A Simple Procedure to Preprocess and Ingest Level-2 Ocean Color Data into Google Earth Engine
Abstract
:1. Introduction
2. Materials and Methods
2.1. Satellite Data
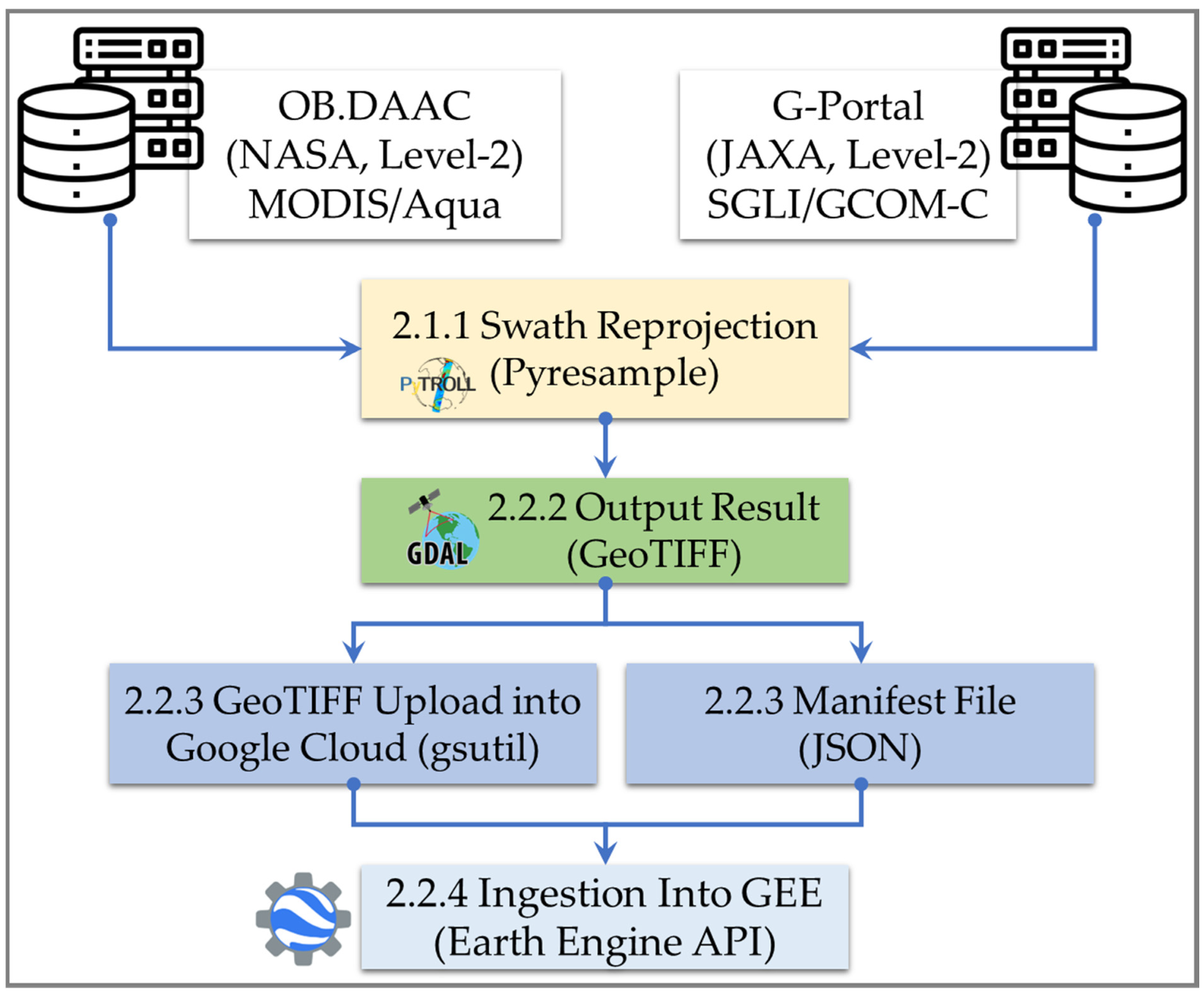
2.2. Data Ingestion Workflow
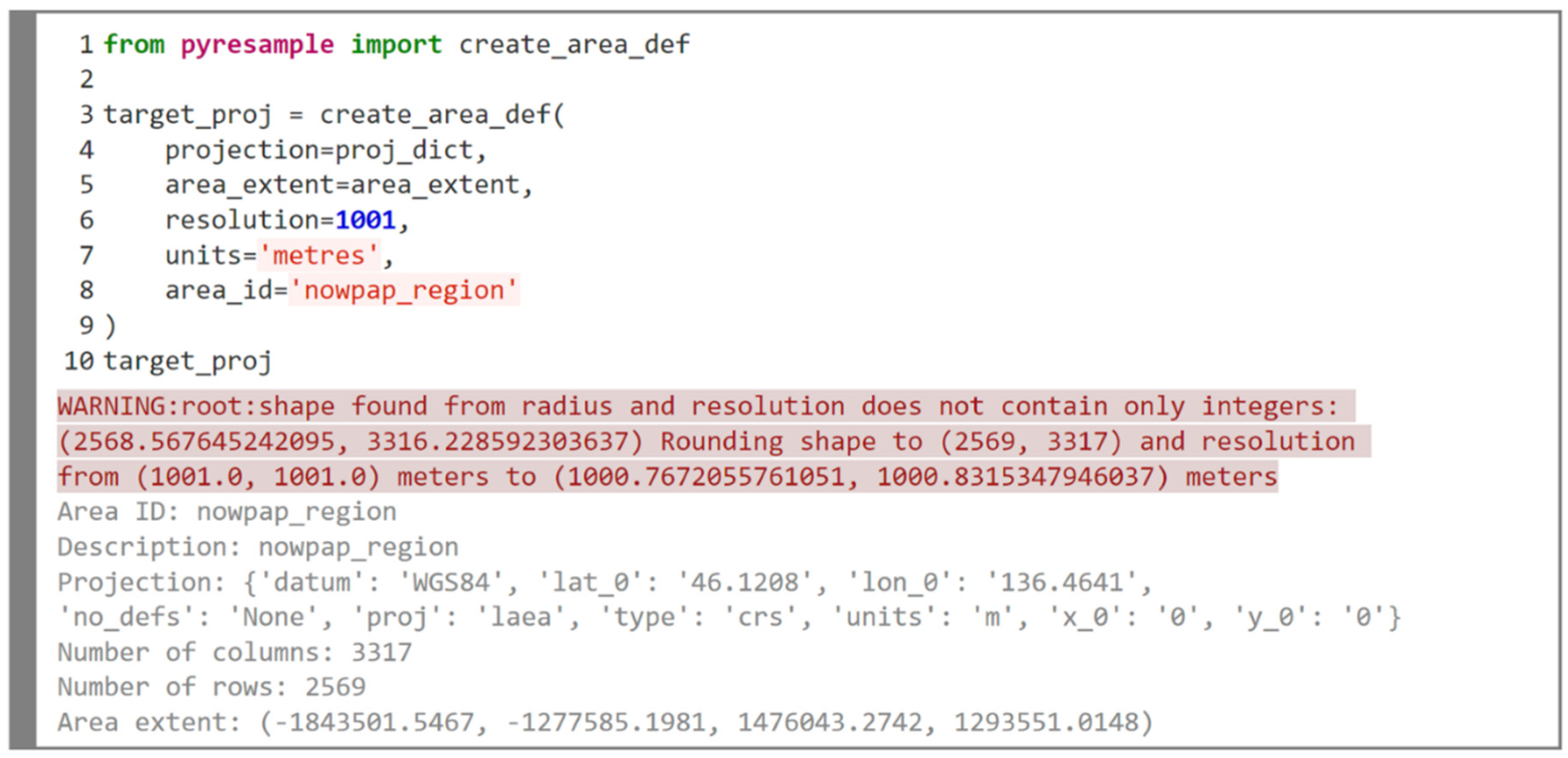
2.2.1. Swath Reprojection
2.2.2. Output Result
2.2.3. GEE Pre-Ingestion Step: Cloud Bucket and Manifest File
2.2.4. Ingestion of the GeoTIFF into GEE
3. Results
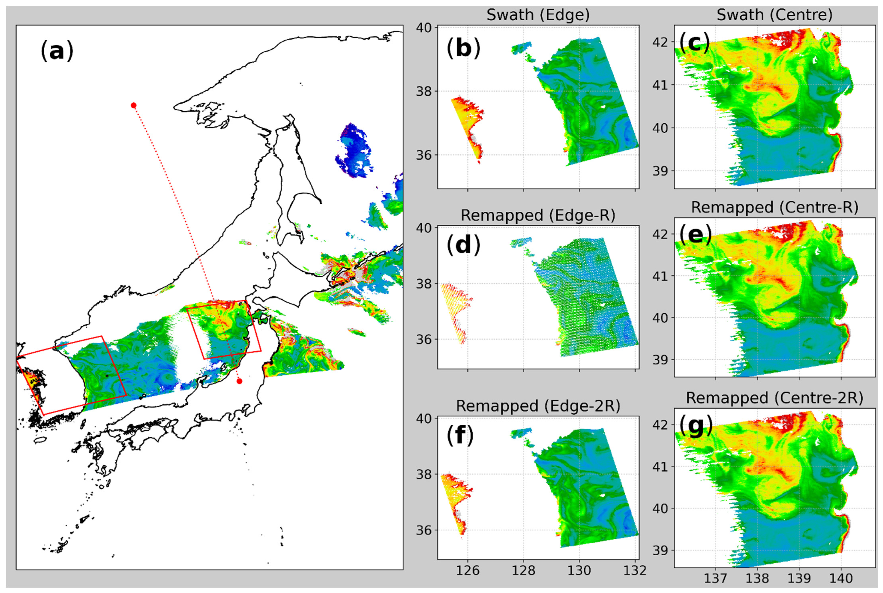
3.1. Mapped Imagery and the Radius of Influence
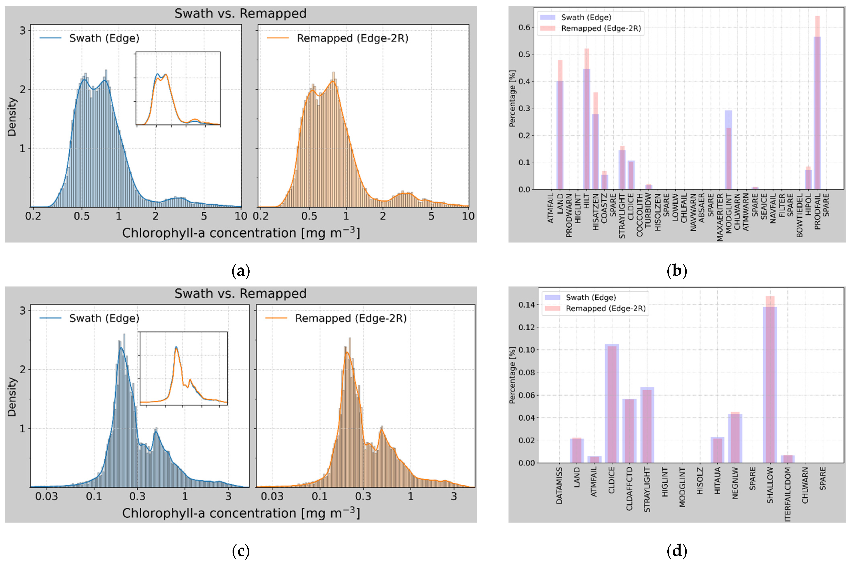
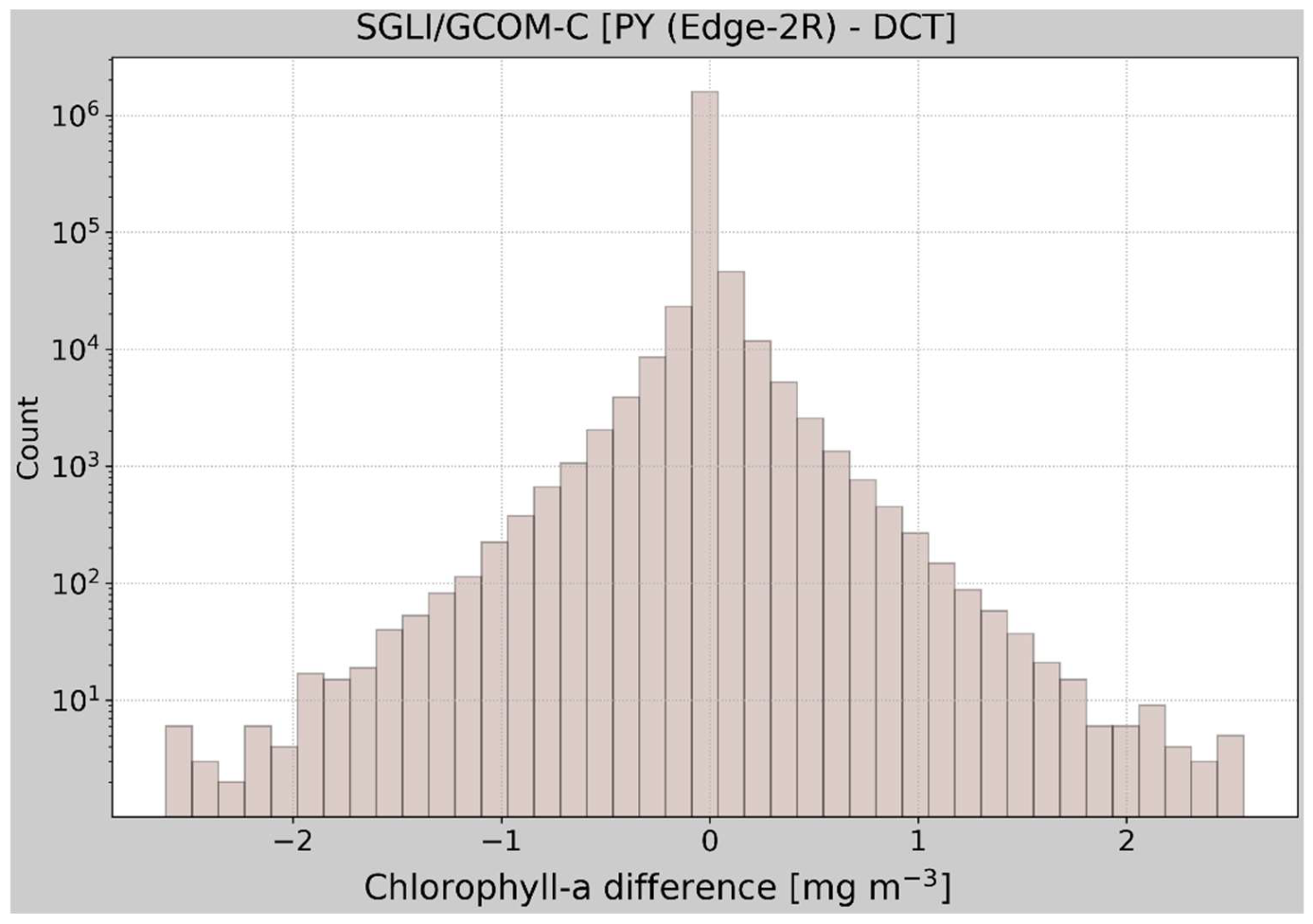
3.2. Verification of the Remapped Results
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A




References
- Groom, S.; Sathyendranath, S.; Ban, Y.; Bernard, S.; Brewin, R.; Brotas, V.; Brockmann, C.; Chauhan, P.; Choi, J.; Chuprin, A.; et al. Satellite Ocean Colour: Current Status and Future Perspective. Front. Mar. Sci. 2019, 6, 485. [Google Scholar] [CrossRef]
- Maúre, E.d.R.; Terauchi, G.; Ishizaka, J.; Clinton, N.; DeWitt, M. Globally Consistent Assessment of Coastal Eutrophication. Nat. Commun. 2021, 12, 6142. [Google Scholar] [CrossRef]
- Feng, C.; Ishizaka, J.; Saitoh, K.; Mine, T.; Zhou, Z. Detection and Tracking of Chattonella Spp. and Skeletonema Spp. Blooms Using Geostationary Ocean Color Imager (GOCI) in Ariake Sea, Japan. J. Geophys. Res. Ocean. 2021, 126, e2020JC016924. [Google Scholar] [CrossRef]
- Stumpf, R.P.; Culver, M.E.; Tester, P.A.; Tomlinson, M.; Kirkpatrick, G.J.; Pederson, B.A.; Truby, E.; Ransibrahmanakul, V.; Soracco, M. Monitoring Karenia Brevis Blooms in the Gulf of Mexico Using Satellite Ocean Color Imagery and Other Data. Harmful Algae 2003, 2, 147–160. [Google Scholar] [CrossRef]
- Cannizzaro, J.P.; Carder, K.L.; Chen, F.R.; Heil, C.A.; Vargo, G.A. A Novel Technique for Detection of the Toxic Dinoflagellate, Karenia Brevis, in the Gulf of Mexico from Remotely Sensed Ocean Color Data. Cont. Shelf Res. 2008, 28, 137–158. [Google Scholar] [CrossRef]
- Siswanto, E.; Ishizaka, J.; Tripathy, S.C.; Miyamura, K. Detection of Harmful Algal Blooms of Karenia Mikimotoi Using MODIS Measurements: A Case Study of Seto-Inland Sea, Japan. Remote Sens. Environ. 2013, 129, 185–196. [Google Scholar] [CrossRef]
- IOCCG. Earth Observations in Support of Global Water Quality Monitoring; Greb, S., Dekker, A., Binding, C., Eds.; IOCCG Report Series, No. 17; International Ocean Colour Coordinating Group: Dartmouth, NS, Canada, 2018; Volume 17. [Google Scholar]
- Huntington, J.L.; Hegewisch, K.C.; Daudert, B.; Morton, C.G.; Abatzoglou, J.T.; McEvoy, D.J.; Erickson, T. Climate Engine: Cloud Computing and Visualization of Climate and Remote Sensing Data for Advanced Natural Resource Monitoring and Process Understanding. Bull. Am. Meteorol. Soc. 2017, 98, 2397–2409. [Google Scholar] [CrossRef]
- Gorelick, N.; Hancher, M.; Dixon, M.; Ilyushchenko, S.; Thau, D.; Moore, R. Google Earth Engine: Planetary-Scale Geospatial Analysis for Everyone. Remote Sens. Environ. 2017, 202, 18–27. [Google Scholar] [CrossRef]
- Tamiminia, H.; Salehi, B.; Mahdianpari, M.; Quackenbush, L.; Adeli, S.; Brisco, B. Google Earth Engine for Geo-Big Data Applications: A Meta-Analysis and Systematic Review. ISPRS J. Photogramm. Remote Sens. 2020, 164, 152–170. [Google Scholar] [CrossRef]
- Zhao, Q.; Yu, L.; Li, X.; Peng, D.; Zhang, Y.; Gong, P. Progress and Trends in the Application of Google Earth and Google Earth Engine. Remote Sens. 2021, 13, 3778. [Google Scholar] [CrossRef]
- Yang, L.; Driscol, J.; Sarigai, S.; Wu, Q.; Chen, H.; Lippitt, C.D. Google Earth Engine and Artificial Intelligence (AI): A Comprehensive Review. Remote Sens. 2022, 14, 3253. [Google Scholar] [CrossRef]
- Kumar, L.; Mutanga, O. Google Earth Engine Applications Since Inception: Usage, Trends, and Potential. Remote Sens. 2018, 10, 1509. [Google Scholar] [CrossRef]
- Mutanga, O.; Kumar, L. Google Earth Engine Applications. Remote Sens. 2019, 11, 591. [Google Scholar] [CrossRef]
- Amani, M.; Ghorbanian, A.; Ahmadi, S.A.; Kakooei, M.; Moghimi, A.; Mirmazloumi, S.M.; Moghaddam, S.H.A.; Mahdavi, S.; Ghahremanloo, M.; Parsian, S.; et al. Google Earth Engine Cloud Computing Platform for Remote Sensing Big Data Applications: A Comprehensive Review. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 5326–5350. [Google Scholar] [CrossRef]
- Hansen, M.C.; Potapov, P.V.; Moore, R.; Hancher, M.; Turubanova, S.A.; Tyukavina, A.; Thau, D.; Stehman, S.V.; Goetz, S.J.; Loveland, T.R.; et al. High-Resolution Global Maps of 21st-Century Forest Cover Change. Science 2013, 342, 850–853. [Google Scholar] [CrossRef]
- Mora-Soto, A.; Palacios, M.; Macaya, E.C.; Gómez, I.; Huovinen, P.; Pérez-Matus, A.; Young, M.; Golding, N.; Toro, M.; Yaqub, M.; et al. A High-Resolution Global Map of Giant Kelp (Macrocystis Pyrifera) Forests and Intertidal Green Algae (Ulvophyceae) with Sentinel-2 Imagery. Remote Sens. 2020, 12, 694. [Google Scholar] [CrossRef]
- Chen, G.; Jin, R.; Ye, Z.; Li, Q.; Gu, J.; Luo, M.; Luo, Y.; Christakos, G.; Morris, J.; He, J.; et al. Spatiotemporal Mapping of Salt Marshes in the Intertidal Zone of China during 1985–2019. J. Remote Sens. 2022, 2022, 9793626. [Google Scholar] [CrossRef]
- Mouw, C.B.; Greb, S.; Aurin, D.; DiGiacomo, P.M.; Lee, Z.; Twardowski, M.; Binding, C.; Hu, C.; Ma, R.; Moore, T.; et al. Aquatic Color Radiometry Remote Sensing of Coastal and Inland Waters: Challenges and Recommendations for Future Satellite Missions. Remote Sens. Environ. 2015, 160, 15–30. [Google Scholar] [CrossRef]
- Luang-on, J.; Ishizaka, J.; Buranapratheprat, A.; Phaksopa, J.; Goes, J.I.; Kobayashi, H.; Hayashi, M.; Maúre, E.d.R.; Matsumura, S. Seasonal and Interannual Variations of MODIS Aqua Chlorophyll—A (2003–2017) in the Upper Gulf of Thailand Influenced by Asian Monsoons. J. Oceanogr. 2022, 78, 209–228. [Google Scholar] [CrossRef]
- Cartwright, P.J.; Fearns, P.R.C.S.; Branson, P.; Cutler, M.V.W.; O’leary, M.; Browne, N.K.; Lowe, R.J. Identifying Metocean Drivers of Turbidity Using 18 Years of Modis Satellite Data: Implications for Marine Ecosystems under Climate Change. Remote Sens. 2021, 13, 3616. [Google Scholar] [CrossRef]
- Lomas, M.W.; Bates, N.R.; Johnson, R.J.; Steinberg, D.K.; Tanioka, T. Adaptive Carbon Export Response to Warming in the Sargasso Sea. Nat. Commun. 2022, 13, 1211. [Google Scholar] [CrossRef]
- Ishizaka, J.; Hirawake, T.; Toratani, M.; Frouin, R. Special Section for Second-Generation Global Imager (SGLI). J. Oceanogr. 2022, 78, 185–186. [Google Scholar] [CrossRef]
- Choi, J.K.; Min, J.E.; Noh, J.H.; Han, T.H.; Yoon, S.; Park, Y.J.; Moon, J.E.; Ahn, J.H.; Ahn, S.M.; Park, J.H. Harmful Algal Bloom (HAB) in the East Sea Identified by the Geostationary Ocean Color Imager (GOCI). Harmful Algae 2014, 39, 295–302. [Google Scholar] [CrossRef]
- Maneewongvatana, S.; Mount, D. Analysis of Approximate Nearest Neighbor Searching with Clustered Point Sets. arXiv 1999, arXiv:cs/9901013. [Google Scholar] [CrossRef]
- Campbell, J.W.; Blaisdell, J.M.; Darzi, M. SeaWiFS Data Products: Spatial and Temporal Binning Algorithms; Hooker, S.B., Firestone, E.R., Acker, J.G., Eds.; SeaWiFS Technical Report Series; Technical Memorandum 104566; NASA Goddard Space Flight Center: Greenbelt, MD, USA, 1995; Volume 32.
- Dorji, P.; Fearns, P. Impact of the Spatial Resolution of Satellite Remote Sensing Sensors in the Quantification of Total Suspended Sediment Concentration: A Case Study in Turbid Waters of Northern Western Australia. PLoS ONE 2017, 12, e0175042. [Google Scholar] [CrossRef]
- Scott, J.P.; Werdell, P.J. Comparing Level-2 and Level-3 Satellite Ocean Color Retrieval Validation Methodologies. Opt. Express 2019, 27, 30140. [Google Scholar] [CrossRef]
- Dong, J.; Xiao, X.; Menarguez, M.A.; Zhang, G.; Qin, Y.; Thau, D.; Biradar, C.; Moore, B. Mapping Paddy Rice Planting Area in Northeastern Asia with Landsat 8 Images, Phenology-Based Algorithm and Google Earth Engine. Remote Sens. Environ. 2016, 185, 142–154. [Google Scholar] [CrossRef]
- Teluguntla, P.; Thenkabail, P.; Oliphant, A.; Xiong, J.; Gumma, M.K.; Congalton, R.G.; Yadav, K.; Huete, A. A 30-m Landsat-Derived Cropland Extent Product of Australia and China Using Random Forest Machine Learning Algorithm on Google Earth Engine Cloud Computing Platform. ISPRS J. Photogramm. Remote Sens. 2018, 144, 325–340. [Google Scholar] [CrossRef]
- Lewkowicz, A.G.; Way, R.G. Extremes of Summer Climate Trigger Thousands of Thermokarst Landslides in a High Arctic Environment. Nat. Commun. 2019, 10, 1329. [Google Scholar] [CrossRef]
- Jahromi, M.N.; Jahromi, M.N.; Zolghadr-Asli, B.; Pourghasemi, H.R.; Alavipanah, S.K. Google Earth Engine and Its Application in Forest Sciences. In Environmental Science and Engineering; Springer: Cham, Switzerland, 2021; pp. 629–649. [Google Scholar]
- Singha, M.; Dong, J.; Sarmah, S.; You, N.; Zhou, Y.; Zhang, G.; Doughty, R.; Xiao, X. Identifying Floods and Flood-Affected Paddy Rice Fields in Bangladesh Based on Sentinel-1 Imagery and Google Earth Engine. ISPRS J. Photogramm. Remote Sens. 2020, 166, 278–293. [Google Scholar] [CrossRef]
- Terauchi, G.; Maúre, E.d.R.; Yu, Z.; Wu, Z.; Kachur, V.; Lee, C.; Ishizaka, J. Assessment of Eutrophication Using Remotely Sensed Chlorophyll-a in the Northwest Pacific Region. In Remote Sensing of the Open and Coastal Ocean and Inland Waters; Frouin, R.J., Murakami, H., Eds.; SPIE: Bellingham WA, USA, 24 October 2018; p. 17. [Google Scholar] [CrossRef]
- Maúre, E.R.; Ishizaka, J.; Sukigara, C.; Mino, Y.; Aiki, H.; Matsuno, T.; Tomita, H.; Goes, J.I.; Gomes, H.R.; Goes, J.I.; et al. Mesoscale Eddies Control the Timing of Spring Phytoplankton Blooms: A Case Study in the Japan Sea. Geophys. Res. Lett. 2017, 44, 11,115–11,124. [Google Scholar] [CrossRef]
- Maúre, E.R.; Ishizaka, J.; Aiki, H.; Mino, Y.; Yoshie, N.; Goes, J.I.; Gomes, H.R.; Tomita, H. One-Dimensional Turbulence-Ecosystem Model Reveals the Triggers of the Spring Bloom in Mesoscale Eddies. J. Geophys. Res. Ocean. 2018, 123, 6841–6860. [Google Scholar] [CrossRef]
| Sensor/Satellite | Data File |
|---|---|
| MODIS/Aqua | A2022125035500.L2_LAC_OC.nc |
| SGLI/GCOM-C | GC1SG1_202205030152F05810_L2SG_IWPRQ_3000.h5 |
| GC1SG1_202205030152F05810_L2SG_NWLRQ_3000.h5 |
| MODIS/Aqua (Edge) | SGLI/GCOM-C (Edge) | |||||
|---|---|---|---|---|---|---|
| Swath | Remapped (R) | Remapped (2R) | Swath | Remapped (R) | Remapped (2R) | |
| Count | 36,591 | 72,827 | 92,072 | 1,313,197 | 1,720,405 | 1,720,405 |
| Mean | 0.740 | 0.741 | 0.795 | 0.293 | 0.307 | 0.307 |
| STD | 1.703 | 1.698 | 1.836 | 2.082 | 2.119 | 2.119 |
| Min | 0.269 | 0.269 | 0.269 | 0.002 | 0.002 | 0.002 |
| 25% | 0.518 | 0.521 | 0.531 | 0.190 | 0.194 | 0.194 |
| 50% | 0.685 | 0.689 | 0.716 | 0.246 | 0.254 | 0.254 |
| 75% | 0.906 | 0.906 | 0.959 | 0.458 | 0.483 | 0.483 |
| Max | 93.196 | 93.196 | 93.196 | 89.963 | 89.963 | 89.963 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maúre, E.d.R.; Ilyushchenko, S.; Terauchi, G. A Simple Procedure to Preprocess and Ingest Level-2 Ocean Color Data into Google Earth Engine. Remote Sens. 2022, 14, 4906. https://doi.org/10.3390/rs14194906
Maúre EdR, Ilyushchenko S, Terauchi G. A Simple Procedure to Preprocess and Ingest Level-2 Ocean Color Data into Google Earth Engine. Remote Sensing. 2022; 14(19):4906. https://doi.org/10.3390/rs14194906
Chicago/Turabian StyleMaúre, Elígio de Raús, Simon Ilyushchenko, and Genki Terauchi. 2022. "A Simple Procedure to Preprocess and Ingest Level-2 Ocean Color Data into Google Earth Engine" Remote Sensing 14, no. 19: 4906. https://doi.org/10.3390/rs14194906
APA StyleMaúre, E. d. R., Ilyushchenko, S., & Terauchi, G. (2022). A Simple Procedure to Preprocess and Ingest Level-2 Ocean Color Data into Google Earth Engine. Remote Sensing, 14(19), 4906. https://doi.org/10.3390/rs14194906








