1. Introduction
Forests cover one-third of our planet and are a key component of the terrestrial carbon cycle, accounting for more than half of the annual carbon flux between the Earth’s surface and atmosphere and sequester 163 tons·ha
−1 of carbon per year [
1]. Forest conservation and sustainable management are crucial for the Earth’s climate. Traditionally, carbon stock capacity measures are based on the relationship between aboveground biomass (AGB) and stored carbon. Quantifying the AGB of forests is also essential for forestry and ecological studies on ecosystems, slope stability, biodiversity, ecological niche, species distributions, and informing fire risk, among many others [
2,
3].
Traditionally, AGB is determined using the tree diameter at breast height (DBH) and the tree height through allometric equations [
4].
Tree DBH and tree height are traditionally measured in field inventories [
5]. However, AGB data are expected to be measured over extensive areas of the globe, especially for the carbon stock estimation. This requires field data from remote and inaccessible areas [
4]. Today, many biomass measurements are estimated, more or less precisely, using remotely sensed data. In fact, in the last 20 years, the use of remote sensing for measuring and monitoring tree parameters has become more frequent [
6,
7]. Tree parameters such as tree height, tree DBH, crown radius, and AGB are derived with mathematical models from optical imagery, synthetic aperture radar (SAR), and Light detection and ranging (LiDAR). In this regard, LiDAR sensors mounted on aerial supports such as airplanes, helicopters, and unmanned aerial systems (UAS) are particularly suitable for forest measurement [
8]. Indeed, the three-dimensional reconstruction of forests with LiDAR point clouds accurately describes the physical characteristics of the forest stand canopy over large areas [
4,
5].
Several methods for estimating biomass from LiDAR data have been developed and published continuously over the last few years with increasing complexity and various techniques. Two main strands can be distinguished from the latest studies on AGB computing from ALS data: empirical methods and modeling methods. The empirical approaches are based on the correlation between the physical characteristics of forest stands (forest structure) and biomass. Such correlations are generally described by regressions with different degrees of complexity. Physical attributes may be regressed against either tree DBH or biomass to obtain general LiDAR-biomass models [
6]. The allometric relations between biomass and LiDAR-derived measures may be defined by linear [
9] and multiparameter equations [
3,
10,
11], or may have stochastic relationships described by machine learning models [
12,
13]. Regardless of how complex the relationship between the input parameters (independent regression variables) and biomass is, empirical methods require data from in situ biomass samples as the dependent variable [
12]. The sample data are then used to compute and calibrate the model. Among the advantages of empirical models is the low cost of calculation and ease of application. Still, they are restricted to specific forest types and geographical areas (i.e., site-specific) [
6,
12,
14].
More recently, AGB quantification procedures have shifted away from the regression between LiDAR-derived characteristics and biomass measurements and increasingly include automated tree top detection and 3D computer vision [
8]. Such modeling methods are based on the computer reconstructions of volumes and geometries based on LiDAR-derived point clouds. Modeling methods typically operate at an individual-tree scale and are applied in both mobile laser scanning (MLS) and ALS data [
15,
16,
17]. Therefore, they require much higher density LiDAR point clouds and higher computational power than those needed by empirical models. On the other hand, modeling methods do not involve field data and are not site-specific. However, particular parameterizations based on the structure of the forests and the shape of the crown (generally related to conifers-broadleaves) are necessary. Frequently, the data retrieved from 3D modeling are then used as inputs in allometric formulas of general value, not site-specific, for the computation of forest metrics. This approach is particularly suitable for point clouds not associated with an in-the-field dataset as it happens, for example, in data collected on extensive areas such as surveys carried out by administrative bodies at the national and regional levels [
2,
12].
One of the main limitations of both the empirical and modeling models is reproducibility over large areas, which is seldom validated. This limitation is exacerbated in multitemporal analysis. In fact, while the estimation of forest parameters with LiDAR instruments has been studied in depth, 3D change detection in forests is still a subject of little attention in the literature due to the challenges introduced by comparing point cloud pairs [
18,
19]. Some attempts have been made in urban vegetation crown shape changes [
15] and forest environments [
19,
20], focusing on the crown height model detection of cuts [
21,
22,
23]. The study by [
18] identified three main problems related to the 3D change detection: (i) the point cloud density; (ii) irregular sampling of point clouds; and (iii) irregular structure of tree crowns. For these reasons, performing a point-to-point comparison between the two epochs is impossible. They found a possible solution by performing the multitemporal analysis on the parameters estimated from the LiDAR data at a plot level instead of directly comparing the point clouds. The tree parameter-based change detection solution is only as good as the input data and the parameters estimation method. In 3D multitemporal analyses, there is a risk of bias induced by the procedure applied for the tree parameter estimation. However, the solution proposed in [
18] allows for the automation of change detection, which in 3D forest is still an unexplored topic. Indeed, by automatically extracting forest parameters at individual tree scale from clouds of different epochs, which have to be co-registered, then the two clouds can be compared automatically.
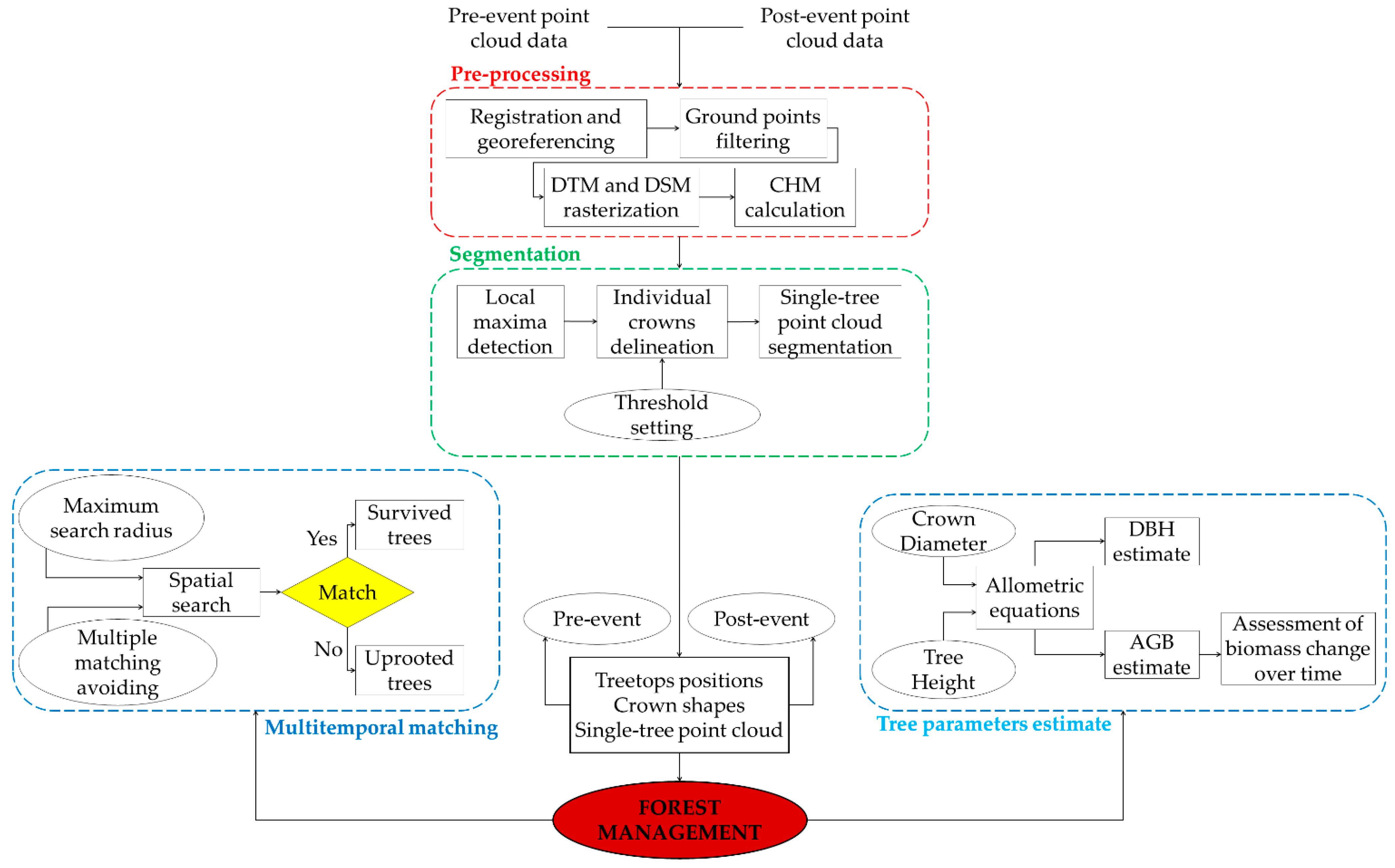
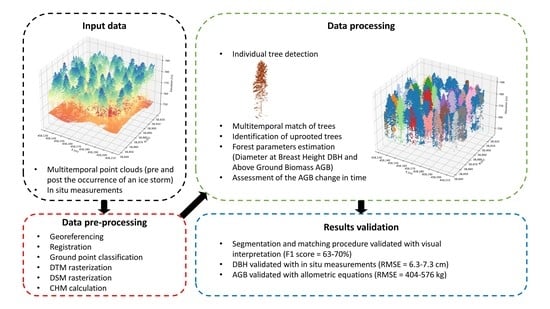
In this paper, we propose a methodology for the automatic 3D change detection of forests by defining the AGB at single-tree level from the airborne LiDAR point cloud. Moreover, we tested: (i) the accuracy of single tree segmentation procedures and allometric relations for the estimation of DBH, height, and crowns diameter; (ii) the role of the allometric variables in terms of error propagation for AGB and DBH estimations; and (iii) the assessment of the AGB changes over time.
The tests were conducted in complex mixed uneven-aged forest stands in the Dinaric Mountains (SW Slovenia), where two ALS flights in 2013 and 2014 were flown. The forest stands were damaged by an ice storm in 2014. In this framework, the automatic AGB change detection has a bearing on the management of the disastrous effects of extreme natural events that have intensified in the study area, as in many other parts of the world, due to climate change.
3. Results
3.1. Segmentation and Multitemporal Matching
The results showed the reliability of the segmentation (
Figure 4a–d) and matching procedures (
Figure 4e,f). For most trees that survived the ice storm, a match was defined with a tree present before the storm (
Figure 4f). Additionally, trees plotted in dark blue in the 2014 time period identified trees that were segmented incorrectly, causing a mismatch with the same tree in the 2013 time period.
Regarding the accuracy of the segmentation procedure, the F1 scores values were equal to 70% and 68% for the pre- and post-ice storm, respectively, and they showed high consistency with each other (
Table 2). With respect to the validation of the multitemporal matching and change analysis algorithm, the F1 score values settled at 69% in the pre-event scenario and 63% post-event (
Table 2). It should be noted that these values cannot be higher than the F1 score values in the segmentation phase since it is not possible to correctly match trees that are not adequately detected simultaneously in the two acquisitions. To further test the reliability of the multitemporal association, it was decided to consider a hypothetical scenario in which we assumed that only during the point cloud segmentation procedure of the data acquired before the ice storm occurrence, no over-segmented trees were recorded. Under these conditions, the F1 score increased, and it reached a value of 80%.
3.2. Error Propagation in the Allometric Equations
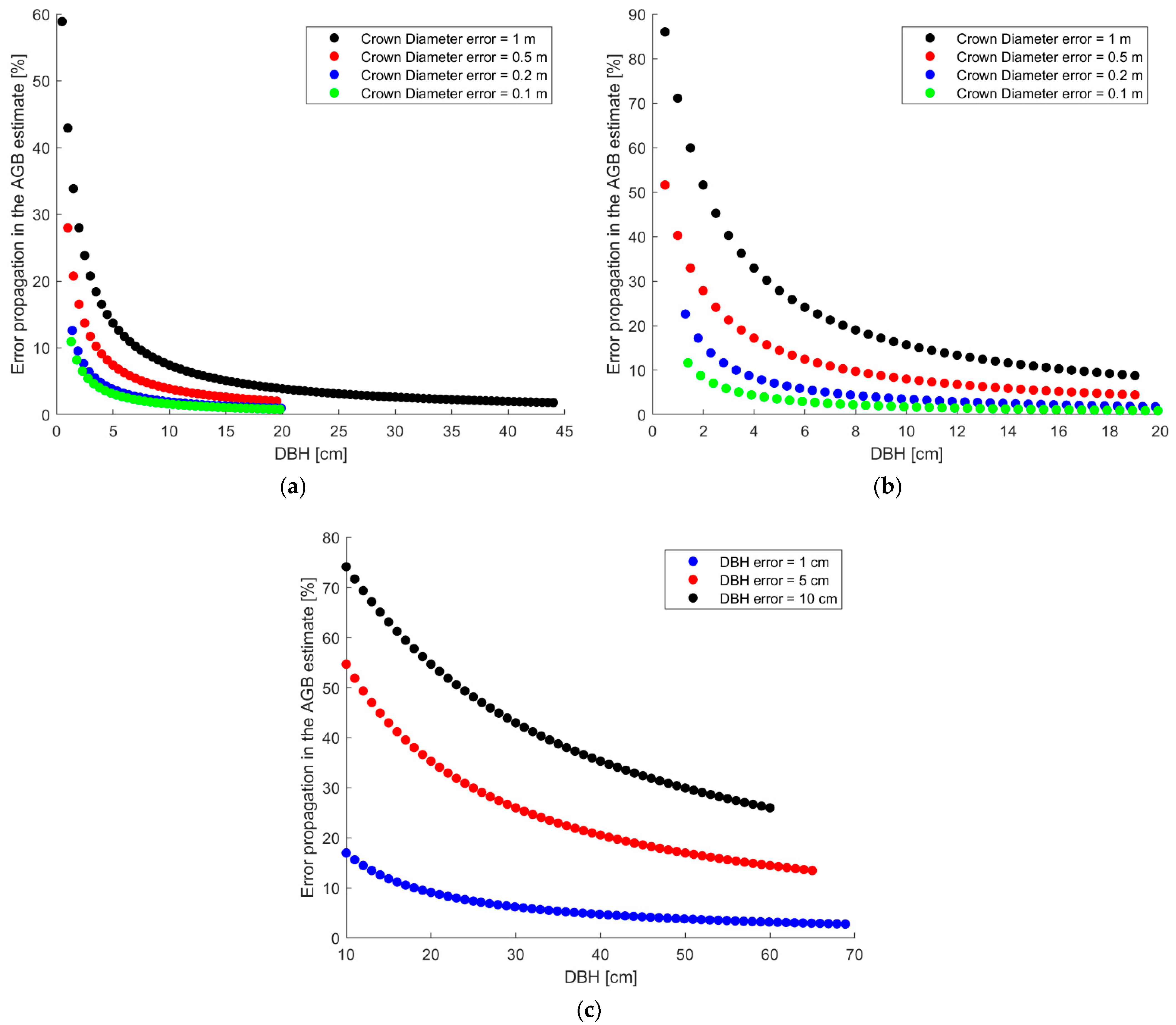
The results of the estimation of the error propagation of Equations (1), (2) and (5) are detailed below. All of the equations assumed an exponential trend in the percentage of error committed, and the propagation of the error in the estimation of the parameters increased as the size of the tree (crown diameter) decreased.
In reference to Equation (1), it was observed (
Figure 5a) that the DBH estimate for trees with a crown described by a diameter between 5 and 14 m (such as those analyzed in the case in question) was affected by a maximum error propagation of about 15% when the error in the evaluation of the diameter of the canopy was one meter. Considering errors of this estimate of less than one meter, the percentage of the error committed was less than 10% for the trees in the study area.
When evaluating the error propagation in the AGB estimation through Equation (2), an error in the estimation of the crown diameter equal to one meter could lead to an overestimation of the biomass of the trees by up to 30%. The overestimation could be reduced and limited to 15% if the overestimation of the CD was halved (
Figure 5b).
Equation (5) is affected by error propagation with the fastest exponential growth compared to the previous equations (
Figure 5c), since the analyzed parameter (the DBH, expressed in centimeters) is squared. For the smallest trees (with DBH between 10 and 20 cm), an error in the estimate of 1 cm caused an error in terms of percentage ABG between 10 and 20%; if the DBH was overestimated by 10 cm, this error could exceed 70%. If we consider the medium/large trees, the percentage error of the ABG was reduced and became reasonable if the DBH was estimated with a maximum error of 1 cm, while it reached values higher than 15–20% for estimates affected by more significant uncertainty (5 cm).
3.3. Tree Parameters
The DBH and the AGB were estimated on the segmented trees. Greater attention was paid to trees for which a multitemporal matching between acquisitions was identified to evaluate the variation of parameters (the AGB) after the natural disaster occurred. Among these trees, those for which no in situ measurements were available (the surveyed area was smaller than the area acquired with the laser scanner) were further discarded.
The trees that had been correctly identified and matched in the overall area totaled 31. Overall, thirty-one trees were correctly identified and matched; of these, 15 were used to evaluate the accuracy of the assessments performed since they were located within the test area for in situ investigations and for which the DBH was measured.
The diameter estimation was carried out using the tree parameters (tree height and crown diameter) obtained from the point cloud acquired before the occurrence of the ice storm. In the case of trees that suffered damage (e.g., the crown had shrunk, the highest branches were broken), there was a risk of underestimating the diameter.
The scatter plots for the DBH estimated with Equation (1) considering Scenario 1 and Scenario 2 (
Figure 6) showed a slight tendency to underestimate the DBH using the allometric equation, for which the bias was between about −9% and −12%, and the RMSE value was between 7.3 cm and 8.3 cm (
Table 3).
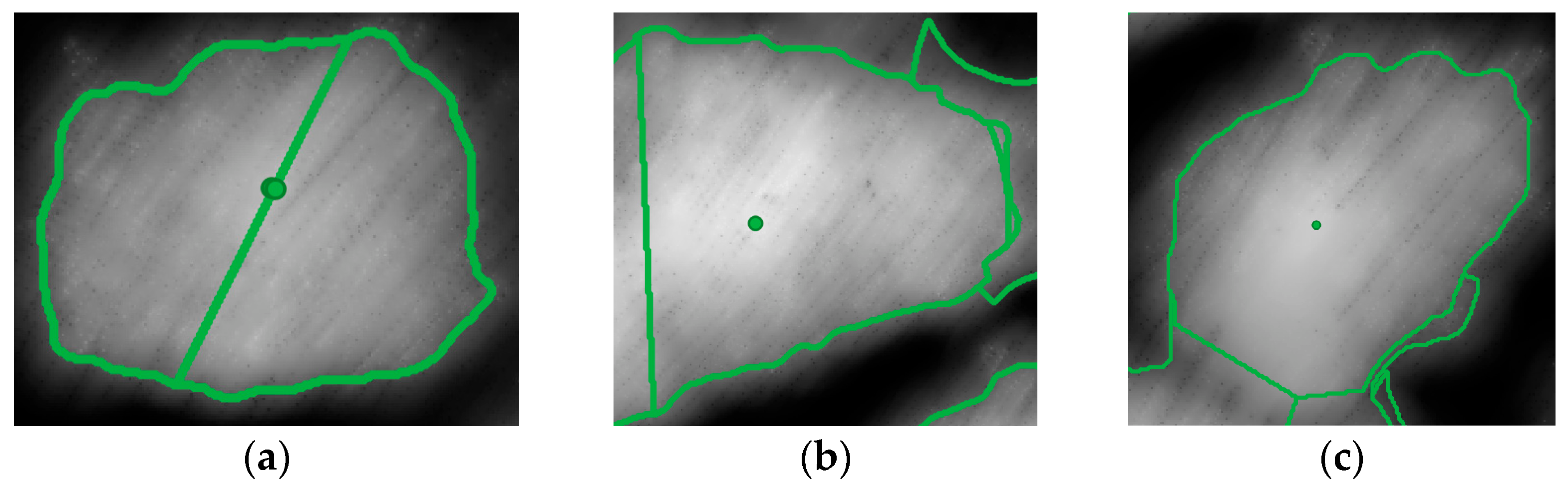
The results in the error metrics showed a lower RMSE and a lower bias when estimating the DBH considering the crown diameter as an output of the automatic single-tree segmentation; In contrast, manual segmentation led to values with a more significant average error and greater variability. In both scenarios, it was observed that Equation (1) seems to lead to underestimating the actual size of the DBH, providing errors up to the order of magnitude of the decimeter. Trees for which the diameter estimate performed the worst were mainly located in particularly dense areas in which the crowns of the trees intersected each other, and the extension of the canopy was not easily identifiable. In the sparser areas, the errors were considerably lower.
The tree biomass assessment was carried out with Equations (2) and (5) according to Scenario 3, Scenario 4, Scenario 5, and Scenario 6 and compared with the estimated ground truth as described in
Section 2.2.5; finally, the metric errors (RMSE and bias) were calculated (
Table 3). The minimum, maximum, and average AGB for each scenario was also calculated (
Table 4)
The metrics (
Table 3) highlight that all of the scenarios considered (Scenario 3, Scenario 4, Scenario 5, and Scenario 6) tend to underestimate the tree AGB with a minimum average percentage of −4.5% and a maximum average percentage of −21.3%. The biomass estimates in the 2014 scenario were, on average, worse when compared to the biomass estimated of the same trees before the natural hazard, both with respect to the RMSE and the bias values.
After having estimated the tree parameters, it was possible to assess the biomass lost in the area under investigation due to the ice storm. At first, we decided to calculate the biomass lost only from the trees that had been correctly segmented and matched in the two multitemporal point clouds. The results (
Table 5) quantify the ground truth AGB loss of these trees as about 4430 kg (about 15% of their total biomass). The estimates resulting from the four scenarios are in line with this quantity, except for Scenario 3, according to which the biomass lost by the matched trees was greater and approximately equal to 29.4%. The scenarios that provided the best results were Scenario 5 and Scenario 6, where Equation (2) was applied.
To the aim of the assessment described above, however, trees that were correctly matched but for which we did not have the in situ measured DBH were excluded. When considering all the trees that survived the hazard (
Table 5), it was shown that the biomass assessment was consistent for all the scenarios, with one considering only matched trees.
Additionally, to account for trees that survived the storm but which were not correctly identified and matched in both acquisitions, we believe that it is necessary to apply a correction factor that increases the amount of biomass estimate loss by about 30%. This percentage was chosen because about 30% of the surviving trees were not correctly multitemporally matched (
Table 2).
Finally, considering the biomass of all of the segmented trees, the estimates were similar to each other, identifying about 40% of the AGB lost due to the ice storm (
Table 5).
4. Discussion
The results of this very first application of the multitemporal analysis of airborne point clouds for the assessment of single tree parameters and the evaluation of their change over time are promising as they are characterized by good accuracies and low errors. In particular, the procedural methodology proposes a new approach for the multitemporal comparison of point clouds in forest environments subjected to strong changes due to natural hazards. The 3D change detection was performed by implementing a single-tree level segmentation algorithm, and subsequently compared the segmented point clouds using forest parameters (treetops and crown diameters); then, the estimation of DBH and AGB was carried out through the application of allometric formulas. Finally, the biomass variation was performed by comparing the AGB estimated before and after the ice storm. A further innovative aspect lies in the complete automation of the entire procedure, which provides the possibility of applying the algorithm to any forest scenario.
The accuracy obtained in the segmentation phase achieved excellent results, with an F1 score of about 70%. In detail, this value appeared to be consistent with those proposed in the literature and obtained with different techniques and algorithms [
32,
33]. In particular, ref. [
32] analyzed how the heterogeneity and density of the investigated area affected the final performance of the segmentation. Moreover, the goodness of the results was in line with the accuracy proposed in the literature [
32]. No cases of simple omission were recorded, which means that all the trees were detected; the main errors were due to under-segmentation and over-segmentation. The main causes of erroneous segmentation were related to the high density of trees and to the spatial heterogeneity typical of non-anthropized environments.
An aspect to be improved is related to the thresholds to be set in the classification algorithm; as described, in this study, they were chosen iteratively; however, for larger areas characterized by more varied forest characteristics, it would be important (i) what quality of segmentation would be obtained with the same thresholds (2) to automate the threshold best fitting procedure to automatically identify those that performed the best segmentation outcomes.
Additionally, it must be considered that the validation, although carried out by exploiting the knowledge of the position of the trees following the in situ measurement campaign, was mainly based on the user-defined visual interpretation of the point cloud and the CHM. This can be affected by discrepancies due to the subjectivity that affects the visual evaluations. Another aspect not to be overlooked is the difficulty in correctly delineating, both automatically and manually, the crown of trees when they are close to each other and the branches intersect each other.
As mentioned, the multitemporal matching procedure between the same trees acquired at different time instants is consistent and unique. As pointed out by [
18], the comparison of forest elements through the use of an instrument that performs irregular sampling of points is complex; nevertheless, after having estimated the position of the tree as a function of the treetop, it is possible to carry out multitemporal analysis based on a unique tree parameter. The positive aspect is that it is possible to estimate the position of the treetops using data acquired from aerial laser scanners (by drones or helicopters). Clearly, the success of the procedure of identifying the same tree in multitemporal acquisitions strictly depends on the reliability of the segmentation procedure itself.
This is a challenging objective regarding the automatic estimation of tree parameters through remote sensing technologies [
34]. If in more regular environments, as in the case of agricultural fields investigated in [
35], good results are obtained even with a lower average density of points, the same is not valid for denser and more irregular areas. In particular, the density of the point cloud is considerably reduced in the description of the stem and of the lower branches, making it more difficult to estimate the DBH directly. The estimates, therefore, were made indirectly, using allometric equations [
4,
10] that made use of more easily estimated tree parameters (H and CD). However, errors in the estimation of forest variables can lead to a systematic error in estimating the parameters. In the proposed case study, the use of the proposed formulas highlighted a systematic underestimation of the DBH and the AGB. The reasons can be sought not only in the estimation of the variables, but also in the applicability of the allometric equations to predict the AGB and the DBH of any tree without making different considerations depending on the tree species.
Finally, with the aboveground biomass estimates, an assessment of the biomass lost in the area under examination due to the storm was carried out. The AGB estimate with Equation (5) was slightly worse than that obtained with Equation (2). This may be caused by the fact that the variable considered in Equation (5) (the DBH) is in turn estimated through Equation (1), which, as highlighted above, is affected in turn by underestimation, as seen previously. Finally, as seen in the DBH estimate, also in the case of the AGB, the best estimates were obtained by considering the scenario with the parameters automatically estimated. Only in the case of the 2014 scenario did manual segmentation seem to be more effective than that performed automatically, thanks to which the RMSE decreased by about 150 kg.
The results highlight the severity of the event, which destroyed about half of the biomass present; depending on the formula considered, it was estimated that the ice storm destroyed between 42% and 45% of the woody biomass, of which about 15–30% was irreversible.
5. Conclusions
In this study, we proposed an automated procedure to predict tree parameters on the single-tree-level using allometric equations that relate the DBH and the AGB to the tree height and the crown diameters. Additionally, we introduced the concept of multitemporal variability, comparing the parameters of the same tree over time; in this way, it is possible to study the evolution of forests at the level of a single tree and quantify the damage caused by a catastrophic natural event. The results are promising and emphasize the algorithm’s validity; however, this study should be considered as the first step in a broader analysis. First, this approach was tested on a relatively small area of interest; further areas with ground truth data will be considered to obtain results that can be considered more statistically robust. It is also necessary to evaluate the method’s accuracy under different forest conditions such as various tree species and forest densities, which can considerably influence the proposed method’s performance. Subsequently, further tests on treetop detection algorithms should be implemented. From the forest management point of view, the proposed approach can be very supportive, allowing for the acquisition of information on large areas in a reasonable amount of time. Moreover, implementing the latest LiDAR technologies on drones would increase the point density and the overall accuracy in smaller areas. Laser scanners equipped on helicopters can still be used to manage more extensive areas with reasonable accuracy, providing a greater quantity of information than other processing (e.g., photogrammetric analysis), thanks to the multiple returns of laser pulses. Moreover, the cost of collecting airborne LiDAR data is justified by the fact that in this way, it is possible to consider areas that are difficult to reach, and the safety of the operators is guaranteed.
Furthermore, among the future objectives, it would be important to be able to quantify the carbon stored within the aboveground biomass; in this way, it would be possible to also quantify the damage caused by a natural hazard from the perspective that the felled trees represent a loss of the capacity of the forest to stock carbon dioxide.