Application of Multi-Source Data for Mapping Plantation Based on Random Forest Algorithm in North China
Abstract
1. Introduction
2. Materials and Methods
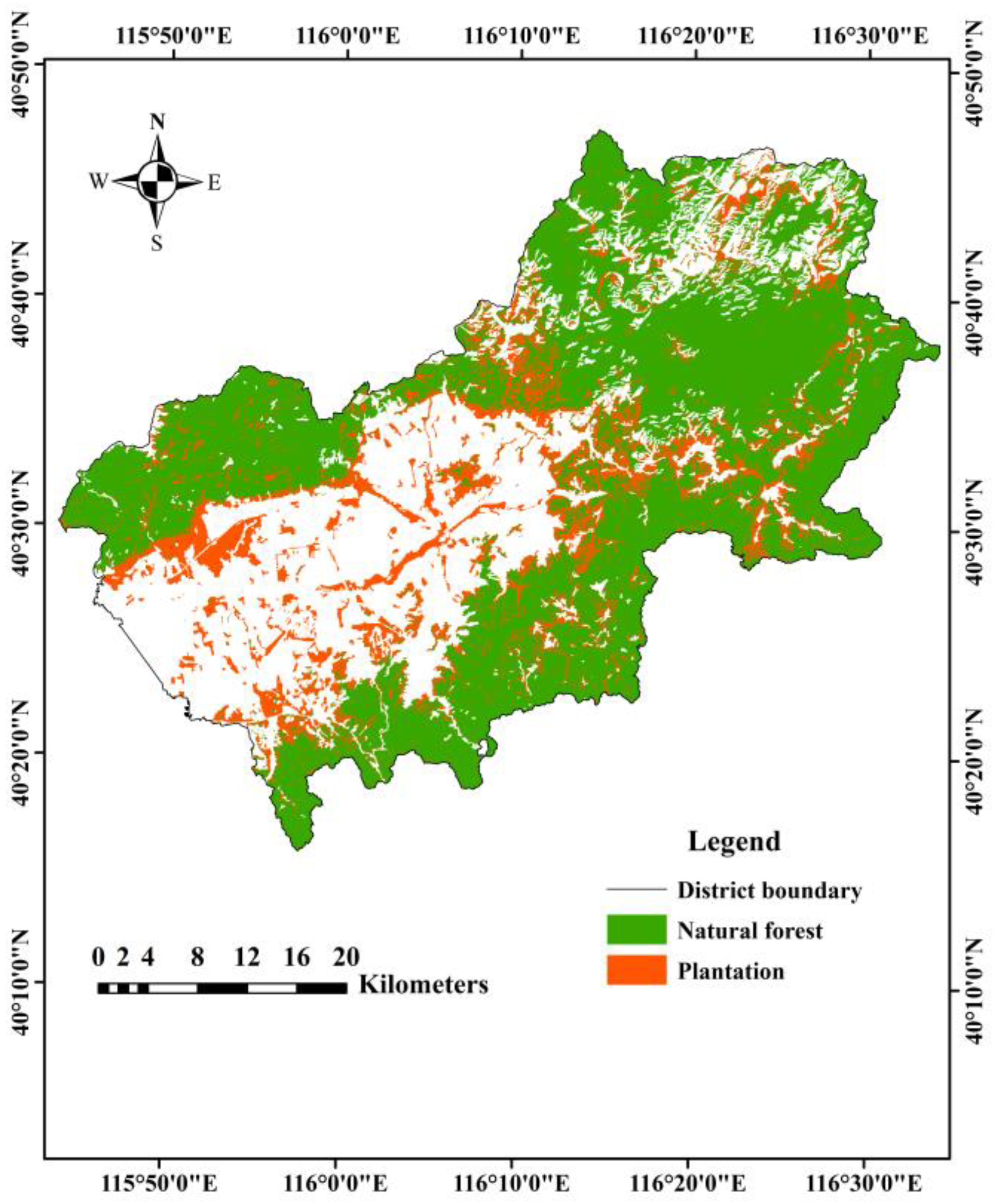
2.1. Study Area
2.2. Data Acquisition and Pre-Processing
2.3. Feature Extraction and Selection
2.3.1. Feature Extraction
2.3.2. Feature Selection
2.4. RF Classification
3. Results
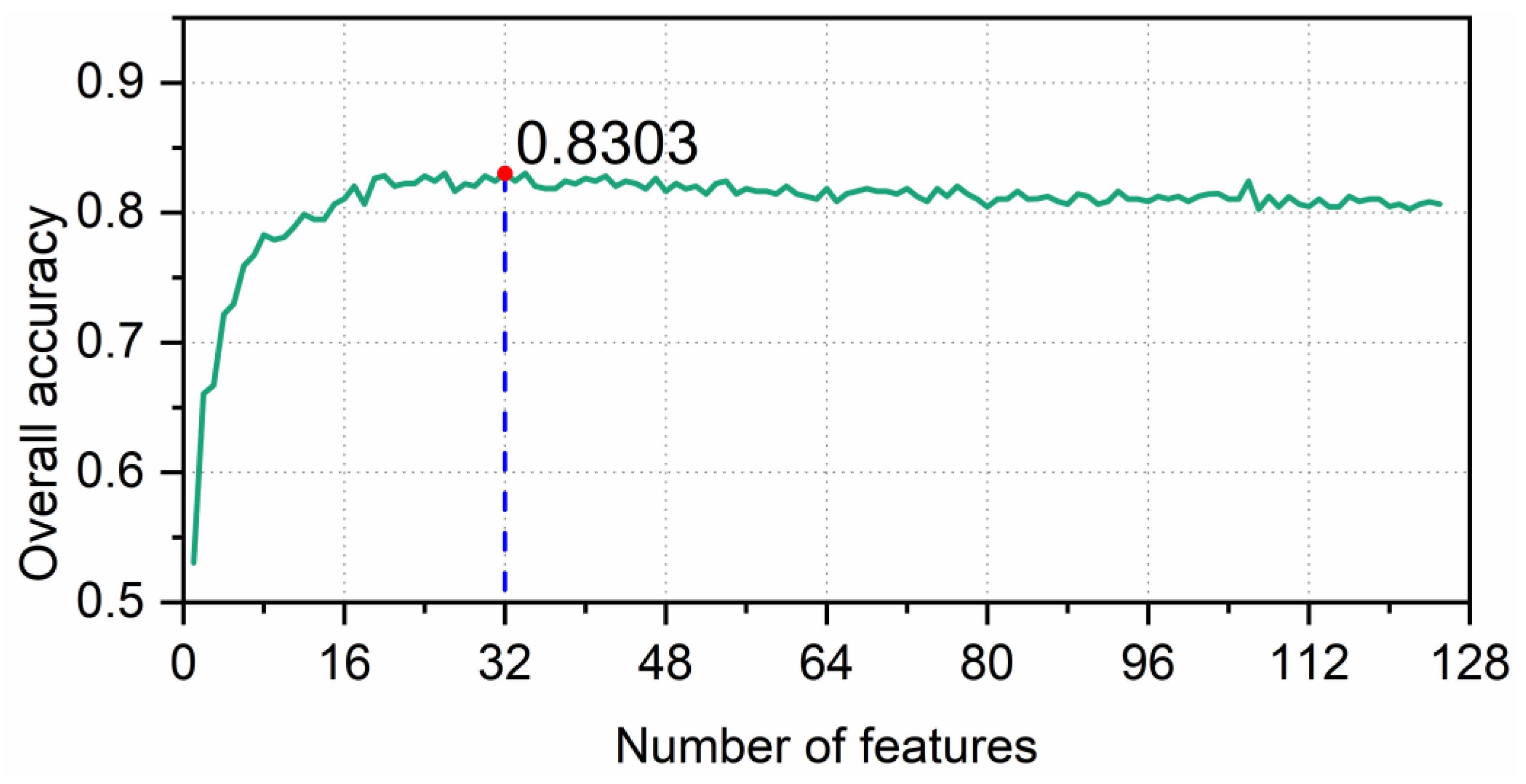
3.1. Feature Analysis and Selection
3.1.1. Feature Analysis
3.1.2. Feature Selection
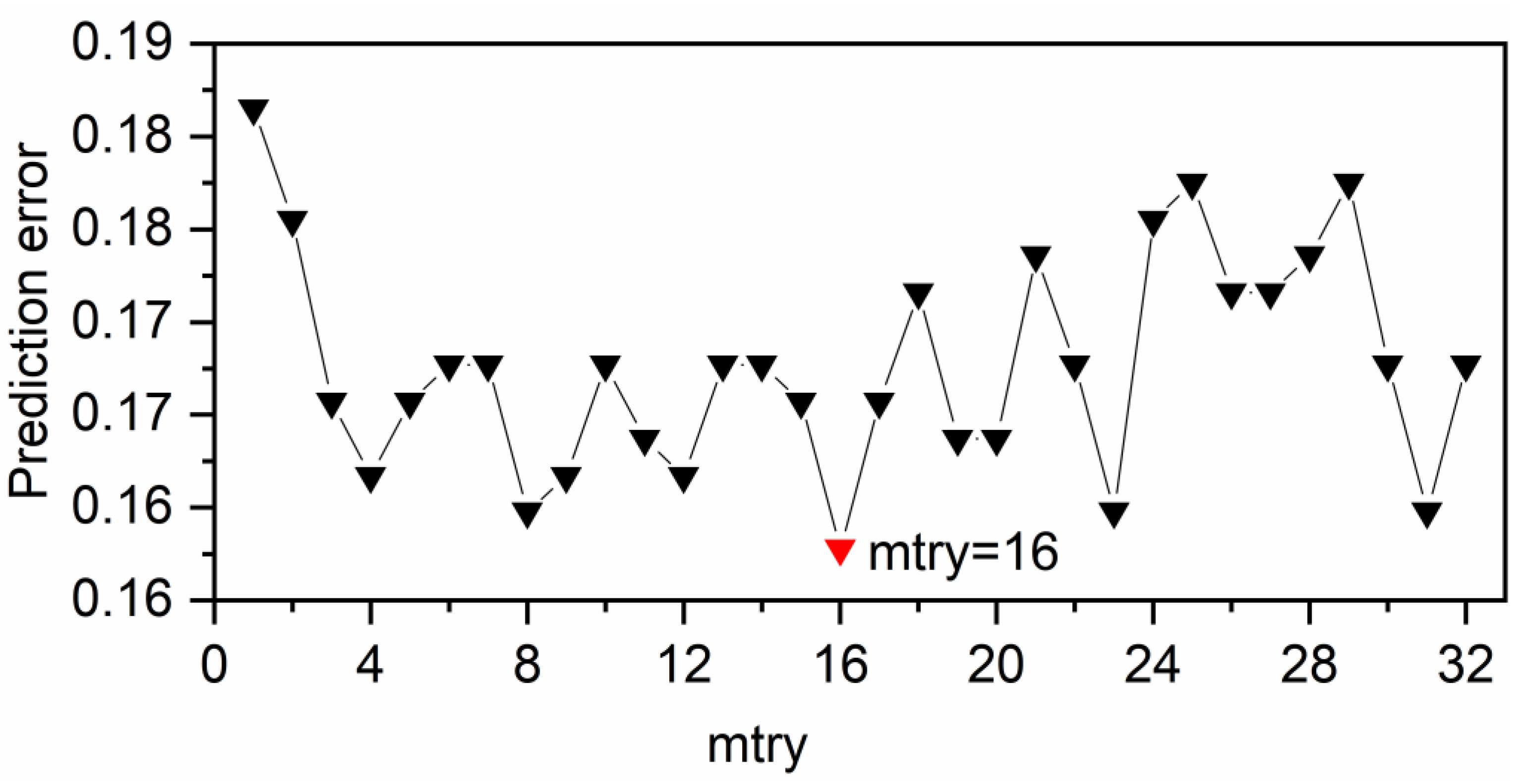
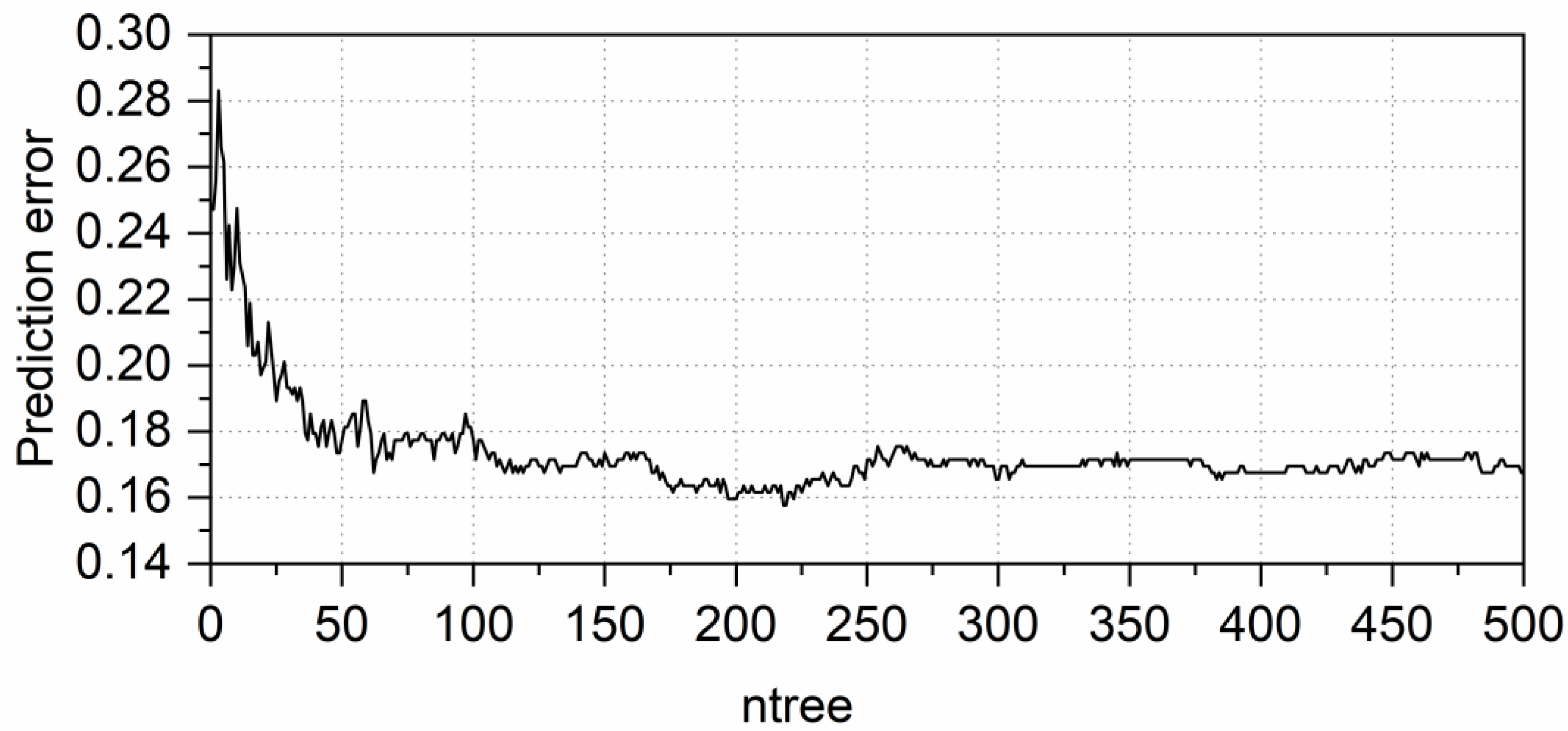
3.2. RF Parameter Optimization
3.3. Accuracy Assessment
3.4. Forest Type Mapping
4. Discussion
4.1. Feature Selection for RF Model
4.2. Potential Application for Multi-Source Data
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- National Forestry and Grassland Administration. China Forestry Statistical Yearbook 2015; Zhang, J., Yan, Z., Liu, J., Eds.; China Forestry Publishing House: Beijing, China, 2016. [Google Scholar]
- National Forestry and Grassland Administration. Report on the Results of the Eighth National Forest Resources Inventory; China Forestry Publishing House: Beijing, China, 2013. [Google Scholar]
- Persson, M.; Lindberg, E.; Reese, H. Tree species classification with multi-temporal Sentinel-2 data. Remote Sens. 2018, 10, 1794. [Google Scholar] [CrossRef]
- Xiao, X.; Biradar, C.M.; Czarnecki, C.; Alabi, T.; Keller, M. A simple algorithm for large-scale mapping of evergreen forests in tropical America, Africa and Asia. Remote Sens. 2009, 1, 355–374. [Google Scholar] [CrossRef]
- Cheng, K.; Wang, J. Forest type classification based on integrated spectral-spatial-temporal features and random forest algorithm-A case study in the Qinling Mountains. Forests 2019, 10, 559. [Google Scholar] [CrossRef]
- Fagan, M.E.; DeFries, R.S.; Sesnie, S.E.; Arroyo-Mora, J.P.; Soto, C.; Singh, A.; Townsend, P.A.; Chazdon, R.L. Mapping species composition of forests and tree plantations in northeastern Costa Rica with an integration of hyperspectral and multitemporal landsat imagery. Remote Sens. 2015, 7, 5660–5696. [Google Scholar] [CrossRef]
- Yu, Y.; Li, M.; Fu, Y. Forest type identification by random forest classification combined with SPOT and multitemporal SAR data. J. For. Res. 2018, 29, 1407–1414. [Google Scholar] [CrossRef]
- Chong, R.; Hongbo, J.; Huaiqing, Z.; Jianwen, H.; Yingxuan, Z. Multi-source data for forest land type precise classication. Sci. Silvae Sin. 2016, 52, 54–65. [Google Scholar]
- Hayes, D.J.; Cohen, W.B.; Sader, S.A.; Irwin, D.E. Estimating proportional change in forest cover as a continuous variable from multi-year MODIS data. Remote Sens. Environ. 2008, 112, 735–749. [Google Scholar] [CrossRef]
- Franklin, S.E.; Maudie, A.J.; Lavlgne, M.B. Using spatial co-occurrence texture to increase forest structure and species composition classification accuracy. Photogramm. Eng. Remote Sens. 2001, 67, 849–855. [Google Scholar]
- Kou, W.; Xiao, X.; Dong, J.; Gan, S.; Zhai, D.; Zhang, G.; Qin, Y.; Li, L. Mapping deciduous rubber plantation areas and stand ages with PALSAR and landsat images. Remote Sens. 2015, 7, 1048–1073. [Google Scholar] [CrossRef]
- Dong, J.; Xiao, X.; Chen, B.; Torbick, N.; Jin, C.; Zhang, G.; Biradar, C. Mapping deciduous rubber plantations through integration of PALSAR and multi-temporal Landsat imagery. Remote Sens. Environ. 2013, 134, 392–402. [Google Scholar] [CrossRef]
- Torbick, N.; Ledoux, L.; Salas, W.; Zhao, M. Regional mapping of plantation extent using multisensor imagery. Remote Sens. 2016, 8, 236. [Google Scholar] [CrossRef]
- Akar, Ö.; Güngör, O. Integrating multiple texture methods and NDVI to the Random Forest classification algorithm to detect tea and hazelnut plantation areas in northeast Turkey. Int. J. Remote Sens. 2015, 36, 442–464. [Google Scholar] [CrossRef]
- Wilson, E.H.; Sader, S.A. Detection of forest harvest type using multiple dates of Landsat TM imagery. Remote Sens. Environ. 2002, 80, 385–396. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Lu, D. Integration of vegetation inventory data and Landsat TM image for vegetation classification in the western Brazilian Amazon. For. Ecol. Manag. 2005, 213, 369–383. [Google Scholar] [CrossRef]
- Ferreira, M.P.; Zortea, M.; Zanotta, D.C.; Shimabukuro, Y.E.; de Souza, C.R.; Filho. Mapping tree species in tropical seasonal semi-deciduous forests with hyperspectral and multispectral data. Remote Sens. Environ. 2016, 179, 66–78. [Google Scholar] [CrossRef]
- Liu, Y.; Gong, W.; Hu, X.; Gong, J. Forest type identification with random forest using Sentinel-1A, Sentinel-2A, multi-temporal Landsat-8 and DEM data. Remote Sens. 2018, 10, 946. [Google Scholar] [CrossRef]
- Gao, T.; Zhu, J.; Zheng, X.; Shang, G.; Huang, L.; Wu, S. Mapping spatial distribution of larch plantations from multi-seasonal landsat-8 OLI imagery and multi-scale textures using random forests. Remote Sens. 2015, 7, 1702–1720. [Google Scholar] [CrossRef]
- Han, P.; Chen, J.; Han, Y.; Yi, L.; Zhang, Y.; Jiang, X. Monitoring rubber plantation distribution on Hainan Island using Landsat OLI imagery. Int. J. Remote Sens. 2018, 39, 2189–2206. [Google Scholar] [CrossRef]
- Kou, W.; Dong, J.; Xiao, X.; Hernandez, A.J.; Qin, Y.; Zhang, G.; Chen, B.; Lu, N.; Doughty, R. Expansion dynamics of deciduous rubber plantations in Xishuangbanna, China during 2000–2010. GIScience Remote Sens. 2018, 55, 905–925. [Google Scholar] [CrossRef]
- Zhou, J.; Proisy, C.; Descombes, X.; le Maire, G.; Nouvellon, Y.; Stape, J.L.; Viennois, G.; Zerubia, J.; Couteron, P. Mapping local density of young Eucalyptus plantations by individual tree detection in high spatial resolution satellite images. For. Ecol. Manag. 2013, 301, 129–141. [Google Scholar] [CrossRef]
- Yun, H.; Chong, H.; He, L.; Qingsheng, L.; Gaohuan, L.; Zhenchao, Z.; Chenchen, Z. Land-cover classification of random forest based on Sentinel-2A image feature optimization. Resour. Sci. 2019, 41, 992–1001. [Google Scholar] [CrossRef]
- Huaipeng, L.; Huijun, A.; Bing, W.; Qiuliang, Z. Tree species classification using WorldView-2 images based on recursive texture feature elimination. J. Beijing For. Univ. 2015, 37, 53–59. [Google Scholar]
- Henareh Khalyani, A.; Falkowski, M.J.; Mayer, A.L. Classification of Landsat images based on spectral and topographic variables for land-cover change detection in Zagros forests. Int. J. Remote Sens. 2012, 33, 6956–6974. [Google Scholar] [CrossRef]
- Jinshui, Z.; Yaozhong, P.; Lijian, H.; Wei, S.; Chunyang, H. Land Use/cover Change Detection with Multi-Source Data. J. Remote Sens. 2007, 11, 12–26. [Google Scholar]
- Bhattarai, R.; Rahimzadeh-Bajgiran, P.; Weiskittel, A.; Meneghini, A.; MacLean, D.A. Spruce budworm tree host species distribution and abundance mapping using multi-temporal Sentinel-1 and Sentinel-2 satellite imagery. ISPRS J. Photogramm. Remote Sens. 2021, 172, 28–40. [Google Scholar] [CrossRef]
- Erinjery, J.J.; Singh, M.; Kent, R. Mapping and assessment of vegetation types in the tropical rainforests of the Western Ghats using multispectral Sentinel-2 and SAR Sentinel-1 satellite imagery. Remote Sens. Environ. 2018, 216, 345–354. [Google Scholar] [CrossRef]
- Hill, M.J. Vegetation index suites as indicators of vegetation state in grassland and savanna: An analysis with simulated SENTINEL 2 data for a North American transect. Remote Sens. Environ. 2013, 137, 94–111. [Google Scholar] [CrossRef]
- Rajah, P.; Odindi, J.; Mutanga, O.; Kiala, Z. The utility of Sentinel-2 Vegetation Indices (VIs) and Sentinel-1 Synthetic Aperture Radar (SAR) for invasive alien species detection and mapping. Nat. Conserv. 2019, 35, 41–61. [Google Scholar] [CrossRef]
- Fan, H.; Fu, X.; Zhang, Z.; Wu, Q. Phenology-based vegetation index differencing for mapping of rubber plantations using landsat OLI data. Remote Sens. 2015, 7, 6041–6058. [Google Scholar] [CrossRef]
- Peña-Barragán, J.M.; Ngugi, M.K.; Plant, R.E.; Six, J. Object-based crop identification using multiple vegetation indices, textural features and crop phenology. Remote Sens. Environ. 2011, 115, 1301–1316. [Google Scholar] [CrossRef]
- Li, G.; Lu, D.; Moran, E.; Hetrick, S. Land-cover classification in a moist tropical region of Brazil with Landsat Thematic Mapper imagery. Int. J. Remote Sens. 2011, 32, 8207–8230. [Google Scholar] [CrossRef] [PubMed]
- Jia, K.; Liang, S.; Zhang, L.; Wei, X.; Yao, Y.; Xie, X. Forest cover classification using Landsat ETM+ data and time series MODIS NDVI data. Int. J. Appl. Earth Obs. Geoinf. 2014, 33, 32–38. [Google Scholar] [CrossRef]
- Senf, C.; Pflugmacher, D.; van der Linden, S.; Hostert, P. Mapping rubber plantations and natural forests in Xishuangbanna (Southwest China) using multi-spectral phenological metrics from modis time series. Remote Sens. 2013, 5, 2795–2812. [Google Scholar] [CrossRef]
- Ling, C.; Wenqian, H.; Deliang, G. The latest applications of optical image texture in forestry. J. Beijing For. Univ. 2015, 37, 10–27. [Google Scholar]
- Zhang, C.; Huang, C.; Li, H.; Liu, Q.; Li, J.; Bridhikitti, A.; Liu, G. Effect of textural features in remote sensed data on rubber plantation extraction at different levels of spatial resolution. Forests 2020, 11, 399. [Google Scholar] [CrossRef]
- Coburn, C.A.; Roberts, A.C.B. A multiscale texture analysis procedure for improved forest stand classification. Int. J. Remote Sens. 2004, 25, 4287–4308. [Google Scholar] [CrossRef]
- Ouma, Y.O.; Tetuko, J.; Tateishi, R. Analysis of co-occurrence and discrete wavelet transform textures for differentiation of forest and non-forest vegetation in very-high-resolution optical-sensor imagery. Int. J. Remote Sens. 2008, 29, 3417–3456. [Google Scholar] [CrossRef]
- Franklin, S.E.; Hall, R.J.; Moskal, L.M.; Maudie, A.J.; Lavigne, M.B. Incorporating texture into classification of forest species composition from airborne multispectral images. Int. J. Remote Sens. 2000, 21, 61–79. [Google Scholar] [CrossRef]
- Johansen, K.; Coops, N.C.; Gergel, S.E.; Stange, Y. Application of high spatial resolution satellite imagery for riparian and forest ecosystem classification. Remote Sens. Environ. 2007, 110, 29–44. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, W.; Zhang, L.; Wu, J. Analysis of correlation between terrain and forest spatial distribution based on DEM. J. North-East For. Univ. 2012, 40, 96–98. [Google Scholar]
- Ma, H.J.; Gao, X.H.; Gu, X.T. Random forest classification of Landsat 8 imagery for the complex terrain area based on the combination of spectral, topographic and texture information. J. Geo-Inf. Sci. 2019, 21, 360–371. [Google Scholar] [CrossRef]
- Zeng, H.; Chen, G.; Yang, Y. Analysis on forest spatial distr ibution based on DEM. Territ. Resour. Study 2005, 3, 85–86. [Google Scholar] [CrossRef]
- Dorren, L.K.A.; Maier, B.; Seijmonsbergen, A.C. Improved Landsat-based forest mapping in steep mountainous terrain using object-based classification. For. Ecol. Manag. 2003, 183, 31–46. [Google Scholar] [CrossRef]
- Strahler, A.H.; Logan, T.L.; Bryant, N.A. Improving forest cover classification accuracy from Landsat by incorporating topographic information. In Proceedings of the 12th International Symposium on Remote Sensing of Environment, Manila, Philippines, 20–26 April 1978. [Google Scholar]
- Pasquarella, V.J.; Holden, C.E.; Woodcock, C.E. Improved mapping of forest type using spectral-temporal Landsat features. Remote Sens. Environ. 2018, 210, 193–207. [Google Scholar] [CrossRef]
- Rodriguez-Galiano, V.F.; Chica-Olmo, M.; Abarca-Hernandez, F.; Atkinson, P.M.; Jeganathan, C. Random Forest classification of Mediterranean land cover using multi-seasonal imagery and multi-seasonal texture. Remote Sens. Environ. 2012, 121, 93–107. [Google Scholar] [CrossRef]
- Song, Q.; Hu, Q.; Zhou, Q.; Hovis, C.; Xiang, M.; Tang, H.; Wu, W. In-season crop mapping with GF-1/WFV data by combining object-based image analysis and random forest. Remote Sens. 2017, 9, 1184. [Google Scholar] [CrossRef]
- Puissant, A.; Rougiera, S.; Stumpf, A. Object-oriented mapping of urban trees using random forestclassifiers. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 235–245. [Google Scholar] [CrossRef]
- Ozigis, M.S.; Kaduk, J.D.; Jarvis, C.H. Mapping terrestrial oil spill impact using machine learning random forest and Landsat 8 OLI imagery: A case site within the Niger Delta region of Nigeria. Environ. Sci. Pollut. Res. 2019, 26, 3621–3635. [Google Scholar] [CrossRef]
- Boonprong, S.; Cao, C.; Chen, W.; Bao, S. Random forest variable importance spectral indices scheme for burnt forest recovery monitoring-multilevel RF-VIMP. Remote Sens. 2018, 10, 807. [Google Scholar] [CrossRef]
- Ruiz Hernandez, I.E.; Shi, W. A Random Forests classification method for urban land-use mapping integrating spatial metrics and texture analysis. Int. J. Remote Sens. 2018, 39, 1175–1198. [Google Scholar] [CrossRef]
- Watts, J.D.; Lawrence, R.L.; Miller, P.R.; Montagne, C. Monitoring of cropland practices for carbon sequestration purposes in north central Montana by Landsat remote sensing. Remote Sens. Environ. 2009, 113, 1843–1852. [Google Scholar] [CrossRef]
- Wen, X.; Zhong, A.; Hu, X. The classification of urban greening tree species based on feature selection of random forest. J. Geo-Inf. Sci. 2018, 20, 1777–1786. [Google Scholar] [CrossRef]
- Chan, J.C.W.; Paelinckx, D. Evaluation of Random Forest and Adaboost tree-based ensemble classification and spectral band selection for ecotope mapping using airborne hyperspectral imagery. Remote Sens. Environ. 2008, 112, 2999–3011. [Google Scholar] [CrossRef]
- Pal, M. Random forest classifier for remote sensing classification. Int. J. Remote Sens. 2005, 26, 217–222. [Google Scholar] [CrossRef]
- Aygun, S.; Gunes, E.O. A benchmarking: Feature extraction and classification of agricultural textures using LBP, GLCM, RBO, Neural Networks, k-NN, and random forest. In Proceedings of the 2017 6th International Conference on Agro-Geoinformatics, Agro-Geoinformatics, Fairfax, VA, USA, 7–10 August 2017. [Google Scholar]
- Qi, H. China High-Resolution Earth Observation System (CHEOS) and Its Latest Development. Available online: http://www.unoosa.org/pdf/pres/stsc2014/tech-47E.pdf (accessed on 5 June 2021).
- Moderate Resolution Imaging Spectroradiometer (MODIS) MOD15A2H Version 6 Product. Available online: https://ladsweb.modaps.eosdis.nasa.gov/ (accessed on 6 July 2020).
- Haralick, R.M.; Shanmugam, K.; Dinstein, I.H. Textural features for image classification. IEEE Trans. Syst. Man Cybern. SMC 1973, 3, 610–621. [Google Scholar] [CrossRef]
- Guan, H.; Yu, J.; Li, J.; Luo, L. Random Forests-Based Feature Selection for Land-Use Classification Using Lidar Data and Orthoimagery. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2012, XXXIX-B7, 203–208. [Google Scholar] [CrossRef]
- Wang, J.; Gao, Y.; Wang, X.; Fu, L. Forest classification based on forest texture in Northwest Yunnan Province. IOP Conf. Ser. Earth Environ. Sci. 2014, 17, 012071. [Google Scholar] [CrossRef]
- Xu, K.J.; Tian, Q.J.; Yue, J.B.; Tang, S.F. Forest tree species identification and its response to spatial scale based on multispectral and multi-resolution remotely sensed data. Chin. J. Appl. Ecol. 2018, 29, 3986–3993. [Google Scholar]
- Pacifici, F.; Chini, M.; Emery, W.J. A neural network approach using multi-scale textural metrics from very high-resolution panchromatic imagery for urban land-use classification. Remote Sens. Environ. 2009, 113, 1276–1292. [Google Scholar] [CrossRef]
- Ouma, Y.O.; Ngigi, T.G.; Tateishi, R. On the optimization and selection of wavelet texture for feature extraction from high-resolution satellite imagery with application towards urban-tree delineation. Int. J. Remote Sens. 2006, 27, 73–104. [Google Scholar] [CrossRef]
- Wei, P.; Zhu, W.; Zhao, Y.; Fang, P.; Zhang, X.; Yan, N.; Zhao, H. Extraction of Kenyan grassland information using PROBA-V based on RFE-RF algorithm. Remote Sens. 2021, 13, 4762. [Google Scholar] [CrossRef]
- Kuhn, M. The Caret Package. Available online: http://cran.r-project.org/web/packages/caret/ (accessed on 17 April 2020).
- Lou, P.; Fu, B.; He, H.; Li, Y.; Tang, T.; Lin, X.; Fan, D.; Gao, E. An optimized object-based random forest algorithm for marsh vegetation mapping using high-spatial-resolution GF-1 and ZY-3 data. Remote Sens. 2020, 12, 1270. [Google Scholar] [CrossRef]
- Xiong, H.; Zhou, X.; Wang, X.; Cui, Y. Mapping the Spatial Distribution of Tea Plantations with 10m Resolution in Fujian Province Using Google Earth Engine. J. Geo-Inf. Sci. 2021, 23, 1325–1337. [Google Scholar] [CrossRef]
- Mellor, A.; Haywood, A.; Stone, C.; Jones, S. The performance of random forests in an operational setting for large area sclerophyll forest classification. Remote Sens. 2013, 5, 2838–2856. [Google Scholar] [CrossRef]
- Zhang, H.R.; He, P.; Li, C.M. Spafio—Temporal Dynamic Analysis and Evaluation of Forest Resources in Yanqing County. For. Eng. 2010, 26, 4–8. [Google Scholar]
- Ying, Z. Classification of Forest Types Based on Multi-Dimensional Features Using Multi-Seasonal Landsat-8 OLI Remote Sensing Images. Doctoral Thesis, Beijing Forestry University, Beijing, China, 2018. [Google Scholar]
- Immitzer, M.; Neuwirth, M.; Böck, S.; Brenner, H.; Vuolo, F.; Atzberger, C. Optimal input features for tree species classification in Central Europe based on multi-temporal Sentinel-2 data. Remote Sens. 2019, 11, 2599. [Google Scholar] [CrossRef]
- Grabska, E.; Hostert, P.; Pflugmacher, D.; Ostapowicz, K. Forest stand species mapping using the sentinel-2 time series. Remote Sens. 2019, 11, 1197. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, R.; Liu, Y.; Zhu, X. The differences between extracting vegetation information from GF1-WFV and Landsat8-OLI. Acta Ecol. Sin. 2020, 40, 3495–3506. [Google Scholar]
- National Forestry and Grassland Administration. China Forest Resources Report (2009–2013); China Forestry Publishing House: Beijing, China, 2014. [Google Scholar]
- Sihan, L. Extraction of Larch Plantations Using Texture Features within High Spatial Resolution Images. Masteral Thesis, Xi’an University of Science and Technology, Xi’an, China, 2020. [Google Scholar]
- Li, M.; Xing, Y.; Liu, M.; Wang, Z.; Yao, S.; Zeng, X.; Xie, J. Identification of forest type with Landsat-8 image based on SVM. J. Cent. South Univ. For. Technol. 2017, 37, 52–58. [Google Scholar] [CrossRef]
- Yu, L.; Gong, P. Google Earth as a virtual globe tool for Earth science applications at the global scale: Progress and perspectives. Int. J. Remote Sens. 2012, 33, 3966–3986. [Google Scholar] [CrossRef]
- Chen, B.; Li, X.; Xiao, X.; Zhao, B.; Dong, J.; Kou, W.; Qin, Y.; Yang, C.; Wu, Z.; Sun, R.; et al. Mapping tropical forests and deciduous rubber plantations in Hainan Island, China by integrating PALSAR 25-m and multi-temporal Landsat images. Int. J. Appl. Earth Obs. Geoinf. 2016, 50, 117–130. [Google Scholar] [CrossRef]
- Luo, H.-X.; Dai, S.-P.; Li, M.-F.; Liu, E.-P.; Zheng, Q.; Hu, Y.-Y.; Yi, X.-P. Comparison of machine learning algorithms for mapping mango plantations based on Gaofen-1 imagery. J. Integr. Agric. 2020, 19, 2815–2828. [Google Scholar] [CrossRef]
- Shang, G.; Zhu, J.; Gao, T.; Zheng, X.; Zhang, J. Using multi-source remote sensing data to classify larch plantations in Northeast China and support the development of multi-purpose silviculture. J. For. Res. 2018, 29, 889–904. [Google Scholar] [CrossRef]
- Lan, Z.; Dong, H.; Liu, J. Monitoring and Evaluation System of Ecological and Environmental Management of Plantation. J. Beijing For. Univ. (Soc. Sci.) 2013, 12, 25–30. [Google Scholar]
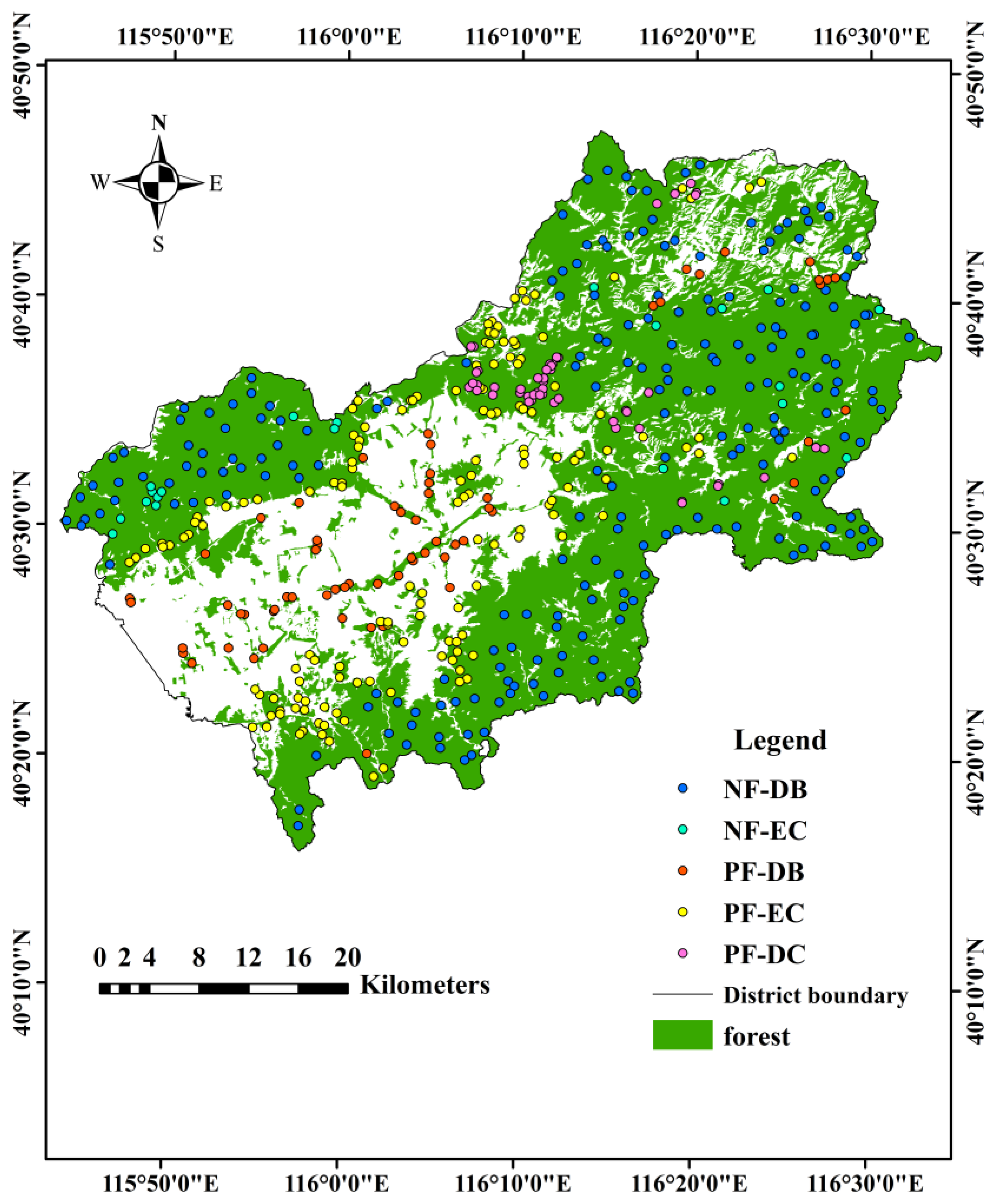
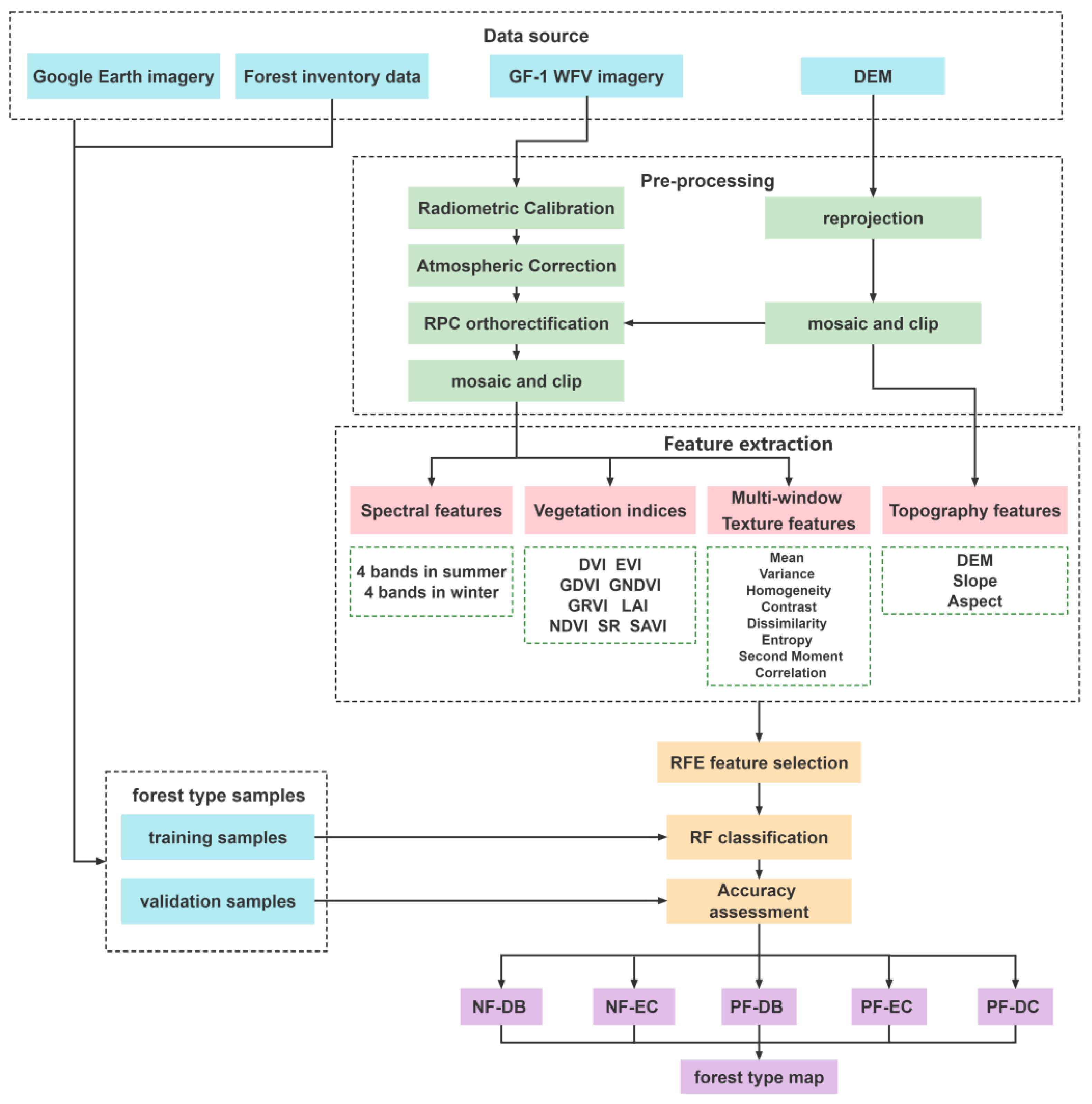
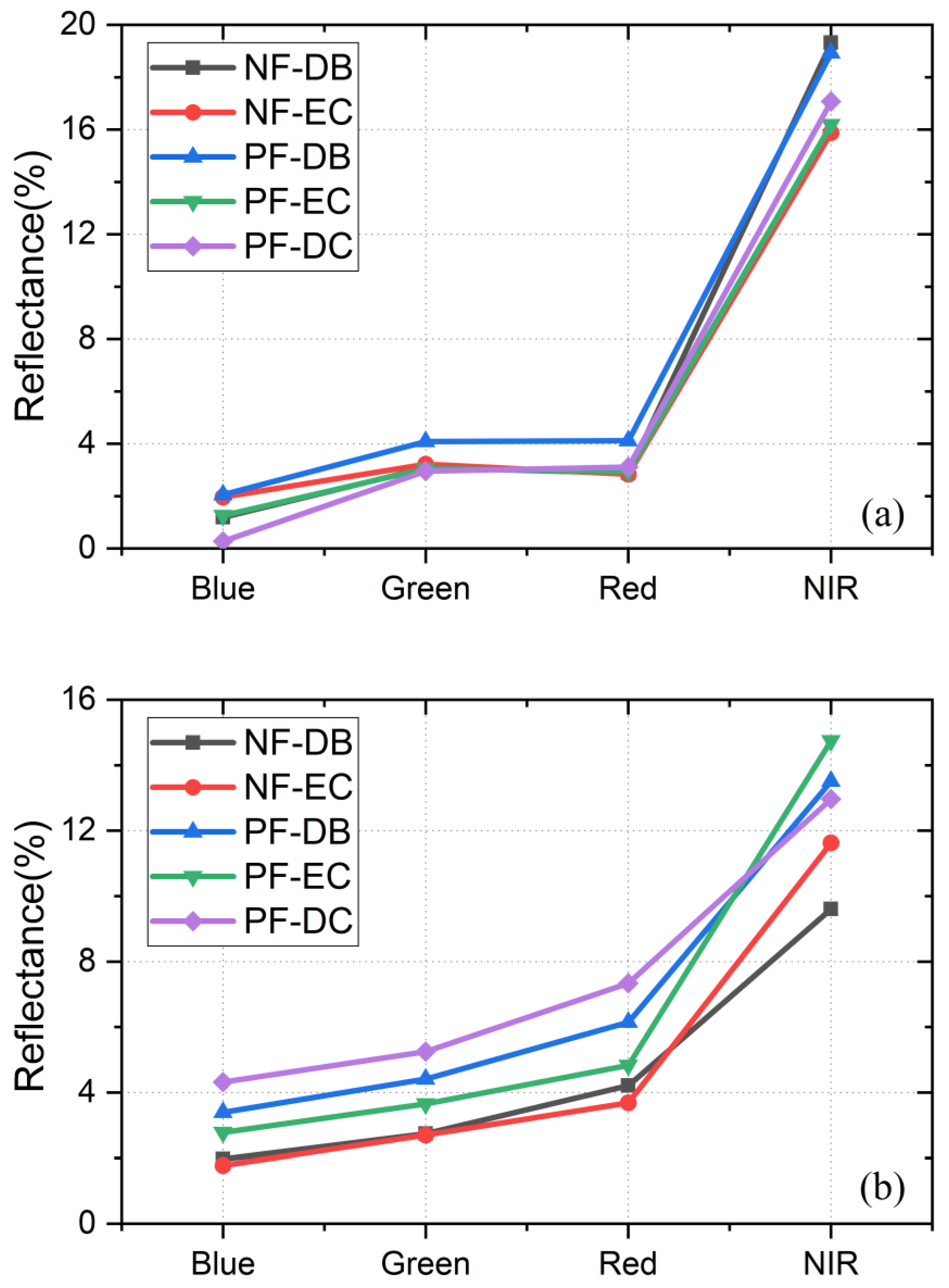
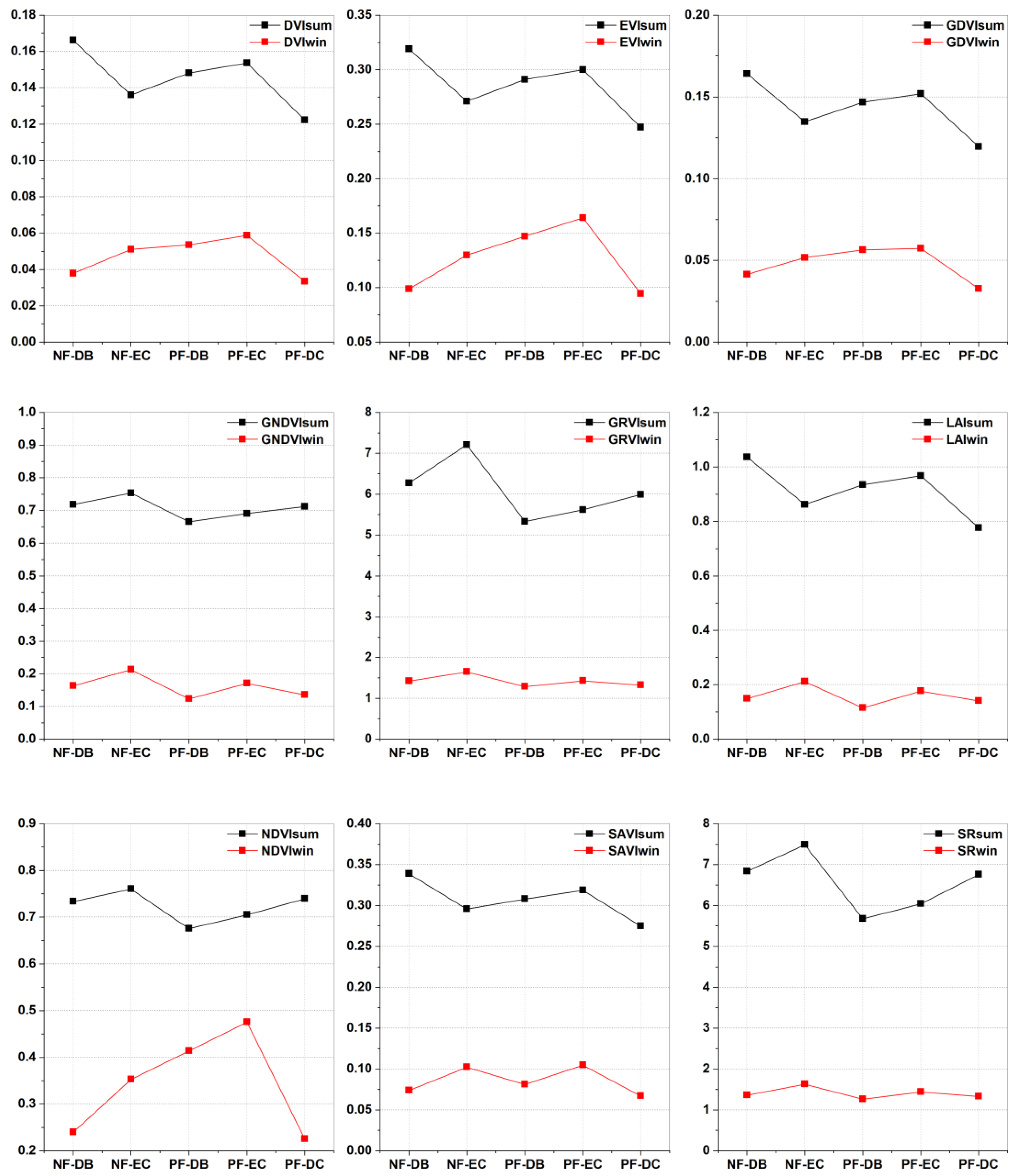


| Forest Type | Abbreviation | Number of Samples | |
|---|---|---|---|
| Natural forest | deciduous broad-leaved | NF-DB | 222 |
| evergreen coniferous | NF-EC | 91 | |
| Plantation | deciduous broad-leaved | PF-DB | 108 |
| evergreen coniferous | PF-EC | 150 | |
| deciduous coniferous | PF-DC | 99 | |
| Reference Data | Classify as | ||||||
|---|---|---|---|---|---|---|---|
| NF-DB | NF-EC | PF-DB | PF-EC | PF-DC | Total | PA (%) | |
| NF-DB | 60 | 4 | 2 | 0 | 0 | 66 | 90.91 |
| NF-EC | 2 | 20 | 0 | 5 | 0 | 27 | 74.07 |
| PF-DB | 3 | 1 | 28 | 1 | 0 | 33 | 84.85 |
| PF-EC | 2 | 4 | 1 | 38 | 0 | 45 | 84.44 |
| PF-DC | 0 | 1 | 0 | 0 | 29 | 30 | 96.67 |
| Total | 67 | 30 | 31 | 44 | 29 | 201 | - |
| UA (%) | 89.55 | 66.67 | 90.32 | 86.36 | 100.00 | - | 87.06 |
| Overall accuracy = 87.06% | |||||||
| Kappa coefficient = 0.833 | |||||||
| Data Type | Classification Results (ha) | Inventory Data (ha) | Area Accuracy (%) |
|---|---|---|---|
| Forest | 132,096 | 112,262 | 82.33 |
| Natural forest | 81,587 | 67,053 | 78.33 |
| Plantation | 50,509 | 45,209 | 88.28 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, F.; Ren, Y.; Wang, X. Application of Multi-Source Data for Mapping Plantation Based on Random Forest Algorithm in North China. Remote Sens. 2022, 14, 4946. https://doi.org/10.3390/rs14194946
Wu F, Ren Y, Wang X. Application of Multi-Source Data for Mapping Plantation Based on Random Forest Algorithm in North China. Remote Sensing. 2022; 14(19):4946. https://doi.org/10.3390/rs14194946
Chicago/Turabian StyleWu, Fan, Yufen Ren, and Xiaoke Wang. 2022. "Application of Multi-Source Data for Mapping Plantation Based on Random Forest Algorithm in North China" Remote Sensing 14, no. 19: 4946. https://doi.org/10.3390/rs14194946
APA StyleWu, F., Ren, Y., & Wang, X. (2022). Application of Multi-Source Data for Mapping Plantation Based on Random Forest Algorithm in North China. Remote Sensing, 14(19), 4946. https://doi.org/10.3390/rs14194946






