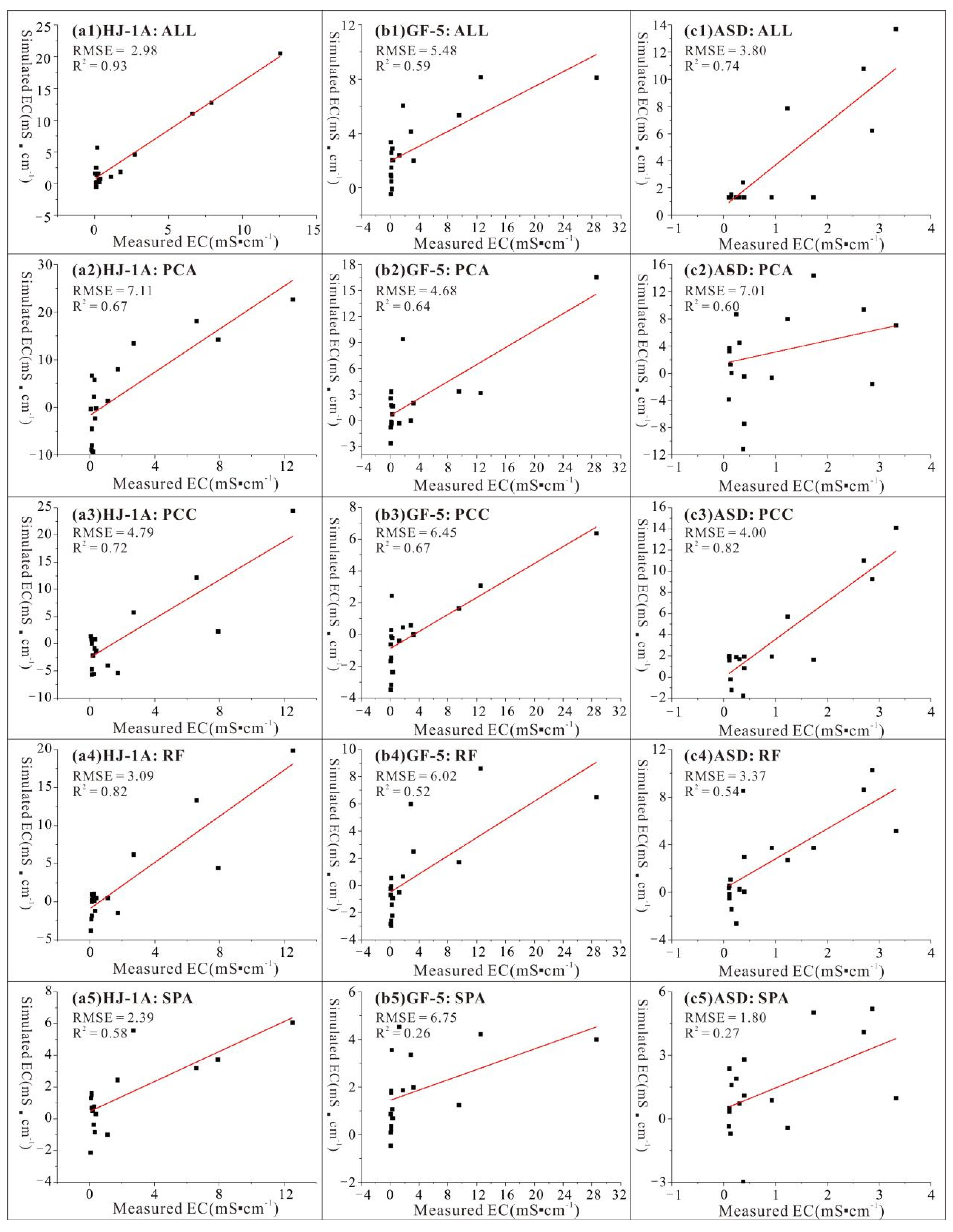
3.4. PSO-BPNN Modeling Results Based on Different Band Screening Methods
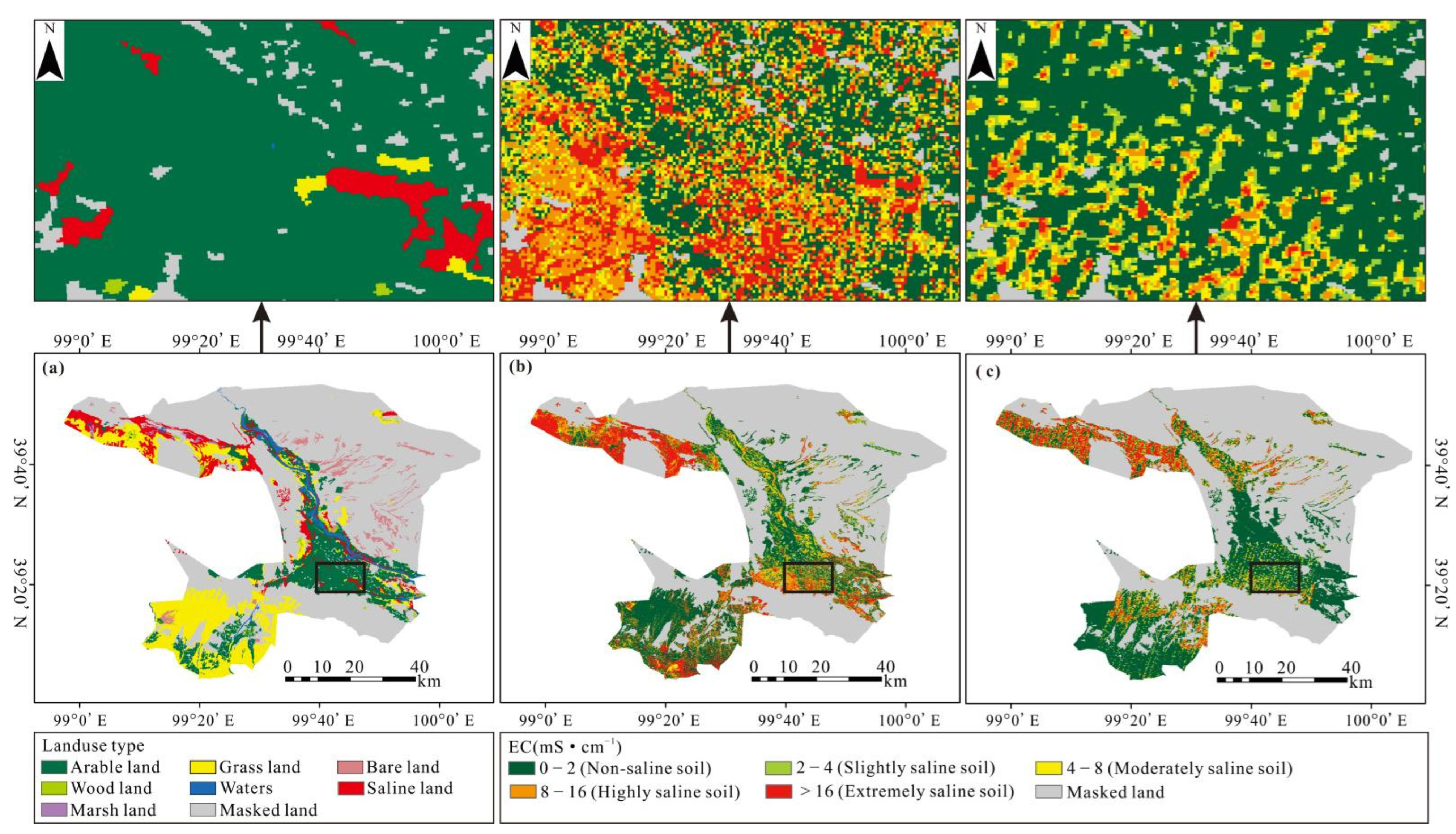
3.5. Distribution of Saline Soil in Gaotai County
4. Discussion
4.1. Comparative Analysis of Different Data Sources
4.2. Spatial Distribution Mapping of Soil Salinity
4.3. Analysis of the Sensitive Bands of Soil Salinization
4.4. Applicability of Machine Learning Algorithms
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hopmans, J.W.; Qureshi, A.S.; Kisekka, I.; Munns, R.; Grattan, S.R.; Rengasamy, P.; Ben-Gal, A.; Assouline, S.; Javaux, M.; Minhas, P.S. Advances in Agronomy; Academic Press: Pittsburgh, PA, USA, 2021. [Google Scholar]
- Salcedo, F.P.; Cutillas, P.P.; Cabañero, J.J.A.; Vivaldi, A.G. Use of remote sensing to evaluate the effects of environmental factors on soil salinity in a semi-arid area. Sci. Total Environ. 2022, 815, 152524. [Google Scholar] [CrossRef]
- Zhu, K.; Sun, Z.; Zhao, F.; Yang, T.; Tian, Z.; Lai, J.; Zhu, W.; Long, B. Relating Hyperspectral Vegetation Indices with Soil Salinity at Different Depths for the Diagnosis of Winter Wheat Salt Stress. Remote Sens. 2021, 13, 250. [Google Scholar] [CrossRef]
- Singh, A. Soil salinization management for sustainable development: A review. J. Environ. Manag. 2021, 277, 111383. [Google Scholar] [CrossRef] [PubMed]
- Mashimbye, Z.E.; Cho, M.A.; Nell, J.P.; De Clercq, W.P.; Van Niekerk, A.; Turner, D.P. Model-Based Integrated Methods for Quantitative Estimation of Soil Salinity from Hyperspectral Remote Sensing Data: A Case Study of Selected South African Soils. Pedosphere 2012, 22, 640–649. [Google Scholar] [CrossRef]
- Wu, W.; Zucca, C.; Muhaimeed, A.S.; Al-Shafie, W.M.; Fadhil Al-Quraishi, A.M.; Nangia, V.; Zhu, M.; Liu, G. Soil salinity prediction and mapping by machine learning regression in Central Mesopotamia, Iraq. Land Degrad. Dev. 2018, 29, 4005–4014. [Google Scholar] [CrossRef]
- Ma, L.; Yang, S.; Simayi, Z.; Gu, Q.; Li, J.; Yang, X.; Ding, J. Modeling variations in soil salinity in the oasis of Junggar Basin, China. Land Degrad. Dev. 2018, 29, 551–562. [Google Scholar] [CrossRef]
- Xu, H.; Chen, C.; Zheng, H.; Luo, G.; Yang, L.; Wang, W.; Wu, S.; Ding, J. AGA-SVR-based selection of feature subsets and optimization of parameter in regional soil salinization monitoring. Int. J. Remote Sens. 2020, 41, 4470–4495. [Google Scholar] [CrossRef]
- Fathizad, H.; Ali Hakimzadeh Ardakani, M.; Sodaiezadeh, H.; Kerry, R.; Taghizadeh-Mehrjardi, R. Investigation of the spatial and temporal variation of soil salinity using random forests in the central desert of Iran. Geoderma 2020, 365, 114233. [Google Scholar] [CrossRef]
- Figueiredo, E.M.N.; Ludermir, T.B. Investigating the use of alternative topologies on performance of the PSO-ELM. Neurocomputing 2014, 127, 4–12. [Google Scholar] [CrossRef]
- Lin, S.; Ying, K.; Chen, S.; Lee, Z. Particle swarm optimization for parameter determination and feature selection of support vector machines. Expert Syst. Appl. 2008, 35, 1817–1824. [Google Scholar] [CrossRef]
- Wei, J.; Jian-qi, Z.; Xiang, Z. Face recognition method based on support vector machine and particle swarm optimization. Expert Syst. Appl. 2011, 38, 4390–4393. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, X.; He, Z. An ACO-based algorithm for parameter optimization of support vector machines. Expert Syst. Appl. 2010, 37, 6618–6628. [Google Scholar] [CrossRef]
- Li, L.; Sun, J.; Tseng, M.; Li, Z. Extreme learning machine optimized by whale optimization algorithm using insulated gate bipolar transistor module aging degree evaluation. Expert Syst. Appl. 2019, 127, 58–67. [Google Scholar] [CrossRef]
- Ab Aziz, M.F.; Mostafa, S.A.; Foozy, C.F.M.; Mohammed, M.A.; Elhoseny, M.; Abualkishik, A.Z. Integrating Elman recurrent neural network with particle swarm optimization algorithms for an improved hybrid training of multidisciplinary datasets. Expert Syst. Appl. 2021, 183, 115441. [Google Scholar] [CrossRef]
- Kushwah, R.; Kaushik, M.; Chugh, K. A modified whale optimization algorithm to overcome delayed convergence in artificial neural networks. Soft Comput. 2021, 25, 10275–10286. [Google Scholar] [CrossRef]
- Liu, X.; Liu, Z.; Liang, Z.; Zhu, S.; Correia, J.A.F.O.; De Jesus, A.M.P. PSO-BP Neural Network-Based Strain Prediction of Wind Turbine Blades. Materials 2019, 12, 1889. [Google Scholar] [CrossRef]
- Das, B.; Manohara, K.K.; Mahajan, G.R.; Sahoo, R.N. Spectroscopy based novel spectral indices, PCA- and PLSR-coupled machine learning models for salinity stress phenotyping of rice. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 229, 117983. [Google Scholar] [CrossRef]
- Wei, Q.; Nurmemet, I.; Gao, M.; Xie, B. Inversion of Soil Salinity Using Multisource Remote Sensing Data and Particle Swarm Machine Learning Models in Keriya Oasis, Northwestern China. Remote Sens. 2022, 14, 512. [Google Scholar] [CrossRef]
- Daniel, K.W.; Tripathi, N.K.; Honda, K. Artificial neural network analysis of laboratory and in situ spectra for the estimation of macronutrients in soils of Lop Buri (Thailand). Soil Res. 2003, 41, 47–59. [Google Scholar] [CrossRef]
- Yu, X.; Chang, C.; Song, J.; Zhuge, Y.; Wang, A. Precise Monitoring of Soil Salinity in China’s Yellow River Delta Using UAV-Borne Multispectral Imagery and a Soil Salinity Retrieval Index. Sensors 2022, 22, 546. [Google Scholar] [CrossRef]
- Taghadosi, M.M.; Hasanlou, M.; Eftekhari, K. Soil salinity mapping using dual-polarized SAR Sentinel-1 imagery. Int. J. Remote Sens. 2019, 40, 237–252. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, L.; Sun, R.; Zhu, D.; Zhang, C.; Chen, Q. Retrieval of the Soil Salinity from Sentinel-1 Dual-Polarized SAR Data Based on Deep Neural Network Regression. IEEE Geosci. Remote Sens. Lett. 2020, 19, 4006905. [Google Scholar] [CrossRef]
- Ma, Y.; Chen, H.; Zhao, G.; Wang, Z.; Wang, D. Spectral Index Fusion for Salinized Soil Salinity Inversion Using Sentinel-2A and UAV Images in a Coastal Area. IEEE Access 2020, 8, 159595–159608. [Google Scholar] [CrossRef]
- Khanna, S.; Maria, J.S.; Ustin, S.L.; Shapiro, K.; Haverkamp, P.J.; Lay, M. Comparing the Potential of Multispectral and Hyperspectral Data for Monitoring Oil Spill Impact. Sensors 2018, 18, 558. [Google Scholar] [CrossRef] [PubMed]
- van Wagtendonk, J.W.; Root, R.R.; Key, C.H. Comparison of AVIRIS and Landsat ETM+ detection capabilities for burn severity. Remote Sens. Environ. 2004, 92, 397–408. [Google Scholar] [CrossRef]
- Teillet, P.M.; Staenz, K.; William, D.J. Effects of spectral, spatial, and radiometric characteristics on remote sensing vegetation indices of forested regions. Remote Sens. Environ. 1997, 61, 139–149. [Google Scholar] [CrossRef]
- Bai, L.; Wang, C.; Zang, S.; Wu, C.; Luo, J.; Wu, Y. Mapping Soil Alkalinity and Salinity in Northern Songnen Plain, China with the HJ-1 Hyperspectral Imager Data and Partial Least Squares Regression. Sensors 2018, 18, 3855. [Google Scholar] [CrossRef]
- Weng, Y.; Gong, P.; Zhu, Z. A Spectral Index for Estimating Soil Salinity in the Yellow River Delta Region of China Using EO-1 Hyperion Data. Pedosphere 2010, 20, 378–388. [Google Scholar] [CrossRef]
- Hu, J.; Peng, J.; Zhou, Y.; Xu, D.; Zhao, R.; Jiang, Q.; Fu, T.; Wang, F.; Shi, Z. Quantitative Estimation of Soil Salinity Using UAV-Borne Hyperspectral and Satellite Multispectral Images. Remote Sens. 2019, 11, 736. [Google Scholar] [CrossRef]
- Hbirkou, C.; Pätzold, S.; Mahlein, A.; Welp, G. Airborne hyperspectral imaging of spatial soil organic carbon heterogeneity at the field-scale. Geoderma 2012, 175–176, 21–28. [Google Scholar] [CrossRef]
- Nouri, M.; Gomez, C.; Gorretta, N.; Roger, J.M. Clay content mapping from airborne hyperspectral Vis-NIR data by transferring a laboratory regression model. Geoderma 2017, 298, 54–66. [Google Scholar] [CrossRef]
- Lin, L.; Gao, Z.; Liu, X. Estimation of soil total nitrogen using the synthetic color learning machine (SCLM) method and hyperspectral data. Geoderma 2020, 380, 114664. [Google Scholar] [CrossRef]
- Pfitzner, K.S.; Harford, A.J.; Whiteside, T.G.; Bartolo, R.E. Mapping magnesium sulfate salts from saline mine discharge with airborne hyperspectral data. Sci. Total Environ. 2018, 640–641, 1259–1271. [Google Scholar] [CrossRef]
- Ghamisi, P.; Couceiro, M.S.; Martins, F.M.L.; Benediktsson, J.A. Multilevel Image Segmentation Based on Fractional-Order Darwinian Particle Swarm Optimization. IEEE Trans. Geosci. Remote Sens. 2014, 52, 2382–2394. [Google Scholar] [CrossRef]
- Hong, Y.; Liu, Y.; Chen, Y.; Liu, Y.; Yu, L.; Liu, Y.; Cheng, H. Application of fractional-order derivative in the quantitative estimation of soil organic matter content through visible and near-infrared spectroscopy. Geoderma 2019, 337, 758–769. [Google Scholar] [CrossRef]
- Tian, A.; Zhao, J.; Xiong, H.; Gan, S.; Fu, C. Application of Fractional Differential Calculation in Pretreatment of Saline Soil Hyperspectral Reflectance Data. J. Sens. 2018, 2018, 8017614. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, F.; Kung, H.; Johnson, V.C. New methods for improving the remote sensing estimation of soil organic matter content (SOMC) in the Ebinur Lake Wetland National Nature Reserve (ELWNNR) in northwest China. Remote Sens. Environ. 2018, 218, 104–118. [Google Scholar] [CrossRef]
- Nawar, S.; Buddenbaum, H.; Hill, J.; Kozak, J. Modeling and Mapping of Soil Salinity with Reflectance Spectroscopy and Landsat Data Using Two Quantitative Methods (PLSR and MARS). Remote Sens. 2014, 6, 10813–10834. [Google Scholar] [CrossRef]
- Meng, L.; Zhou, S.; Zhang, H.; Bi, X. Estimating soil salinity in different landscapes of the Yellow River Delta through Landsat OLI/TIRS and ETM+ Data. J. Coast. Conserv. 2016, 20, 271–279. [Google Scholar] [CrossRef]
- Wang, F.; Yang, S.; Wei, Y.; Shi, Q.; Ding, J. Characterizing soil salinity at multiple depth using electromagnetic induction and remote sensing data with random forests: A case study in Tarim River Basin of southern Xinjiang, China. Sci. Total Environ. 2021, 754, 142030. [Google Scholar] [CrossRef]
- Shi, H.; Hellwich, O.; Luo, G.; Chen, C.; He, H.; Ochege, F.U.; Van de Voorde, T.; Kurban, A.; de Maeyer, P. A Global Meta-Analysis of Soil Salinity Prediction Integrating Satellite Remote Sensing, Soil Sampling, and Machine Learning. IEEE Trans. Geosci. Remote Sens. 2021, 60, 4505815. [Google Scholar] [CrossRef]
- Ding, Y.X.; Peng, S.Z. Spatiotemporal trends and attribution of drought across China from 1901–2100. Sustainability 2020, 12, 477. [Google Scholar] [CrossRef]
- Ding, Y.X.; Peng, S.Z. Spatiotemporal change and attribution of potential evapotranspiration over China from 1901 to 2100. Theor. Appl. Climatol. 2021, 145, 79–94. [Google Scholar] [CrossRef]
- Peng, S.Z.; Gang, C.; Cao, Y.; Chen, Y.M. Assessment of climate change trends over the loess plateau in China from 1901 to 2100. Int. J. Climatol. 2017, 38, 2250–2264. [Google Scholar] [CrossRef]
- Peng, S.Z.; Ding, Y.X.; Liu, W.Z.; Li, Z. 1 km monthly temperature and precipitation dataset for China from 1901 to 2017. Earth Syst. Sci. Data 2019, 11, 1931–1946. [Google Scholar] [CrossRef]
- Peng, S.Z. 1 km Monthly Potential Evapotranspiration Dataset in China (1990–2021); National Tibetan Plateau Data Center: Beijing, China, 2022. [Google Scholar]
- Peng, S.Z. 1-km Monthly Maximum Temperature Dataset for China (1901–2021); National Tibetan Plateau Data Center: Beijing, China, 2020. [Google Scholar]
- Peng, S.Z. 1-km Monthly Precipitation Dataset for China (1901–2021); National Tibetan Plateau Data Center: Beijing, China, 2020. [Google Scholar]
- Peng, S.Z.; Ding, Y.X.; Wen, Z.M.; Chen, Y.M.; Cao, Y.; Ren, J.Y. Spatiotemporal change and trend analysis of potential evapotranspiration over the Loess Plateau of China during 2011–2100. Agric. For. Meteorol. 2020, 233, 183–194. [Google Scholar] [CrossRef]
- Wang, Q.; Li, P.; Chen, X. Modeling salinity effects on soil reflectance under various moisture conditions and its inverse application: A laboratory experiment. Geoderma 2012, 170, 103–111. [Google Scholar] [CrossRef]
- Adeline, K.R.M.; Gomez, C.; Gorretta, N.; Roger, J.M. Predictive ability of soil properties to spectral degradation from laboratory Vis-NIR spectroscopy data. Geoderma 2017, 288, 143–153. [Google Scholar] [CrossRef]
- Farifteh, J.; van der Meer, F.; van der Meijde, M.; Atzberger, C. Spectral characteristics of salt-affected soils: A laboratory experiment. Geoderma 2008, 145, 196–206. [Google Scholar] [CrossRef]
- Tian, Y.; Zhang, J.; Yao, X.; Cao, W.; Zhu, Y. Laboratory assessment of three quantitative methods for estimating the organic matter content of soils in China based on visible/near-infrared reflectance spectra. Geoderma 2013, 202–203, 161–170. [Google Scholar] [CrossRef]
- Nawar, S.; Buddenbaum, H.; Hill, J.; Kozak, J.; Mouazen, A.M. Estimating the soil clay content and organic matter by means of different calibration methods of vis-NIR diffuse reflectance spectroscopy. Soil Tillage Res. 2016, 155, 510–522. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Tian, H.; Cao, C.; Xu, M.; Zhu, Z.; Liu, D.; Wang, X.; Cui, S. Estimation of chlorophyll-a concentration in coastal waters with HJ-1A HSI data using a three-band bio-optical model and validation. Int. J. Remote Sens. 2014, 35, 5984–6003. [Google Scholar] [CrossRef]
- Yan, J.; Zhou, K.; Liu, D.; Wang, J.; Wang, L.; Liu, H. Alteration information extraction using improved relative absorption band-depth images, from HJ-1A HSI data: A case study in Xinjiang Hatu gold ore district. Int. J. Remote Sens. 2014, 35, 6728–6741. [Google Scholar] [CrossRef]
- Kennard, R.W.; Stone, L.A. Computer aided design of experiments. Technometrics 1969, 11, 137–148. [Google Scholar] [CrossRef]
- Wang, J.; Wang, W.; Hu, Y.; Tian, S.; Liu, D. Soil Moisture and Salinity Inversion Based on New Remote Sensing Index and Neural Network at a Salina-Alkaline Wetland. Water 2021, 13, 2762. [Google Scholar] [CrossRef]
- Bao, Z.Y.; Yu, J.Z.; Yang, S. Intelligent Optimization Algorithm and Its MATLAB Example; Electronics Industry Press: Beijing, China, 2018. [Google Scholar]
- Wang, X.C.; Shi, F.; Yu, L.; Li, Y. Analysis of 43 Cases of Neural Network in MATLAB; Beihang University Press: Beijing, China, 2013. [Google Scholar]
- Norouz, M. Visual-Inertial State Estimation Based on PSO-BPNN UKF. In Proceedings of the 2020 28th Iranian Conference on Electrical Engineering (ICEE), Tabriz, Iran, 4–6 August 2020; IEEE: New York, NY, USA, 2020. [Google Scholar]
- Shi, Z.; Wang, Q.L.; Peng, J.; Ji, W.J.; Liu, H.X.; Li, X.; Rossel, R.A.V. Development of a national VNIR soil-spectral library for soil classification and prediction of organic matter concentrations. Sci. China Earth Sci. 2014, 44, 978–988. [Google Scholar] [CrossRef]
- Dai, C.D.; Jiang, X.G.; Tang, L.L. Application Processing and Analysis of Remote Sensing Image; Tsinghua University Press: Beijing, China, 2003. [Google Scholar]
- Ivushkin, K.; Bartholomeus, H.; Bregt, A.K.; Pulatov, A.; Kempen, B.; de Sousa, L. Global mapping of soil salinity change. Remote Sens. Environ. 2019, 231, 111260. [Google Scholar] [CrossRef]
- Yin, F.; Wu, M.; Liu, L.; Zhu, Y.; Feng, J.; Yin, D.; Yin, C.; Yin, C. Predicting the abundance of copper in soil using reflectance spectroscopy and GF5 hyperspectral imagery. Int. J. Appl. Earth Obs. 2021, 102, 102420. [Google Scholar] [CrossRef]
- Hong, Y.; Chen, S.; Chen, Y.; Linderman, M.; Mouazen, A.M.; Liu, Y.; Guo, L.; Yu, L.; Liu, Y.; Cheng, H.; et al. Comparing laboratory and airborne hyperspectral data for the estimation and mapping of topsoil organic carbon: Feature selection coupled with random forest. Soil Tillage Res. 2020, 199, 104589. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, M.; Zheng, L.; Qin, Q.; Lee, W.S. Spectral features extraction for estimation of soil total nitrogen content based on modified ant colony optimization algorithm. Geoderma 2019, 333, 23–34. [Google Scholar] [CrossRef]
- Lu, B.; He, Y.; Dao, P.D. Comparing the Performance of Multispectral and Hyperspectral Images for Estimating Vegetation Properties. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2019, 12, 1784–1797. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, F.; Ding, J.; Kung, H.T.; Latif, A.; Johnson, V.C. Estimation of soil salt content (SSC) in the Ebinur Lake Wetland National Nature Reserve (ELWNNR), Northwest China, based on a Bootstrap-BP neural network model and optimal spectral indices. Sci. Total Environ. 2018, 615, 918–930. [Google Scholar] [CrossRef]
- Franceschini, M.H.D.; Demattê, J.A.M.; Da Silva Terra, F.; Vicente, L.E.; Bartholomeus, H.; de Souza Filho, C.R. Prediction of soil properties using imaging spectroscopy: Considering fractional vegetation cover to improve accuracy. Int. J. Appl. Earth Obs. 2015, 38, 358–370. [Google Scholar] [CrossRef]
- Guo, L.; Zhang, H.; Shi, T.; Chen, Y.; Jiang, Q.; Linderman, M. Prediction of soil organic carbon stock by laboratory spectral data and airborne hyperspectral images. Geoderma 2019, 337, 32–41. [Google Scholar] [CrossRef]
- Lu, P.; Wang, L.; Niu, Z.; Li, L.; Zhang, W. Prediction of soil properties using laboratory VIS–NIR spectroscopy and Hyperion imagery. J. Geochem. Explor. 2013, 132, 26–33. [Google Scholar] [CrossRef]
- Han, L.; Liu, Z.; Ning, Y.; Zhao, Z. Research Process of Remote Sensing Inversion of Soil Heavy Metal Pollution in Mining Area. Conserv. Util. Miner. Resour. 2019, 39, 109–117. [Google Scholar]
- Allbed, A.; Kumar, L.; Aldakheel, Y.Y. Assessing soil salinity using soil salinity and vegetation indices derived from IKONOS high-spatial resolution imageries: Applications in a date palm dominated region. Geoderma 2014, 230, 1–8. [Google Scholar] [CrossRef]
- Allbed, A.; Kumar, L.; Sinha, P. Mapping and Modelling Spatial Variation in Soil Salinity in the Al Hassa Oasis Based on Remote Sensing Indicators and Regression Techniques. Remote Sens. 2014, 6, 1137–1157. [Google Scholar] [CrossRef]
- Douaoui, A.E.K.; Nicolas, H.; Walter, C. Detecting salinity hazards within a semiarid context by means of combining soil and remote-sensing data. Geoderma 2006, 134, 217–230. [Google Scholar] [CrossRef]
- Wang, J.; Ding, J.; Yu, D.; Teng, D.; He, B.; Chen, X.; Ge, X.; Zhang, Z.; Wang, Y.; Yang, X.; et al. Machine learning-based detection of soil salinity in an arid desert region, Northwest China: A comparison between Landsat-8 OLI and Sentinel-2 MSI. Sci. Total Environ. 2020, 707, 136092. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhang, F.; Liu, C.; Kung, H.; Johnson, V.C. Soil salinity inversion based on novel spectral index. Environ. Earth Sci. 2021, 80, 501. [Google Scholar] [CrossRef]
- Xu, X.; Chen, Y.; Wang, M.; Wang, S.; Li, K.; Li, Y. Improving Estimates of Soil Salt Content by Using Two-Date Image Spectral Changes in Yinbei, China. Remote Sens. 2021, 13, 4165. [Google Scholar] [CrossRef]
- Liu, P.; Liu, Z.; Hu, Y.; Shi, Z.; Pan, Y.; Wang, L.; Wang, G. Integrating a Hybrid Back Propagation Neural Network and Particle Swarm Optimization for Estimating Soil Heavy Metal Contents Using Hyperspectral Data. Sustainability 2019, 11, 419. [Google Scholar] [CrossRef]
- Liu, X.; Fu, H. PSO-based support vector machine with cuckoo search technique for clinical disease diagnoses. Sci. World J. 2014, 2014, 548483. [Google Scholar] [CrossRef]
- Zhou, J.; Zhu, S.; Qiu, Y.; Armaghani, D.J.; Zhou, A.; Yong, W. Predicting tunnel squeezing using support vector machine optimized by whale optimization algorithm. Acta Geotech. 2022, 17, 1343–1366. [Google Scholar] [CrossRef]
- Fu, C.; Gan, S.; Yuan, X.; Xiong, H.; Tian, A.A. Determination of Soil Salt Content Using a Probability Neural Network Model Based on Particle Swarm Optimization in Areas Affected and Non-Affected by Human Activities. Remote Sens. 2018, 10, 1387. [Google Scholar] [CrossRef]


| Parameter | Value |
|---|---|
| Transfer functions for hidden layer | Logsig |
| Transfer function for output layer | Purelin |
| Training function | Traingdx |
| Neural network creation function | Newff |
| Learning rate | 0.01 |
| Maximum epochs | 1000 |
| Performance goal | 0.00001 |
| Population size | 20 |
| Number of input layer node | The dimension of the input data |
| Number of hidden layer node | 10 |
| Number of output layer node | The dimension of the output data |
| Data | Sample Numbers | Maximum (mS·cm−1) | Minimum (mS·cm−1) | Mean (mS·cm−1) | Median (mS·cm−1) | SD (mS·cm−1) | CV (%) |
|---|---|---|---|---|---|---|---|
| All Datasets | 50 | 57.40 | 0.05 | 4.77 | 1.09 | 10.54 | 221 |
| Calibration Datasets | 33 | 57.40 | 0.06 | 5.57 | 1.10 | 12.02 | 216 |
| Validation Datasets | 17 | 28.60 | 0.05 | 3.23 | 0.40 | 6.87 | 212 |
| Study Area | Data Source | Algorithm | R2 | RMSE | References |
|---|---|---|---|---|---|
| The northwest of Zhongtiao Mountain in Shanxi Province, China | GF-5 spectra | P-PLSR | 0.60 | 80.78 mg/kg | [67] |
| Laboratory hyperspectral data | P-PLSR | 0.77 | 24.43 mg/kg | ||
| Southern Ontario, Canada | A hyperspectral image on a manned helicopter | RF | 0.85 | 12 µg/cm2 | [70] |
| A modified camera-based three-band image | RF | 0.43 | 24 µg/cm2 | ||
| A RedEdge sensor-based five-band image | RF | 0.82 | 13 µg/cm2 | ||
| Southeastern Iowa, the United States | Laboratory hyperspectral data | RF | 0.81 | 0.18% | [68] |
| Airborne hyperspectral data | RF | 0.49 | 0.30% | ||
| Ebinur Lake Wetland National Nature Reserve (ELWNNR), China | Field hyperspectral data | Bootstrap-BPNN | 0.76 | 6.97 g/kg | [71] |
| HJ-B CCD | Bootstrap-BPNN | 0.86 | 5.72 g/kg | ||
| Landsat OLI | Bootstrap-BPNN | 0.65 | 5.42 g/kg |
| Algorithm | Validation Indicators | References |
|---|---|---|
| BPNN | R2 = 0.54, RMSE = 1.45 g/kg | [19] |
| SVR | R2 = 0.53, RMSE = 1.25 g/kg | |
| PSO-BPNN | R2 = 0.78, RMSE = 3.67 g/kg | |
| PSO-SVR | R2 = 0.73, RMSE = 1.47 g/kg | |
| GA-SVM | Precision = 76.09%, Recall = 87.50% | [83] |
| PSO-SVM | Precision = 76.09%, Recall = 87.50% | |
| CS-PSO-SVM | Precision = 81.82%, Recall = 90.00% | |
| BPNN | R2 = 0.39, relative root mean square error (RRMSE) = 34.05% | [82] |
| PSO-BPNN | R2 = 0.76, RRMSE = 12.04% | |
| WOA-SVM | Precision = 1.00, Recall = 1.00 | [84] |
| SVM | Precision = 0.63, Recall = 1.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, X.; Xue, X. Comparing Gaofen-5, Ground, and Huanjing-1A Spectra for the Monitoring of Soil Salinity with the BP Neural Network Improved by Particle Swarm Optimization. Remote Sens. 2022, 14, 5719. https://doi.org/10.3390/rs14225719
Jiang X, Xue X. Comparing Gaofen-5, Ground, and Huanjing-1A Spectra for the Monitoring of Soil Salinity with the BP Neural Network Improved by Particle Swarm Optimization. Remote Sensing. 2022; 14(22):5719. https://doi.org/10.3390/rs14225719
Chicago/Turabian StyleJiang, Xiaofang, and Xian Xue. 2022. "Comparing Gaofen-5, Ground, and Huanjing-1A Spectra for the Monitoring of Soil Salinity with the BP Neural Network Improved by Particle Swarm Optimization" Remote Sensing 14, no. 22: 5719. https://doi.org/10.3390/rs14225719
APA StyleJiang, X., & Xue, X. (2022). Comparing Gaofen-5, Ground, and Huanjing-1A Spectra for the Monitoring of Soil Salinity with the BP Neural Network Improved by Particle Swarm Optimization. Remote Sensing, 14(22), 5719. https://doi.org/10.3390/rs14225719









