Detection of Peanut Leaf Spot Disease Based on Leaf-, Plant-, and Field-Scale Hyperspectral Reflectance
Abstract
1. Introduction
2. Materials and Methods
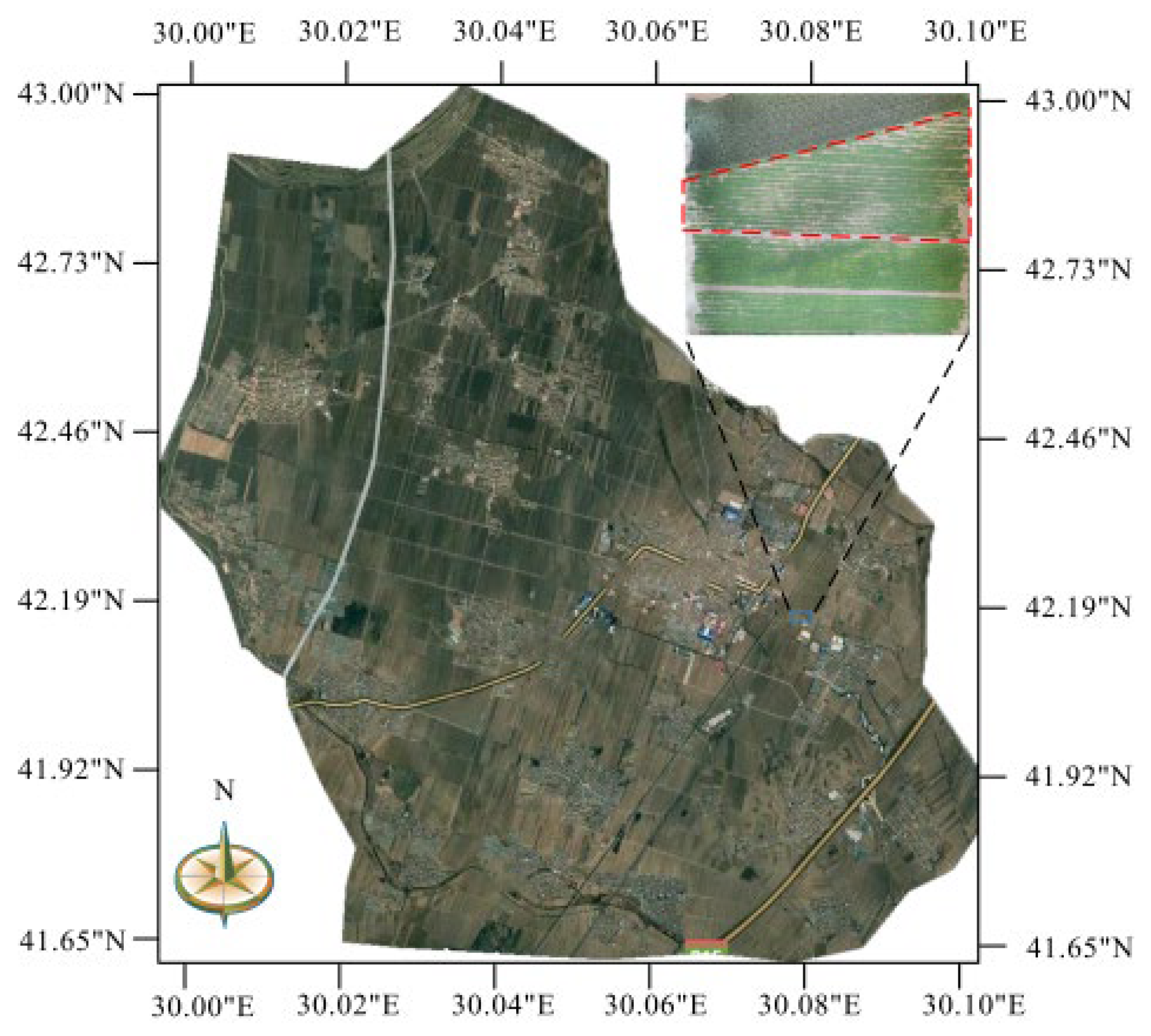
2.1. Overview of the Experiment Site
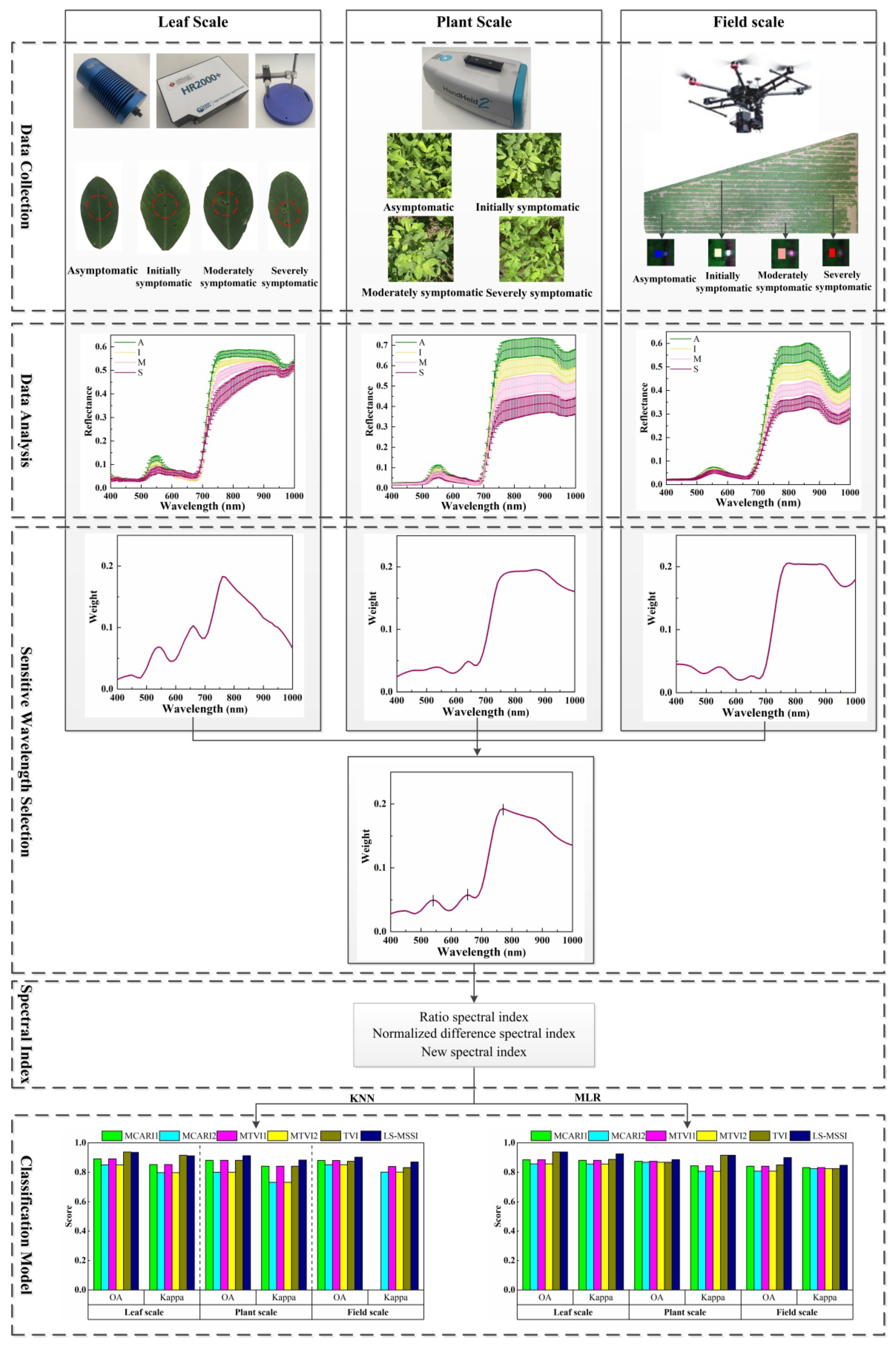
2.2. Data Collection
2.2.1. Disease Severity Assessment
2.2.2. Leaf-Scale Spectral Collection
2.2.3. Plant-Scale Spectral Collection
2.2.4. Field-Scale Spectral Collection
2.3. Sensitive Wavelength Selection
2.4. SI Construction
2.5. Classification Methods
2.5.1. KNN
2.5.2. Multinomial Logistic Regression
2.6. Flow of the Study
3. Evaluation Indicators
3.1. Evaluation Indicators of Wavelength
3.2. Evaluation Indicators of SI
4. Results
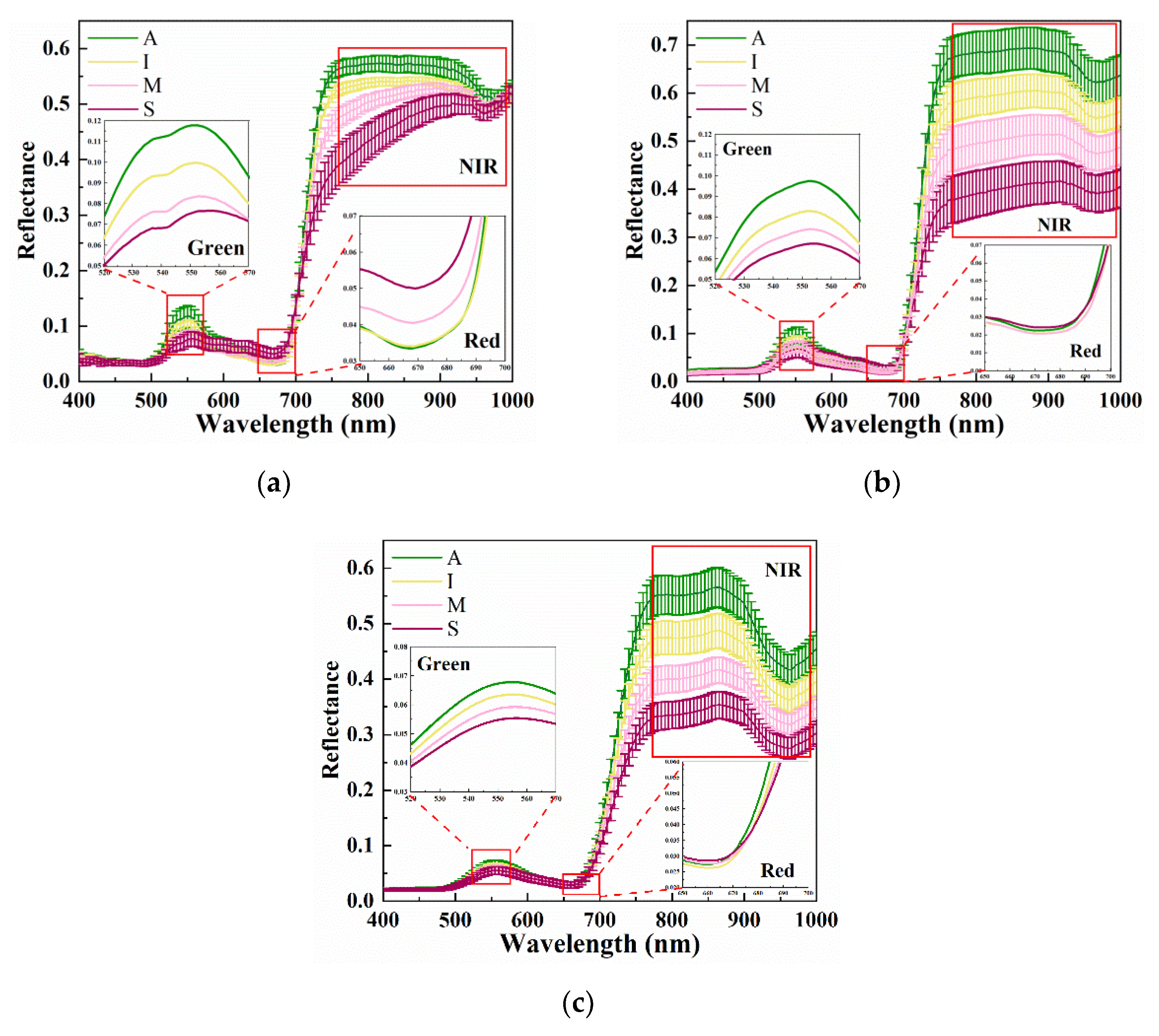
4.1. Multi-Scale Spectral Features
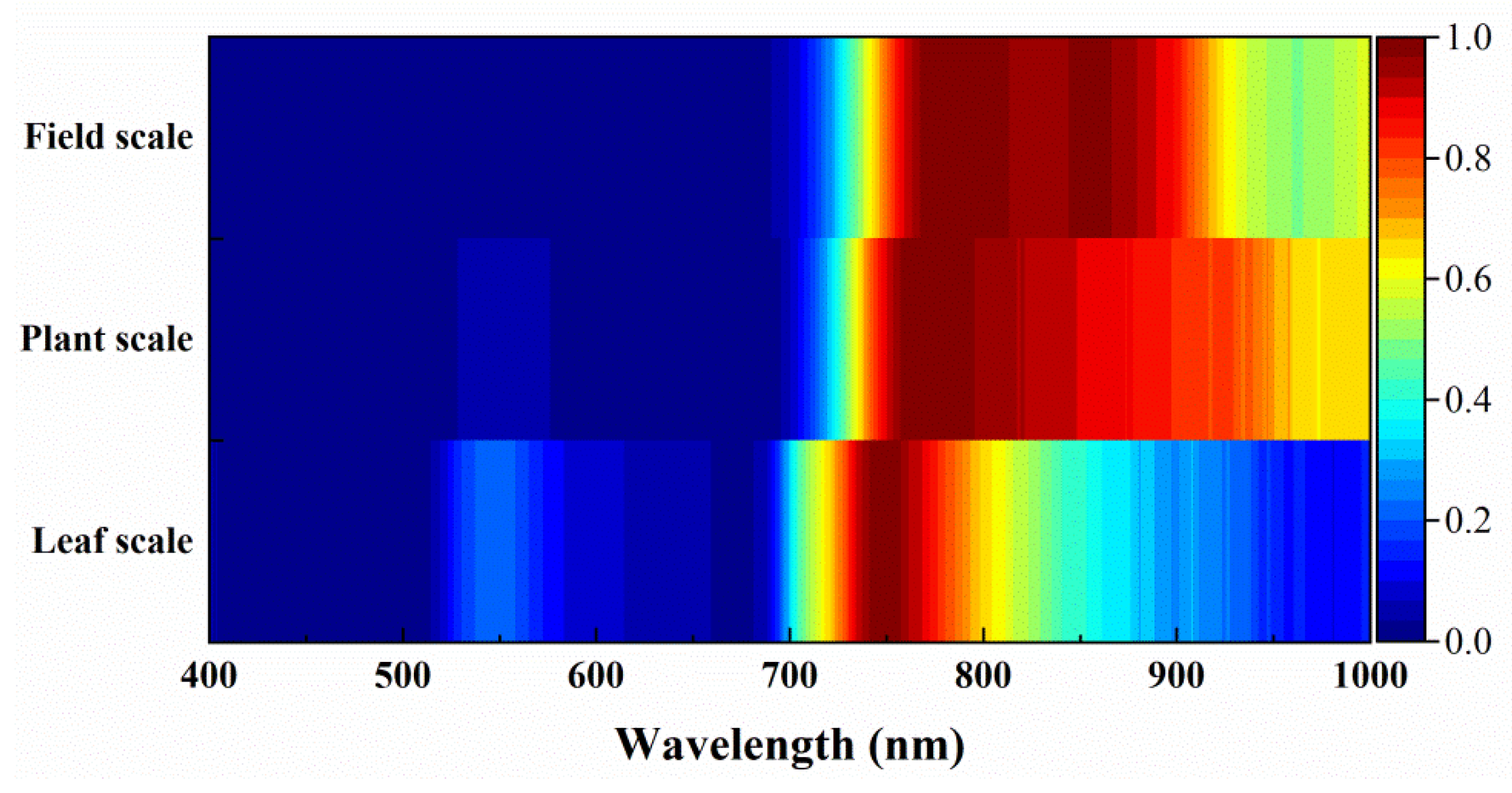
4.2. Multi-Scale Spectral Separability
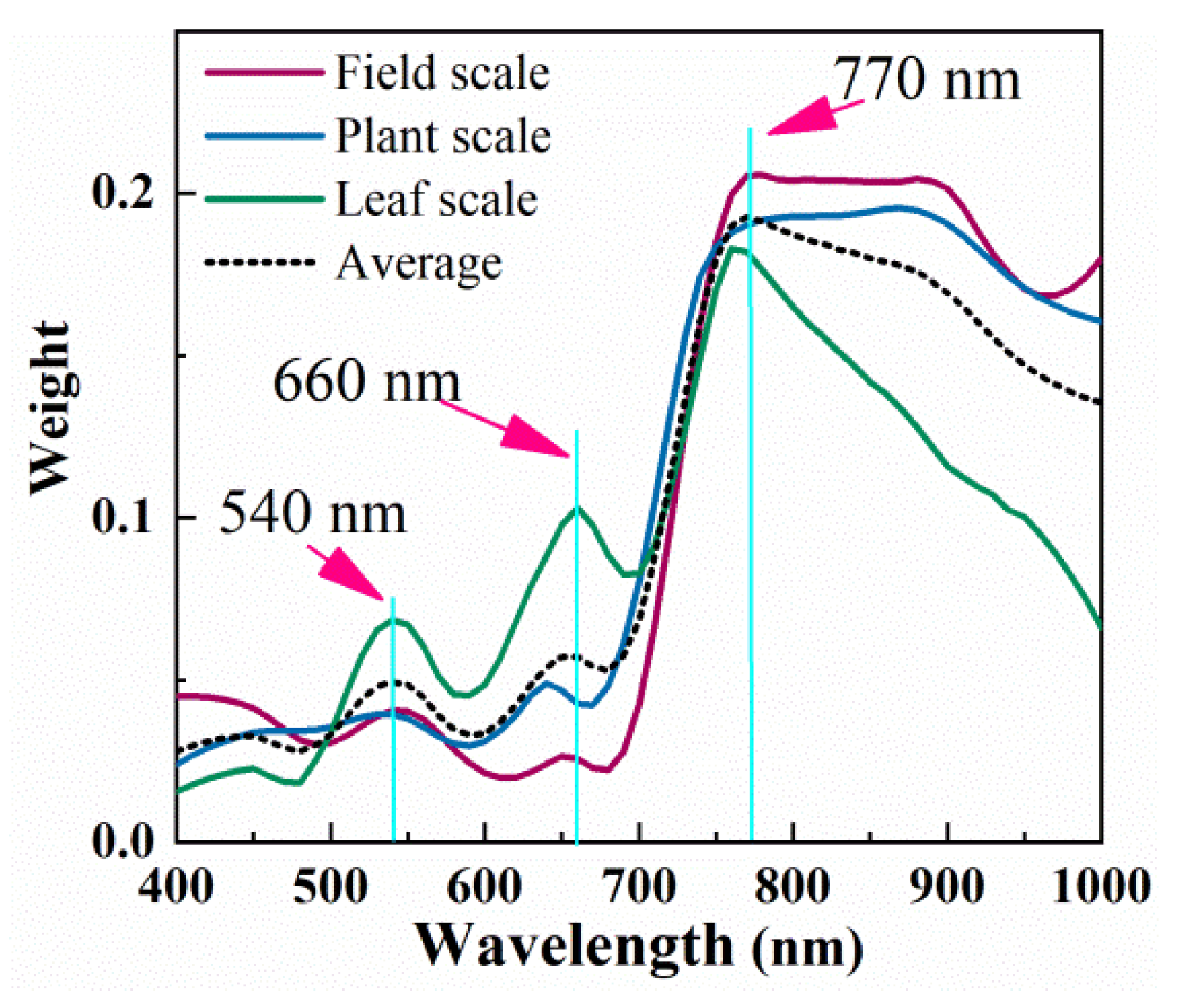
4.3. Multi-Scale Sensitive Wavelength Selection
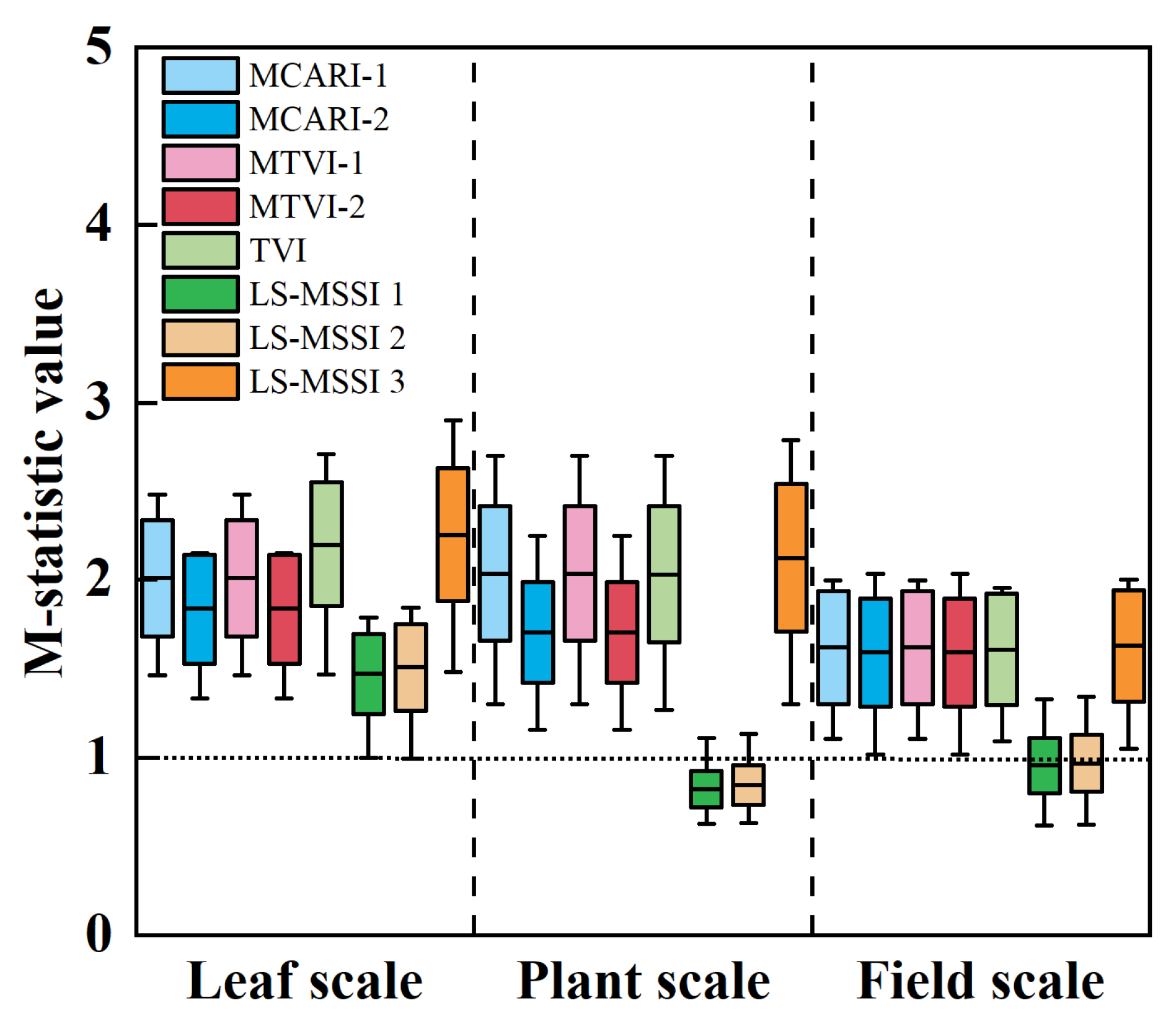
4.4. Comparison of New SI and Traditional SI
4.5. Classification Results
5. Discussion
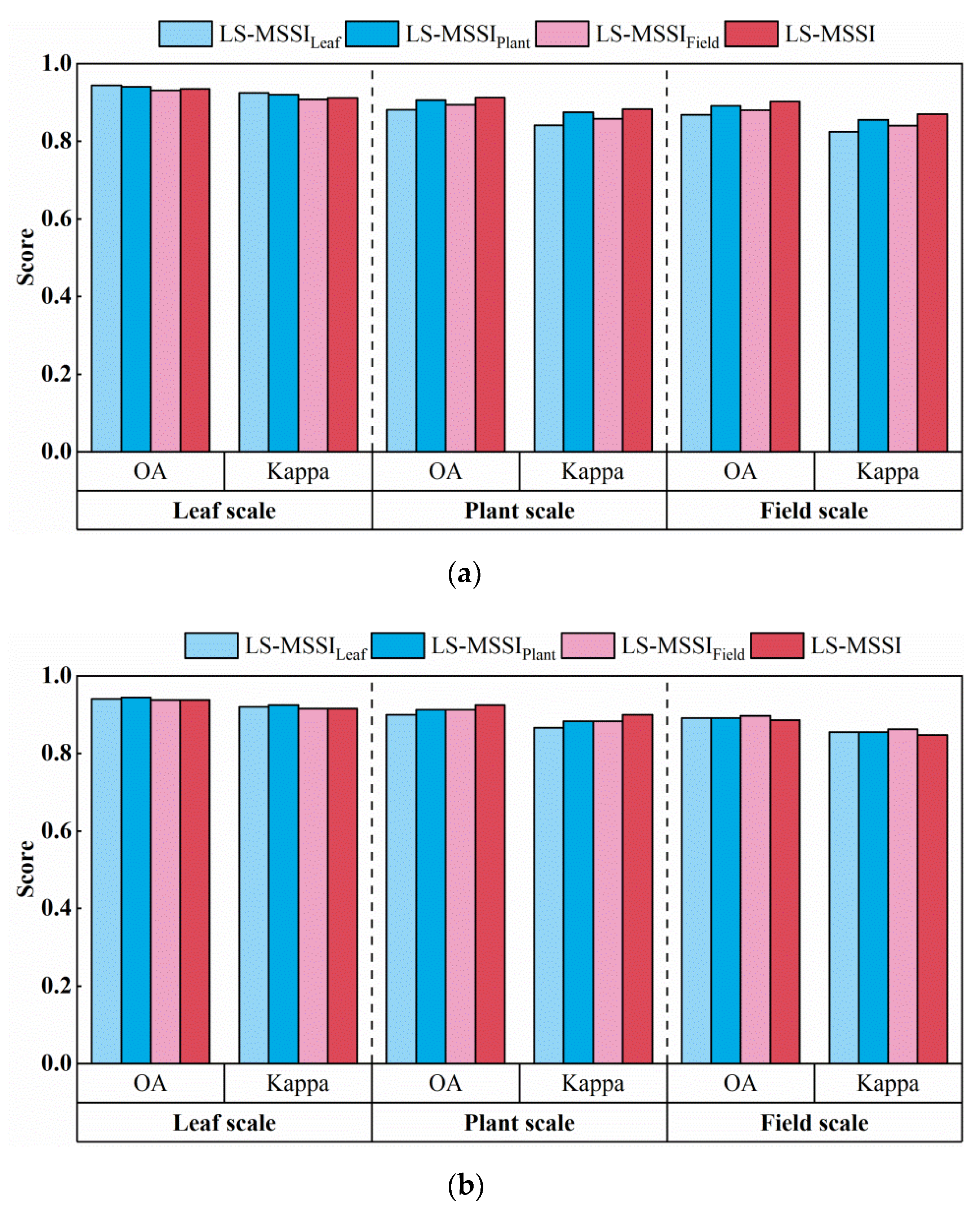
5.1. Analysis of Individual-Scale SI and Multi-Scale SI
5.2. Analysis of the Proposed New SI and the Traditional SI
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Khera, P.; Pandey, M.K.; Wang, H.; Feng, S.; Qiao, L.; Culbreath, A.K.; Kale, S.; Wang, J.; Holbrook, C.C.; Zhuang, W.J.P.O. Mapping quantitative trait loci of resistance to tomato spotted wilt virus and leaf spots in a recombinant inbred line population of peanut (Arachis hypogaea L.) from SunOleic 97R and NC94022. PLoS ONE 2016, 11, e0158452. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Yang, G.; Pan, Y.; Yang, X.; Chen, L.; Zhao, C. A Review of Advanced Technologies and Development for Hyperspectral-Based Plant Disease Detection in the Past Three Decades. Remote Sens. 2020, 12, 3188. [Google Scholar] [CrossRef]
- Gremillion, S.; Culbreath, A.; Gorbet, D.; Mullinix, B.; Pittman, R.; Stevenson, K.; Todd, J.; Condori, M. Response of progeny bred from Bolivian and North American cultivars in integrated management systems for leaf spot of peanut (Arachis hypogaea). Crop Prot. 2011, 30, 698–704. [Google Scholar] [CrossRef]
- Chiteka, Z.A.; Gorbet, D.W.; Shokes, F.M.; Kucharek, T.A.; Knauft, D.A. Components of Resistance to Late Leafspot in Peanut. I. Levels and Variability—Implications for Selection1. Peanut Sci. 1988, 15, 25–30. [Google Scholar] [CrossRef]
- Partel, V.; Charan Kakarla, S.; Ampatzidis, Y. Development and evaluation of a low-cost and smart technology for precision weed management utilizing artificial intelligence. Comput. Electron. Agric. 2019, 157, 339–350. [Google Scholar] [CrossRef]
- Mueller, D.S.; Bradley, C.A.; Grau, C.R.; Gaska, J.M.; Kurle, J.E.; Pedersen, W.L. Application of thiophanate-methyl at different host growth stages for management of sclerotinia stem rot in soybean. Crop Prot. 2004, 23, 983–988. [Google Scholar] [CrossRef]
- Reen, D.J. Enzyme-Linked Immunosorbent Assay (ELISA). In Basic Protein and Peptide Protocols; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 1994; pp. 461–466. [Google Scholar] [CrossRef]
- Rahman, M.T.; Uddin, M.S.; Sultana, R.; Moue, A.; Setu, M. Polymerase Chain Reaction (PCR): A Short Review. Anwer Khan Mod. Med. Coll. J. 2013, 4, 30–36. [Google Scholar] [CrossRef]
- Weiss, M.; Jacob, F.; Duveiller, G. Remote sensing for agricultural applications: A meta-review. Remote Sens. Environ. 2020, 236, 111402. [Google Scholar] [CrossRef]
- Pantazi, X.-E.; Moshou, D.; Bravo, C. Active learning system for weed species recognition based on hyperspectral sensing. Biosyst. Eng. 2016, 146, 193–202. [Google Scholar] [CrossRef]
- Vaglio Laurin, G.; Chen, Q.; Lindsell, J.A.; Coomes, D.A.; Frate, F.D.; Guerriero, L.; Pirotti, F.; Valentini, R. Above ground biomass estimation in an African tropical forest with lidar and hyperspectral data. ISPRS J. Photogramm. Remote Sens. 2014, 89, 49–58. [Google Scholar] [CrossRef]
- Zhao, D.; Feng, S.; Cao, Y.; Yu, F.; Guan, Q.; Li, J.; Zhang, G.; Xu, T. Study on the Classification Method of Rice Leaf Blast Levels Based on Fusion Features and Adaptive-Weight Immune Particle Swarm Optimization Extreme Learning Machine Algorithm. Front. Plant Sci. 2022, 13, 879668. [Google Scholar] [CrossRef]
- Cao, X.; Luo, Y.; Zhou, Y.; Fan, J.; Xu, X.; West, J.S.; Duan, X.; Cheng, D.J.P.O. Detection of powdery mildew in two winter wheat plant densities and prediction of grain yield using canopy hyperspectral reflectance. PLoS ONE 2015, 10, e0121462. [Google Scholar] [CrossRef]
- Lu, J.; Ehsani, R.; Shi, Y.; de Castro, A.I.; Wang, S. Detection of multi-tomato leaf diseases (late blight, target and bacterial spots) in different stages by using a spectral-based sensor. Sci. Rep. 2018, 8, 2793. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, Q.; Lin, F.; Yin, X.; Gu, C.; Qiao, H. Development and Evaluation of a New Spectral Disease Index to Detect Wheat Fusarium Head Blight Using Hyperspectral Imaging. Sensors 2020, 20, 2260. [Google Scholar] [CrossRef]
- Mahlein, A.-K. Plant Disease Detection by Imaging Sensors—Parallels and Specific Demands for Precision Agriculture and Plant Phenotyping. Plant Dis. 2015, 100, 241–251. [Google Scholar] [CrossRef]
- Zheng, Q.; Huang, W.; Cui, X.; Dong, Y.; Shi, Y.; Ma, H.; Liu, L. Identification of Wheat Yellow Rust Using Optimal Three-Band Spectral Indices in Different Growth Stages. Sensors 2019, 19, 35. [Google Scholar] [CrossRef]
- Huang, L.; Ding, W.; Liu, W.; Zhao, J.; Huang, W.; Xu, C.; Zhang, D.; Liang, D. Identification of wheat powdery mildew using in-situ hyperspectral data and linear regression and support vector machines. J. Plant Pathol. 2019, 101, 1035–1045. [Google Scholar] [CrossRef]
- Yuan, L.; Yan, P.; Han, W.; Huang, Y.; Wang, B.; Zhang, J.; Zhang, H.; Bao, Z. Detection of anthracnose in tea plants based on hyperspectral imaging. Comput. Electron. Agric. 2019, 167, 105039. [Google Scholar] [CrossRef]
- Abdulridha, J.; Ampatzidis, Y.; Kakarla, S.C.; Roberts, P. Detection of target spot and bacterial spot diseases in tomato using UAV-based and benchtop-based hyperspectral imaging techniques. Precis. Agric. 2020, 21, 955–978. [Google Scholar] [CrossRef]
- Abdulridha, J.; Ampatzidis, Y.; Roberts, P.; Kakarla, S.C. Detecting powdery mildew disease in squash at different stages using UAV-based hyperspectral imaging and artificial intelligence. Biosyst. Eng. 2020, 197, 135–148. [Google Scholar] [CrossRef]
- Abdulridha, J.; Batuman, O.; Ampatzidis, Y. UAV-Based Remote Sensing Technique to Detect Citrus Canker Disease Utilizing Hyperspectral Imaging and Machine Learning. Remote Sens. 2019, 11, 1373. [Google Scholar] [CrossRef]
- Abdulridha, J.; Ampatzidis, Y.; Qureshi, J.; Roberts, P. Laboratory and UAV-Based Identification and Classification of Tomato Yellow Leaf Curl, Bacterial Spot, and Target Spot Diseases in Tomato Utilizing Hyperspectral Imaging and Machine Learning. Remote Sens. 2020, 12, 2732. [Google Scholar] [CrossRef]
- Thomas, S.; Kuska, M.T.; Bohnenkamp, D.; Brugger, A.; Alisaac, E.; Wahabzada, M.; Behmann, J.; Mahlein, A.-K. Benefits of hyperspectral imaging for plant disease detection and plant protection: A technical perspective. J. Plant Dis. Prot. 2018, 125, 5–20. [Google Scholar] [CrossRef]
- Li, Z.; Li, Z.; Fairbairn, D.; Li, N.; Xu, B.; Feng, H.; Yang, G. Multi-LUTs method for canopy nitrogen density estimation in winter wheat by field and UAV hyperspectral. Comput. Electron. Agric. 2019, 162, 174–182. [Google Scholar] [CrossRef]
- Liu, S.; Li, L.; Gao, W.; Zhang, Y.; Liu, Y.; Wang, S.; Lu, J. Diagnosis of nitrogen status in winter oilseed rape (Brassica napus L.) using in-situ hyperspectral data and unmanned aerial vehicle (UAV) multispectral images. Comput. Electron. Agric. 2018, 151, 185–195. [Google Scholar] [CrossRef]
- Bauriegel, E.; Giebel, A.; Geyer, M.; Schmidt, U.; Herppich, W.B. Early detection of Fusarium infection in wheat using hyper-spectral imaging. Comput. Electron. Agric. 2011, 75, 304–312. [Google Scholar] [CrossRef]
- Chen, T.; Yang, W.; Zhang, H.; Zhu, B.; Zeng, R.; Wang, X.; Wang, S.; Wang, L.; Qi, H.; Lan, Y.; et al. Early detection of bacterial wilt in peanut plants through leaf-level hyperspectral and unmanned aerial vehicle data. Comput. Electron. Agric. 2020, 177, 105708. [Google Scholar] [CrossRef]
- Huang, W.; Lamb, D.W.; Niu, Z.; Zhang, Y.; Liu, L.; Wang, J. Identification of yellow rust in wheat using in-situ spectral reflectance measurements and airborne hyperspectral imaging. Precis. Agric. 2007, 8, 187–197. [Google Scholar] [CrossRef]
- Kira, K.; Rendell, L.A. A Practical Approach to Feature Selection. In Machine Learning Proceedings 1992; Sleeman, D., Edwards, P., Eds.; Morgan Kaufmann: San Francisco, CA, USA, 1992; pp. 249–256. [Google Scholar] [CrossRef]
- Robnik-Šikonja, M.; Kononenko, I. Theoretical and Empirical Analysis of ReliefF and RReliefF. Mach. Learn. 2003, 53, 23–69. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, Y.; Reddy, K.N.; Wang, B. Assessing crop damage from dicamba on non-dicamba-tolerant soybean by hyperspectral imaging through machine learning. Pest Manag. Sci. 2019, 75, 3260–3272. [Google Scholar] [CrossRef]
- Huang, W.; Guan, Q.; Luo, J.; Zhang, J.; Zhao, J.; Liang, D.; Huang, L.; Zhang, D. New Optimized Spectral Indices for Identifying and Monitoring Winter Wheat Diseases. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2516–2524. [Google Scholar] [CrossRef]
- Penuelas, J.; Pinol, J.; Ogaya, R.; Filella, I. Estimation of plant water concentration by the reflectance Water Index WI (R900/R970). Int. J. Remote Sens. 1997, 18, 2869–2875. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Pattey, E.; Zarco-Tejada, P.J.; Strachan, I.B. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Carter, G.A. Ratios of leaf reflectances in narrow wavebands as indicators of plant stress. Int. J. Remote Sens. 1994, 15, 697–703. [Google Scholar] [CrossRef]
- Rouse, J.W., Jr.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring Vegetation Systems in the Great Plains with ERTS. In Proceedings of the 3rd Earth Resources Technology Satellite-1 Symposium, Washington, DC, USA, 10–14 December 1973; Volume 351, p. 309. [Google Scholar]
- Mirik, M.; Michels, G.J., Jr.; Kassymzhanova-Mirik, S.; Elliott, N.C.; Bowling, R. Hyperspectral Spectrometry as a Means to Differentiate Uninfested and Infested Winter Wheat by Greenbug (Hemiptera: Aphididae). J. Econ. Entomol. 2006, 99, 1682–1690. [Google Scholar] [CrossRef]
- Pen¯Uelas, J.; Filella, I.; Lloret, P.; Mun¯Oz, F.; Vilajeliu, M. Reflectance assessment of mite effects on apple trees. Int. J. Remote Sens. 1995, 16, 2727–2733. [Google Scholar] [CrossRef]
- Mirik, M.; Michels, G.J.; Kassymzhanova-Mirik, S.; Elliott, N.C.; Catana, V.; Jones, D.B.; Bowling, R. Using digital image analysis and spectral reflectance data to quantify damage by greenbug (Hemitera: Aphididae) in winter wheat. Comput. Electron. Agric. 2006, 51, 86–98. [Google Scholar] [CrossRef]
- Barnes, J.D.; Balaguer, L.; Manrique, E.; Elvira, S.; Davison, A.W. A reappraisal of the use of DMSO for the extraction and determination of chlorophylls a and b in lichens and higher plants. Environ. Exp. Bot. 1992, 32, 85–100. [Google Scholar] [CrossRef]
- Read, J.J.; Tarpley, L.; McKinion, J.M.; Reddy, K.R. Narrow-Waveband Reflectance Ratios for Remote Estimation of Nitrogen Status in Cotton. J. Environ. Qual. 2002, 31, 1442–1452. [Google Scholar] [CrossRef]
- Zhao, D.; Reddy, K.R.; Kakani, V.G.; Read, J.J.; Koti, S. Selection of Optimum Reflectance Ratios for Estimating Leaf Nitrogen and Chlorophyll Concentrations of Field-Grown Cotton. Agron. J. 2005, 97, 89–98. [Google Scholar] [CrossRef]
- Chappelle, E.W.; Kim, M.S.; McMurtrey, J.E. Ratio analysis of reflectance spectra (RARS): An algorithm for the remote estimation of the concentrations of chlorophyll A, chlorophyll B, and carotenoids in soybean leaves. Remote Sens. Environ. 1992, 39, 239–247. [Google Scholar] [CrossRef]
- Prabhakar, M.; Prasad, Y.G.; Thirupathi, M.; Sreedevi, G.; Dharajothi, B.; Venkateswarlu, B. Use of ground based hyperspectral remote sensing for detection of stress in cotton caused by leafhopper (Hemiptera: Cicadellidae). Comput. Electron. Agric. 2011, 79, 189–198. [Google Scholar] [CrossRef]
- Wang, Z.J.; Wang, J.H.; Liu, L.Y.; Huang, W.J.; Zhao, C.J.; Wang, C.Z. Prediction of grain protein content in winter wheat (Triticum aestivum L.) using plant pigment ratio (PPR). Field Crops Res. 2004, 90, 311–321. [Google Scholar] [CrossRef]
- Gamon, J.A.; Peñuelas, J.; Field, C.B. A narrow-waveband spectral index that tracks diurnal changes in photosynthetic efficiency. Remote Sens. Environ. 1992, 41, 35–44. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Tremblay, N.; Zarco-Tejada, P.J.; Dextraze, L. Integrated narrow-band vegetation indices for prediction of crop chlorophyll content for application to precision agriculture. Remote Sens. Environ. 2002, 81, 416–426. [Google Scholar] [CrossRef]
- Merzlyak, M.N.; Gitelson, A.A.; Chivkunova, O.B.; Rakitin, V.Y.U. Non-destructive optical detection of pigment changes during leaf senescence and fruit ripening. Physiol. Plant. 1999, 106, 135–141. [Google Scholar] [CrossRef]
- Peñuelas, J.; Isla, R.; Filella, I.; Araus, J.L. Visible and Near-Infrared Reflectance Assessment of Salinity Effects on Barley. Crop Sci. 1997, 37, 198–202. [Google Scholar] [CrossRef]
- Broge, N.H.; Leblanc, E. Comparing prediction power and stability of broadband and hyperspectral vegetation indices for estimation of green leaf area index and canopy chlorophyll density. Remote Sens. Environ. 2001, 76, 156–172. [Google Scholar] [CrossRef]
- Babar, M.A.; Reynolds, M.P.; van Ginkel, M.; Klatt, A.R.; Raun, W.R.; Stone, M.L. Spectral Reflectance to Estimate Genetic Variation for In-Season Biomass, Leaf Chlorophyll, and Canopy Temperature in Wheat. Crop Sci. 2006, 46, 1046–1057. [Google Scholar] [CrossRef]
- Peñuelas, J.; Gamon, J.A.; Fredeen, A.L.; Merino, J.; Field, C.B. Reflectance indices associated with physiological changes in nitrogen- and water-limited sunflower leaves. Remote Sens. Environ. 1994, 48, 135–146. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N.; Chivkunova, O.B. Optical Properties and Nondestructive Estimation of Anthocyanin Content in Plant Leaves. Photochem. Photobiol. 2001, 74, 38–45. [Google Scholar] [CrossRef]
- Blackburn, G.A. Spectral indices for estimating photosynthetic pigment concentrations: A test using senescent tree leaves. Int. J. Remote Sens. 1998, 19, 657–675. [Google Scholar] [CrossRef]
- Chen, T.; Zhang, J.; Chen, Y.; Wan, S.; Zhang, L. Detection of peanut leaf spots disease using canopy hyperspectral reflectance. Comput. Electron. Agric. 2019, 156, 677–683. [Google Scholar] [CrossRef]
- Omran, E.-S.E. Early sensing of peanut leaf spot using spectroscopy and thermal imaging. Arch. Agron. Soil Sci. 2017, 63, 883–896. [Google Scholar] [CrossRef]
- Abdullah, M.Z.; Guan, L.C.; Mohd Azemi, B.M.N. Stepwise Discriminant Analysis for Colour Grading of Oil Palm Using Machine Vision System. Food Bioprod. Process. 2001, 79, 223–231. [Google Scholar] [CrossRef]
- Böhning, D. Multinomial logistic regression algorithm. Ann. Inst. Stat. Math. 1992, 44, 197–200. [Google Scholar] [CrossRef]
- Fukunada, K. Introduction to Statistical Pattern Recognition; Computer Science and Scientific Computing; Academic Press: Cambridge, MA, USA, 1990. [Google Scholar]
- Kaufman, Y.J.; Remer, L.A. Detection of forests using mid-IR reflectance: An application for aerosol studies. IEEE Trans. Geosci. Remote Sens. 1994, 32, 672–683. [Google Scholar] [CrossRef]
- Smith, A.M.S.; Drake, N.A.; Wooster, M.J.; Hudak, A.T.; Holden, Z.A.; Gibbons, C.J. Production of Landsat ETM+ reference imagery of burned areas within Southern African savannahs: Comparison of methods and application to MODIS. Int. J. Remote Sens. 2007, 28, 2753–2775. [Google Scholar] [CrossRef]
- De Castro, A.I.; Ehsani, R.; Ploetz, R.; Crane, J.H.; Abdulridha, J. Optimum spectral and geometric parameters for early detection of laurel wilt disease in avocado. Remote Sens. Environ. 2015, 171, 33–44. [Google Scholar] [CrossRef]
- Ashourloo, D.; Mobasheri, M.R.; Huete, A. Developing Two Spectral Disease Indices for Detection of Wheat Leaf Rust (Pucciniatriticina). Remote Sens. 2014, 6, 4723–4740. [Google Scholar] [CrossRef]
- Mahlein, A.K.; Rumpf, T.; Welke, P.; Dehne, H.W.; Plümer, L.; Steiner, U.; Oerke, E.C. Development of spectral indices for detecting and identifying plant diseases. Remote Sens. Environ. 2013, 128, 21–30. [Google Scholar] [CrossRef]
- Devadas, R.; Lamb, D.W.; Simpfendorfer, S.; Backhouse, D. Evaluating ten spectral vegetation indices for identifying rust infection in individual wheat leaves. Precis. Agric. 2009, 10, 459–470. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Stark, R.; Rundquist, D. Novel algorithms for remote estimation of vegetation fraction. Remote Sens. Environ. 2002, 80, 76–87. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Baret, F. PROSPECT: A model of leaf optical properties spectra. Remote Sens. Environ. 1990, 34, 75–91. [Google Scholar] [CrossRef]
- Malthus, T.J.; Madeira, A.C. High resolution spectroradiometry: Spectral reflectance of field bean leaves infected by Botrytis fabae. Remote Sens. Environ. 1993, 45, 107–116. [Google Scholar] [CrossRef]
- Franke, J.; Menz, G. Multi-temporal wheat disease detection by multi-spectral remote sensing. Precis. Agric. 2007, 8, 161–172. [Google Scholar] [CrossRef]
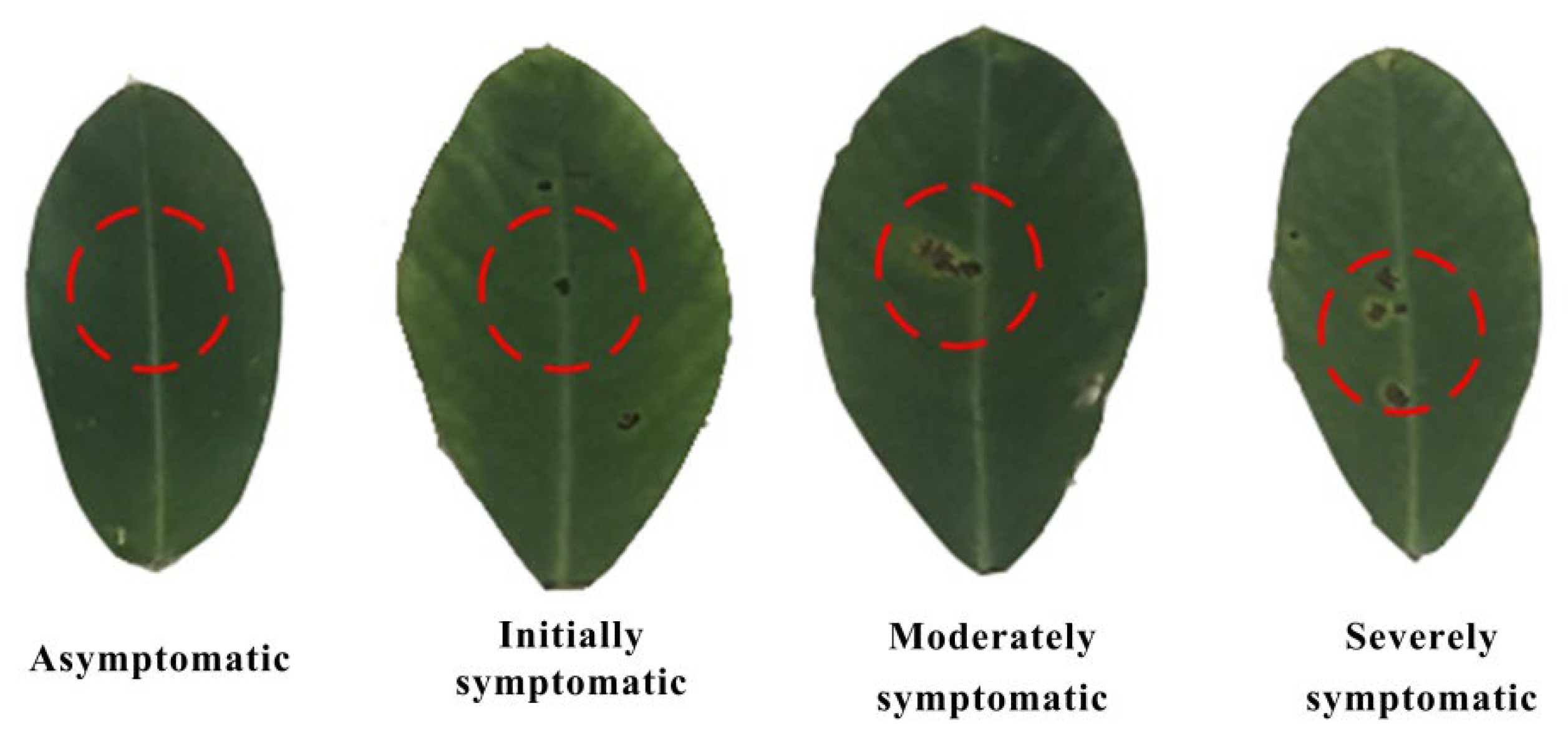
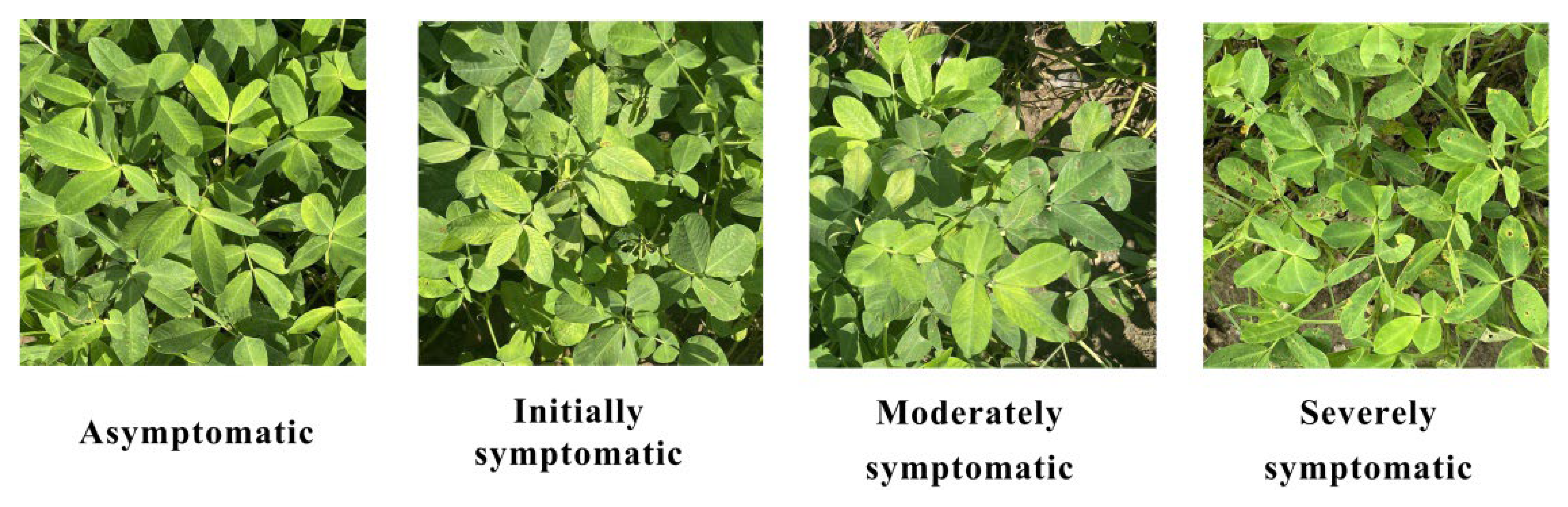

| Level | Disease Severity | Area Ratio |
|---|---|---|
| A | Asymptomatic | |
| I | Initially symptomatic | |
| M | Moderately symptomatic | |
| S | Severely symptomatic |
| Disease Severity | Leaf-Scale Samples | Plant-Scale Samples | Field-Scale Samples |
|---|---|---|---|
| Asymptomatic | 261 | 136 | 148 |
| Initially symptomatic | 338 | 148 | 157 |
| Moderately symptomatic | 242 | 138 | 138 |
| Severely symptomatic | 230 | 112 | 143 |
| Total | 1071 | 534 | 586 |
| Method | Calculation Formula | Reference |
|---|---|---|
| Ratio spectral index (RSI) | [32] | |
| Normalized difference spectral index (NDSI) | [32] | |
| New spectral index (NSI) | [33] |
| NO. | SI | Calculation Formula | Reference |
|---|---|---|---|
| 1 | Simple ratio (SR) | [36] | |
| 2 | Normalized difference vegetation index (NDVI) | [37] | |
| 3 | Aphid index (AI) | [38] | |
| 4 | Structural independent pigment index (SIPI) | [39] | |
| 5 | Damage-sensitive spectral index-1 (DSSI-1) | [40] | |
| 6 | Damage-sensitive spectral index-2 (DSSI-2) | [40] | |
| 7 | Chlorophyll index (CI) | [41] | |
| 8 | Chl stress index-1 (Chl SI-1) | [42] | |
| 9 | Chl stress index-2 (Chl SI-2) | [43] | |
| 10 | Chl stress index-3 (Chl SI-3) | [43] | |
| 11 | Ratio analysis of reflectance spectral chlorophyll a (RARSa) | [44] | |
| 12 | Ratio analysis of reflectance spectral chlorophyll b (RARSb) | [44] | |
| 13 | Ratio analysis of reflectance spectra chlorophyll c (RARSc) | [44] | |
| 14 | Leaf hopper index (LHI) | [45] | |
| 15 | Nitrogen stress index-1 (NSI-1) | [42] | |
| 16 | Nitrogen stress index-2 (NSI-2) | [43] | |
| 17 | Plant pigment ratio (PPR) | [46] | |
| 18 | Physiological reflectance index (PRI) | [47] | |
| 19 | Transformed chlorophyll absorption and reflectance index (TCARI) | [48] | |
| 20 | Modified chlorophyll absorption in reflectance index-1 (MCARI-1) | [35] | |
| 21 | Modified chlorophyll absorption in reflectance index-2 (MCARI-2) | [35] | |
| 22 | Modified triangular vegetation index-1 (MTVI-1) | [35] | |
| 23 | Modified triangular vegetation index-2 (MTVI-2) | [35] | |
| 24 | Plant senescence reflectance index (PSRI) | [49] | |
| 25 | Water index (WI) | [50] | |
| 26 | Triangle vegetation index (TVI) | [51] | |
| 27 | Normalized water index-1 (NWI-1) | [52] | |
| 28 | Normalized water index-2 (NWI-2) | [52] | |
| 29 | Normalized pigment chlorophyll ratio index (NPCI) | [53] | |
| 30 | Anthocyanin reflectance index (ARI) | [54] | |
| 31 | Pigment-specific simple ratio (PSSRa) | [55] | |
| 32 | Pigment-specific simple ratio (PSSRb) | [55] | |
| 33 | Leaf spot index (LSI) | [56] | |
| 34 | Early leaf spot index (ELSI) | [57] | |
| 35 | Late leaf spot index (LLSI) | [57] | |
| 36 | Leaf spot multi-scale spectral index 1 (LS-MSSI 1) | This study | |
| 37 | Leaf spot multi-scale spectral index 2 (LS-MSSI 2) | This study | |
| 38 | Leaf spot multi-scale spectral index 3 (LS-MSSI 3) | This study |
| Actual Class | Predicted Class | |
|---|---|---|
| Positive | Negative | |
| Positive | TP | FN |
| Negative | FP | TN |
| Scale | Wavelengths (nm) | KNN | MLR | ||
|---|---|---|---|---|---|
| OA (%) | Kappa (%) | OA (%) | Kappa (%) | ||
| Leaf | 540, 660, 760 | 89.72 | 86.06 | 93.83 | 90.32 |
| 530, 640, 770 | 91.59 | 88.62 | 93.46 | 91.21 | |
| 540, 650, 770 | 89.10 | 85.21 | 92.21 | 89.50 | |
| 540, 660, 770 | 89.41 | 85.62 | 92.21 | 89.48 | |
| All | 98.44 | 97.91 | 95.95 | 94.56 | |
| Plant | 540, 660, 760 | 84.38 | 78.96 | 90.00 | 86.58 |
| 530, 640, 770 | 86.88 | 82.35 | 90.62 | 87.43 | |
| 540, 650, 770 | 83.13 | 77.25 | 92.50 | 89.94 | |
| 540, 660, 770 | 85.62 | 80.64 | 91.87 | 89.10 | |
| All | 91.87 | 89.13 | 90.00 | 86.65 | |
| Field | 540, 660, 760 | 87.43 | 83.24 | 88.57 | 84.75 |
| 530, 640, 770 | 88.00 | 84.02 | 88.00 | 83.97 | |
| 540, 650, 770 | 87.43 | 83.22 | 88.57 | 84.74 | |
| 540, 660, 770 | 87.43 | 83.22 | 89.14 | 85.52 | |
| All | 89.71 | 86.27 | 89.14 | 85.50 | |
| NO. | SI | Leaf Scale | Plant Scale | Field Scale |
|---|---|---|---|---|
| 1 | SR | 0.57 | 0.47 | −0.66 |
| 2 | NDVI | −0.83 ** | −0.70 | −0.79 |
| 3 | AI | 0.91 ** | 0.77 | 0.55 |
| 4 | SIPI | −0.76 | −0.64 | −0.80 ** |
| 5 | DSSI-1 | −0.19 | 0.52 | 0.68 |
| 6 | DSSI-2 | −0.03 | −0.56 | −0.04 |
| 7 | CI | −0.26 | −0.11 | 0.10 |
| 8 | Chl SI-1 | −0.59 | −0.45 | 0.65 |
| 9 | Chl SI-2 | −0.02 | 0.44 | 0.38 |
| 10 | Chl SI-3 | −0.61 | 0.22 | 0.62 |
| 11 | RARSa | 0.59 | 0.65 | 0.83 ** |
| 12 | RARSb | −0.10 | 0.49 | 0.71 |
| 13 | RARSc | −0.62 | −0.62 | −0.82 ** |
| 14 | LHI | −0.47 | −0.51 | −0.72 |
| 15 | NSI-1 | −0.01 | 0.17 | 0.82 ** |
| 16 | NSI-2 | −0.23 | −0.02 | −0.47 |
| 17 | PPR | −0.55 | −0.50 | −0.54 |
| 18 | PRI | 0.76 | −0.31 | −0.19 |
| 19 | TCARI | −0.65 | −0.76 | −0.53 |
| 20 | MCARI-1 | −0.93 ** | −0.94 ** | −0.92 ** |
| 21 | MCARI-2 | −0.93 ** | −0.94 ** | −0.91 ** |
| 22 | MTVI-1 | −0.93 ** | −0.94 ** | −0.92 ** |
| 23 | MTVI-2 | −0.93 ** | −0.94 ** | −0.91 ** |
| 24 | PSRI | 0.77 | 0.02 | 0.11 |
| 25 | WI | 0.88 ** | 0.51 | 0.58 |
| 26 | TVI | −0.95 ** | −0.94 ** | −0.92 ** |
| 27 | NWI-1 | 0.88 ** | 0.51 | 0.58 |
| 28 | NWI-2 | 0.93 ** | 0.68 | 0.65 |
| 29 | NPCI | 0.78 | 0.75 | 0.05 |
| 30 | ARI | 0.92 ** | 0.82 ** | 0.27 |
| 31 | PSSRa | −0.83 ** | −0.70 | −0.76 |
| 32 | PSSRb | −0.70 | −0.59 | −0.80 ** |
| 33 | LSI | 0.92 ** | 0.70 | 0.59 |
| 34 | ELSI | 0.36 | 0.20 | 0.07 |
| 35 | LLSI | 0.87 ** | 0.80 ** | −0.65 |
| 36 | LS-MSSI 1 | −0.91 ** | −0.83 ** | −0.81 ** |
| 37 | LS-MSSI 2 | −0.91 ** | −0.83 ** | −0.81 ** |
| 38 | LS-MSSI 3 | −0.95 ** | −0.94 ** | −0.92 ** |
| Method | VIs | Leaf Scale | Plant Scale | Field Scale | |||
|---|---|---|---|---|---|---|---|
| OA (%) | Kappa (%) | OA (%) | Kappa (%) | OA (%) | Kappa (%) | ||
| KNN | MCARI-1 | 89.10 | 85.23 | 88.13 | 84.10 | 88.00 | 83.97 |
| MCARI-2 | 85.05 | 79.78 | 80.00 | 73.18 | 85.14 | 80.17 | |
| MTVI-1 | 89.10 | 85.23 | 88.13 | 84.10 | 88.00 | 83.97 | |
| MTVI-2 | 85.05 | 79.78 | 80.00 | 73.18 | 85.14 | 80.17 | |
| TVI | 93.77 | 91.59 | 88.13 | 84.10 | 87.43 | 83.23 | |
| LS-MSSI | 93.46 | 91.15 | 91.25 | 88.29 | 90.29 | 87.04 | |
| MLR | MCARI-1 | 88.47 | 84.38 | 88.13 | 84.11 | 87.43 | 83.22 |
| MCARI-2 | 85.67 | 80.65 | 85.63 | 80.75 | 86.86 | 82.45 | |
| MTVI-1 | 88.47 | 84.38 | 88.13 | 84.11 | 87.43 | 83.22 | |
| MTVI-2 | 85.67 | 80.65 | 85.63 | 80.75 | 86.86 | 82.45 | |
| TVI | 93.77 | 91.59 | 88.75 | 84.95 | 86.86 | 82.47 | |
| LS-MSSI | 93.77 | 91.59 | 92.50 | 89.97 | 88.57 | 84.75 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guan, Q.; Song, K.; Feng, S.; Yu, F.; Xu, T. Detection of Peanut Leaf Spot Disease Based on Leaf-, Plant-, and Field-Scale Hyperspectral Reflectance. Remote Sens. 2022, 14, 4988. https://doi.org/10.3390/rs14194988
Guan Q, Song K, Feng S, Yu F, Xu T. Detection of Peanut Leaf Spot Disease Based on Leaf-, Plant-, and Field-Scale Hyperspectral Reflectance. Remote Sensing. 2022; 14(19):4988. https://doi.org/10.3390/rs14194988
Chicago/Turabian StyleGuan, Qiang, Kai Song, Shuai Feng, Fenghua Yu, and Tongyu Xu. 2022. "Detection of Peanut Leaf Spot Disease Based on Leaf-, Plant-, and Field-Scale Hyperspectral Reflectance" Remote Sensing 14, no. 19: 4988. https://doi.org/10.3390/rs14194988
APA StyleGuan, Q., Song, K., Feng, S., Yu, F., & Xu, T. (2022). Detection of Peanut Leaf Spot Disease Based on Leaf-, Plant-, and Field-Scale Hyperspectral Reflectance. Remote Sensing, 14(19), 4988. https://doi.org/10.3390/rs14194988








