Tree Crowns Cause Border Effects in Area-Based Biomass Estimations from Remote Sensing
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset and Allometric Assumptions
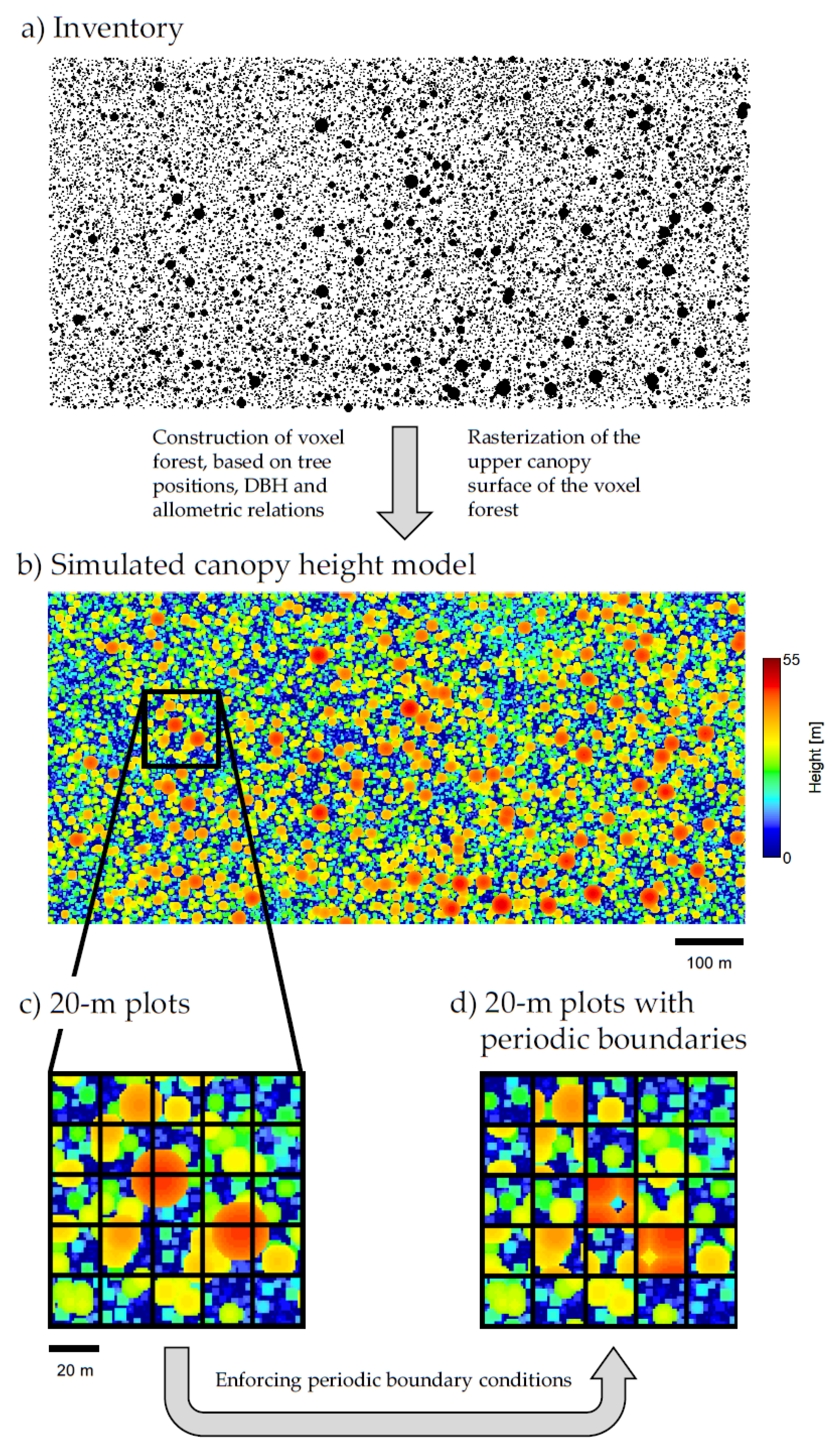
2.2. Voxel Forest Generation
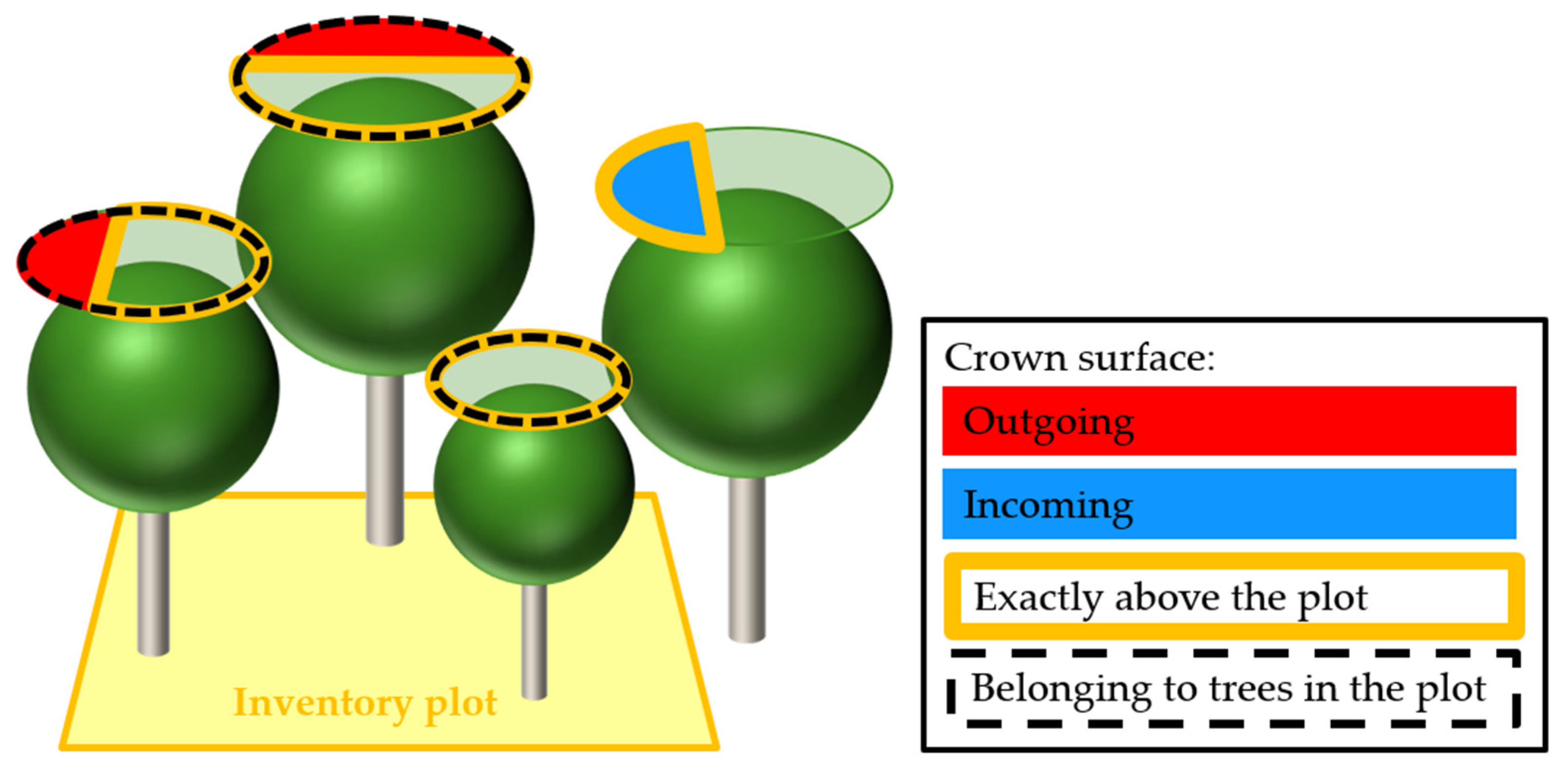
2.3. Crown Space Quantification
2.4. Biomass Estimation Using Square Plots and CHM
2.5. Biomass Estimation Using Large-Footprint Lidar
2.6. Uncertainty Quantification
2.7. Periodic Boundary Conditions
3. Results
3.1. Proportions of Incoming and Outgoing Crowns
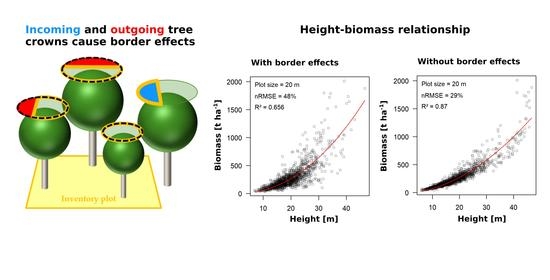
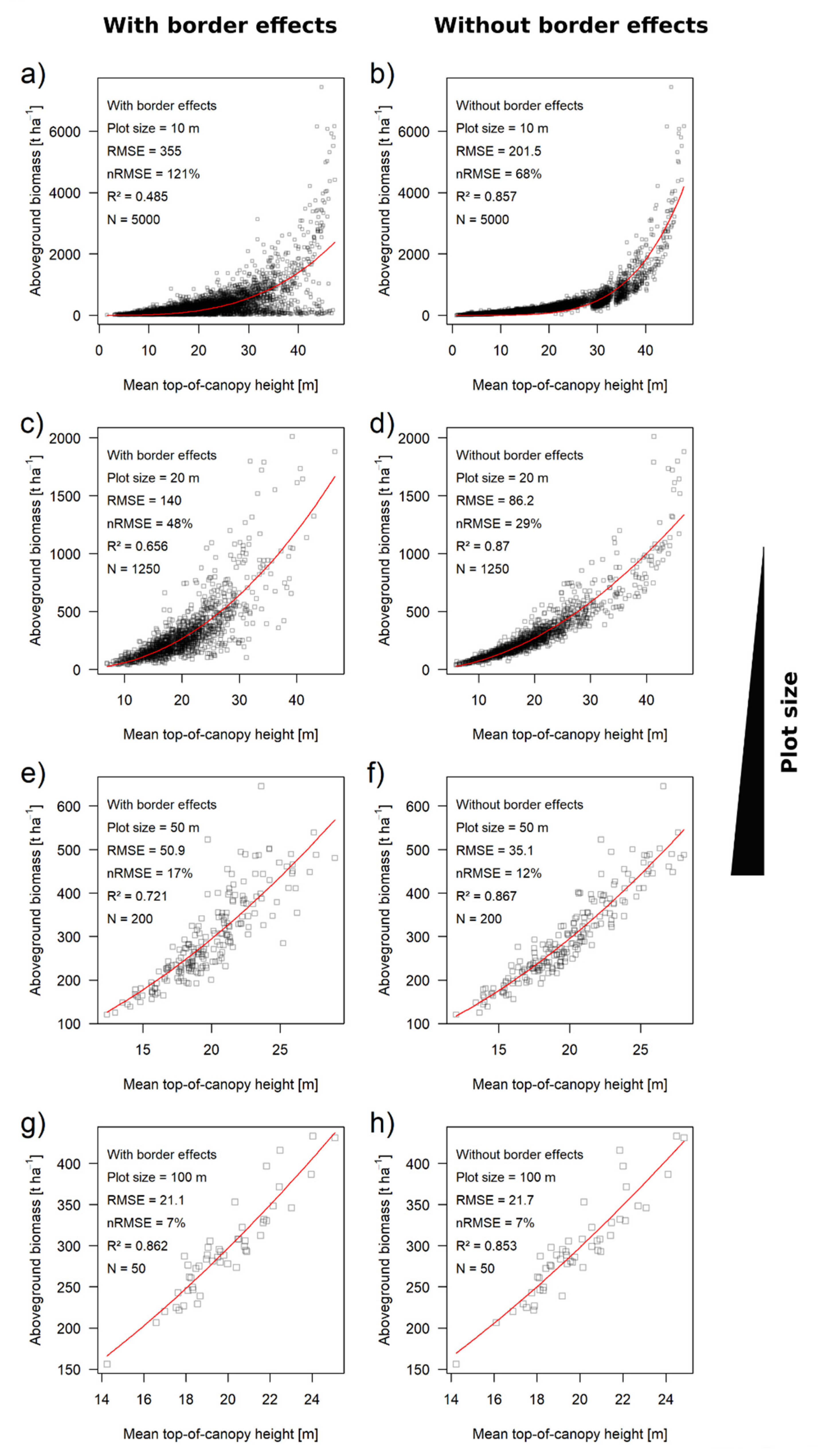
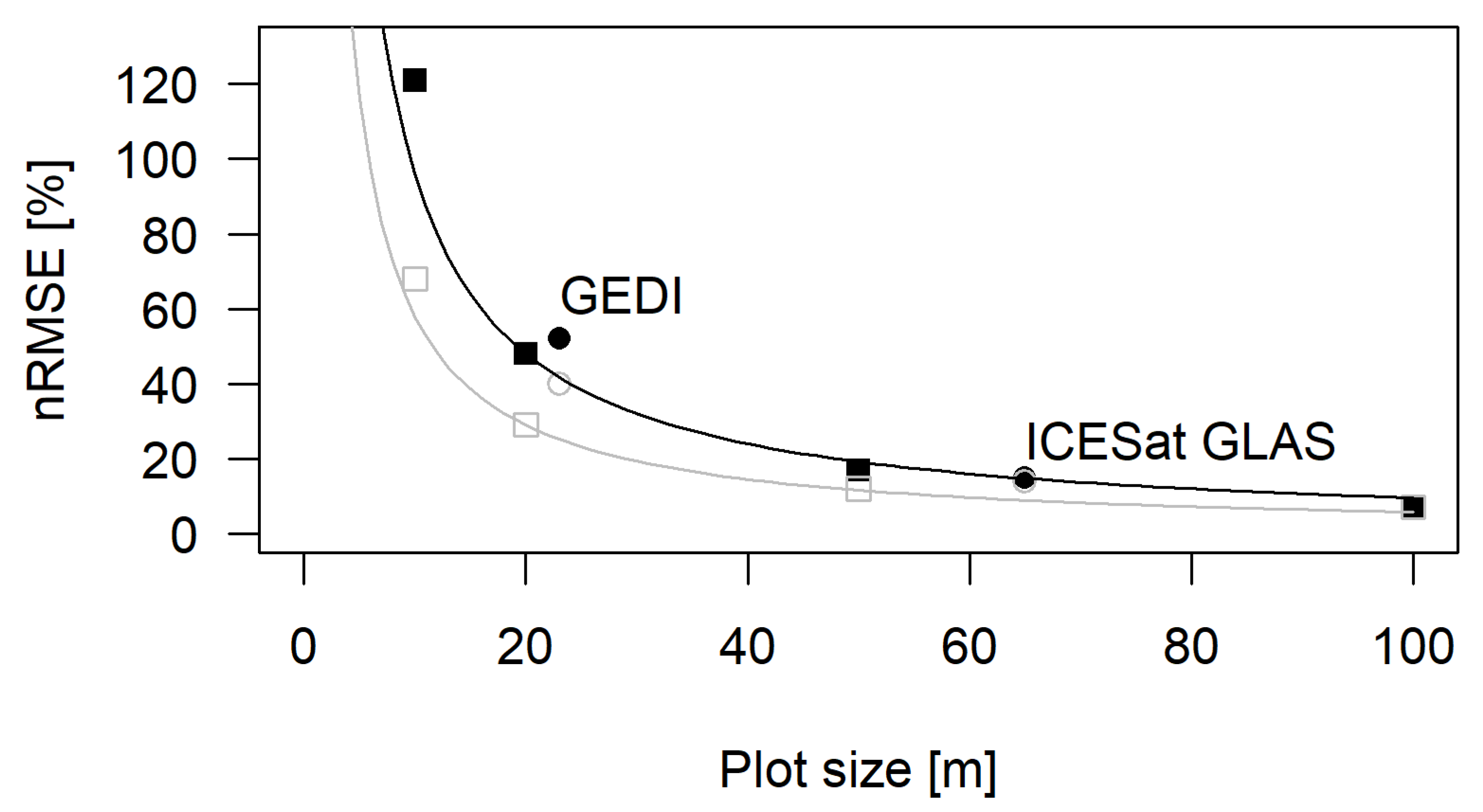
3.2. Biomass Estimation Results Using Square Plots and CHM
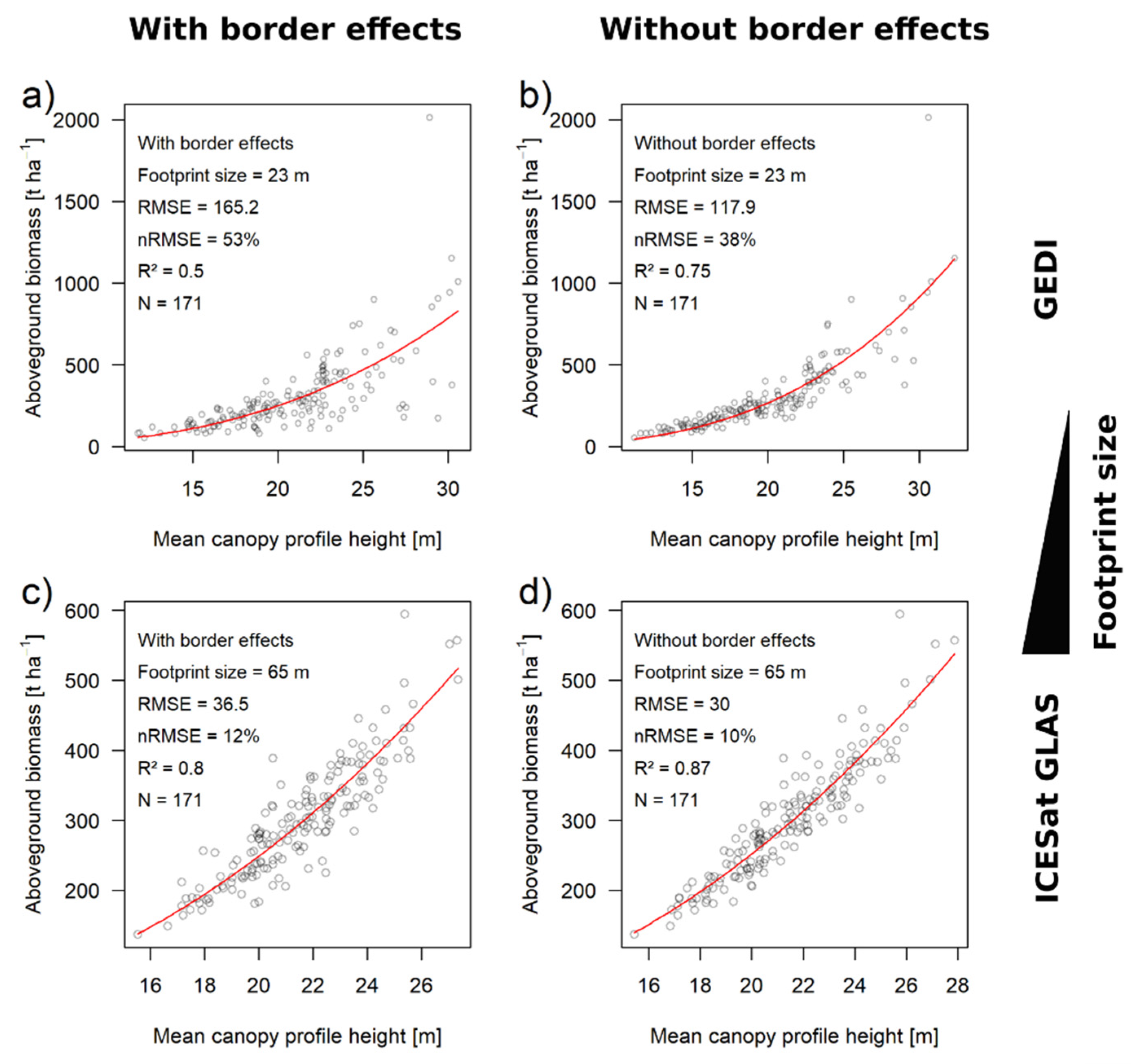
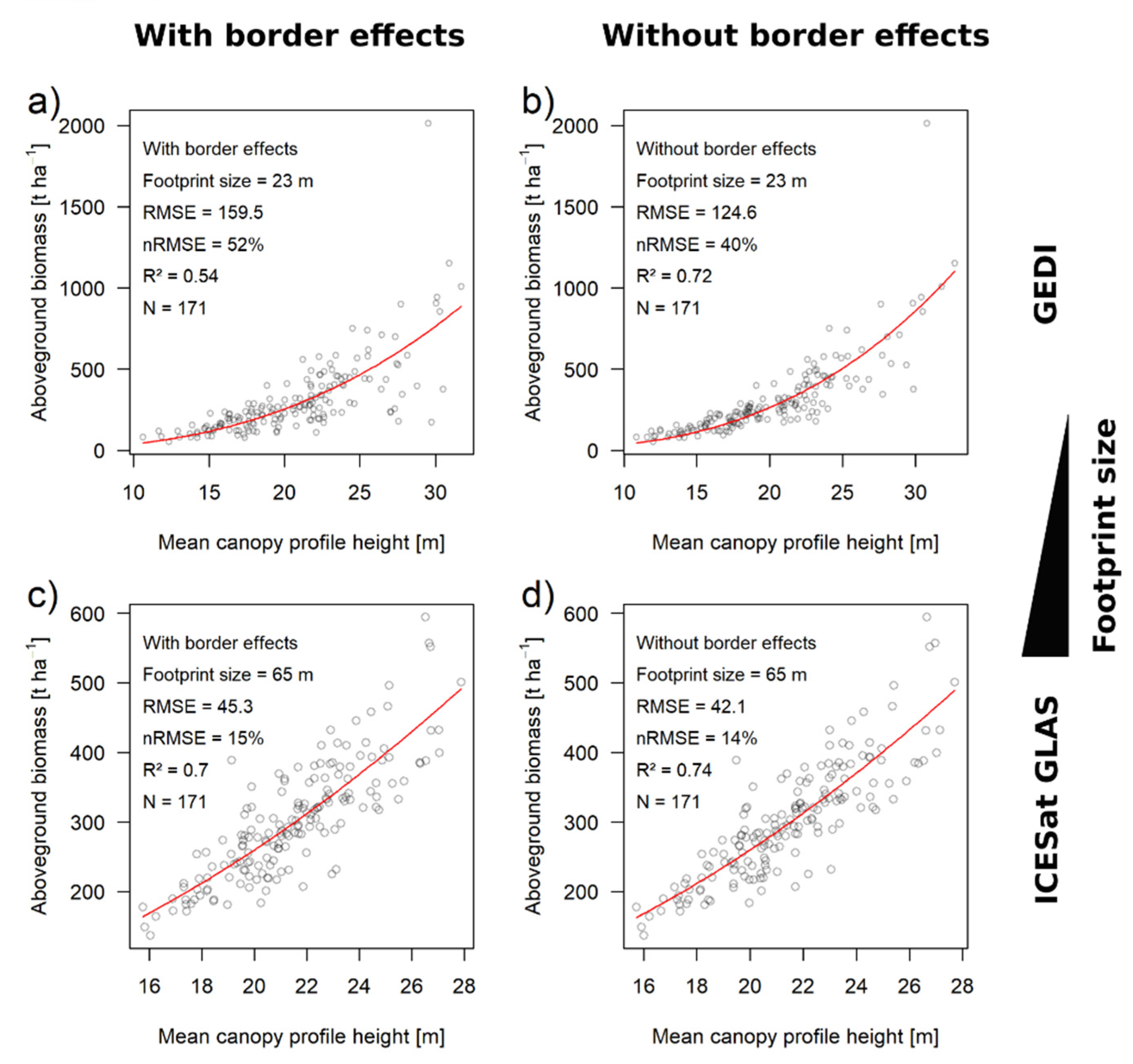
3.3. Biomass Estimation Results Using Large-Footprint Lidar
3.4. Effects of the Gaussian Energy Distribution
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

| Standard Deviation | ||||
|---|---|---|---|---|
| Plot Side Length [m] | 10 | 20 | 50 | 100 |
| CVin/CVp | 0.22 | 0.11 | 0.03 | 0.01 |
| CVin/CVt | 1 | 0.19 | 0.04 | 0.01 |
| CVout/CVt | 0.14 | 0.08 | 0.03 | 0.01 |
| CSin/plot area | 0.22 | 0.1 | 0.03 | 0.01 |
| CSout/plot area | 0.45 | 0.14 | 0.03 | 0.01 |
References
- Mitchard, E.T.A. The tropical forest carbon cycle and climate change. Nat. Cell Biol. 2018, 559, 527–534. [Google Scholar] [CrossRef]
- Lu, D.; Chen, Q.; Wang, G.; Liu, L.; Li, G.; Moran, E. A survey of remote sensing-based aboveground biomass estimation methods in forest ecosystems. Int. J. Digit. Earth 2014, 9, 63–105. [Google Scholar] [CrossRef]
- Goetz, S.; Dubayah, R. Advances in remote sensing technology and implications for measuring and monitoring forest carbon stocks and change. Carbon Manag. 2011, 2, 231–244. [Google Scholar] [CrossRef]
- Coomes, D.A.; Dalponte, M.; Jucker, T.; Asner, G.P.; Banin, L.F.; Burslem, D.F.; Lewis, S.L.; Nilus, R.; Phillips, O.L.; Phua, M.-H.; et al. Area-based vs tree-centric approaches to mapping forest carbon in Southeast Asian forests from airborne laser scanning data. Remote Sens. Environ. 2017, 194, 77–88. [Google Scholar] [CrossRef]
- Réjou-Méchain, M.; Muller-Landau, H.C.; Detto, M.; Thomas, S.C.; Le Toan, T.; Saatchi, S.S.; Barreto-Silva, J.S.; Bourg, N.A.; Bunyavejchewin, S.; Butt, N.; et al. Local spatial structure of forest biomass and its consequences for remote sensing of carbon stocks. Biogeosciences 2014, 11, 6827–6840. [Google Scholar] [CrossRef]
- Mascaro, J.; Detto, M.; Asner, G.P.; Muller-Landau, H.C. Evaluating uncertainty in mapping forest carbon with airborne LiDAR. Remote Sens. Environ. 2011, 115, 3770–3774. [Google Scholar] [CrossRef]
- Frazer, G.; Magnussen, S.; Wulder, M.; Niemann, K. Simulated impact of sample plot size and co-registration error on the accuracy and uncertainty of LiDAR-derived estimates of forest stand biomass. Remote Sens. Environ. 2011, 115, 636–649. [Google Scholar] [CrossRef]
- Brinck, K.; Fischer, R.; Groeneveld, J.; Lehmann, S.; De Paula, M.D.; Pütz, S.; Sexton, J.O.; Song, D.; Huth, A. High resolution analysis of tropical forest fragmentation and its impact on the global carbon cycle. Nat. Commun. 2017, 8, 14855. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Sun, G.; Dubayah, R.; Cook, B.; Montesano, P.; Ni, W.; Zhang, Z. Mapping biomass change after forest disturbance: Applying LiDAR footprint-derived models at key map scales. Remote Sens. Environ. 2013, 134, 319–332. [Google Scholar] [CrossRef]
- Zolkos, S.; Goetz, S.; Dubayah, R. A meta-analysis of terrestrial aboveground biomass estimation using lidar remote sensing. Remote Sens. Environ. 2013, 128, 289–298. [Google Scholar] [CrossRef]
- Knapp, N.; Fischer, R.; Huth, A. Linking lidar and forest modeling to assess biomass estimation across scales and disturbance states. Remote Sens. Environ. 2018, 205, 199–209. [Google Scholar] [CrossRef]
- Fischer, F.J.; Labrière, N.; Vincent, G.; Hérault, B.; Alonso, A.; Memiaghe, H.; Bissiengou, P.; Kenfack, D.; Saatchi, S.; Chave, J. A simulation method to infer tree allometry and forest structure from airborne laser scanning and forest inventories. Remote Sens. Environ. 2020, 251, 112056. [Google Scholar] [CrossRef]
- Durrett, R.; Levin, S.A. Stochastic spatial models: A user’s guide to ecological applications. Philos. Trans. R. Soc. B: Biol. Sci. 1994, 343, 329–350. [Google Scholar] [CrossRef]
- Schutz, B.E.; Zwally, H.J.; Shuman, C.A.; Hancock, D.; DiMarzio, J.P. Overview of the ICESat mission. Geophys. Res. Lett. 2005, 32, 14. [Google Scholar] [CrossRef]
- Los, S.O.; Rosette, J.A.B.; Kljun, N.; North, P.R.J.; Chasmer, L.; Suárez, J.C.; Hopkinson, C.; Hill, R.A.; Van Gorsel, E.; Mahoney, C.; et al. Vegetation height and cover fraction between 60° S and 60° N from ICESat GLAS data. Geosci. Model Dev. 2012, 5, 413–432. [Google Scholar] [CrossRef]
- Dubayah, R.; Blair, J.B.; Goetz, S.; Fatoyinbo, L.; Hansen, M.; Healey, S.; Hofton, M.; Hurtt, G.; Kellner, J.; Luthcke, S.; et al. The global ecosystem dynamics investigation: High-resolution laser ranging of the Earth’s forests and topography. Sci. Remote Sens. 2020, 1, 100002. [Google Scholar] [CrossRef]
- Condit, R. Tropical Forest Census Plots; Springer: Berlin, Germany; R. G. Landes Company: George Town, TX, USA, 1998; ISBN 3540641440. [Google Scholar]
- Hubbell, S.P.; Foster, R.B.; O’Brien, S.T.; Harms, K.E.; Condit, R.; Wechsler, B.; Wright, S.J.; De Lao, S.L. Light-gap disturbances, recruitment limitation, and tree diversity in a neotropical forest. Science 1999, 283, 554–557. [Google Scholar] [CrossRef] [PubMed]
- Condit, R.; Hubbell, S.P.; Foster, R.B. Mortality rates of 205 neotropical tree and shrub species and the impact of a severe drought. Ecol. Monogr. 1995, 65, 419–439. [Google Scholar] [CrossRef]
- Condit, R.; Lao, S.; Pérez, R.; Dolins, S.B.; Foster, R.; Hubbell, S. Barro colorado forest census plot data (version 2012). Cent. Trop. For. Sci. Databases 2013. [Google Scholar] [CrossRef]
- Condit, R.; Pérez, R.; Aguilar, S.; Lao, S.; Foster, R.; Hubbell, S. Complete Data from the Barro Colorado 50-ha Plot: 423617 Trees, 35 Years; Dryad: Walnut Creek, CA, USA, 2019. [Google Scholar] [CrossRef]
- Meyer, V.; Saatchi, S.S.; Chave, J.; Dalling, J.W.; Bohlman, S.A.; Fricker, G.A.; Robinson, C.B.; Neumann, M.M.; Hubbell, S.P. Detecting tropical forest biomass dynamics from repeated airborne lidar measurements. Biogeosciences 2013, 10, 5421–5438. [Google Scholar] [CrossRef]
- Lobo, E.; Dalling, J.W. Spatial scale and sampling resolution affect measures of gap disturbance in a lowland tropical forest: Implications for understanding forest regeneration and carbon storage. Proc. R. Soc. B: Boil. Sci. 2014, 281, 20133218. [Google Scholar] [CrossRef]
- Knapp, N.; Fischer, R.; Cazcarra-Bes, V.; Huth, A. Structure metrics to generalize biomass estimation from lidar across forest types from different continents. Remote Sens. Environ. 2020, 237, 111597. [Google Scholar] [CrossRef]
- Jucker, T.; Caspersen, J.; Chave, J.; Antin, C.; Barbier, N.; Bongers, F.; Dalponte, M.; Van Ewijk, K.Y.; Forrester, D.I.; Haeni, M.; et al. Allometric equations for integrating remote sensing imagery into forest monitoring programmes. Glob. Chang. Biol. 2016, 23, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Chave, J.; Réjou-Méchain, M.; Búrquez, A.; Chidumayo, E.; Colgan, M.S.; Delitti, W.B.; Duque, A.; Eid, T.; Fearnside, P.M.; Goodman, R.C.; et al. Improved allometric models to estimate the aboveground biomass of tropical trees. Glob. Chang. Biol. 2014, 20, 3177–3190. [Google Scholar] [CrossRef] [PubMed]
- CTFS Wood Density Database. Available online: http://ctfs.si.edu/Public/Datasets/CTFSWoodDensity/ (accessed on 24 May 2017).
- Roussel, J.-R.; Auty, D.; Coops, N.C.; Tompalski, P.; Goodbody, T.R.; Meador, A.S.; Bourdon, J.-F.; de Boissieu, F.; Achim, A. lidR: An R package for analysis of Airborne Laser Scanning (ALS) data. Remote Sens. Environ. 2020, 251, 112061. [Google Scholar] [CrossRef]
- A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2020.
- Hancock, S.; Armston, J.; Hofton, M.; Sun, X.; Tang, H.; Duncanson, L.I.; Kellner, J.R.; Dubayah, R. The GEDI simulator: A large-footprint waveform lidar simulator for calibration and validation of spaceborne missions. Earth Space Sci. 2019, 6, 294–310. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Ganguly, S.; Zhang, G.; Hofton, M.A.; Nelson, R.F.; Dubayah, R. Characterizing leaf area index (LAI) and Vertical Foliage Profile (VFP) characterizing leaf area index (LAI) and vertical foliage profile (VFP) over the United States. BGD Biogeosciences Discuss 2015, 12, 13675–13710. [Google Scholar] [CrossRef]
- Asner, G.P.; Powell, G.V.N.; Mascaro, J.; Knapp, D.E.; Clark, J.K.; Jacobson, J.; Kennedy-Bowdoin, T.; Balaji, A.; Paez-Acosta, G.; Victoria, E.; et al. High-resolution forest carbon stocks and emissions in the Amazon. Proc. Natl. Acad. Sci. USA 2010, 107, 16738–16742. [Google Scholar] [CrossRef]
- Hernández-Stefanoni, J.L.; Reyes-Palomeque, G.; Castillo-Santiago, M. Ángel; George-Chacón, S.P.; Huechacona-Ruiz, A.H.; Tun-Dzul, F.; Rondon-Rivera, D.; Dupuy, J.M. Effects of sample plot size and GPS location errors on aboveground biomass estimates from LiDAR in tropical dry forests. Remote Sens. 2018, 10, 1586. [Google Scholar] [CrossRef]
- Hernández-Stefanoni, J.L.; Dupuy, J.M.; Johnson, K.D.; Birdsey, R.; Tun-Dzul, F.; Peduzzi, A.; Caamal-Sosa, J.P.; Sánchez-Santos, G.; López-Merlín, D. Improving species diversity and biomass estimates of tropical dry forests using airborne LiDAR. Remote Sens. 2014, 6, 4741–4763. [Google Scholar] [CrossRef]
- Dalponte, M.; Jucker, T.; Liu, S.; Frizzera, L.; Gianelle, D. Characterizing forest carbon dynamics using multi-temporal lidar data. Remote Sens. Environ. 2019, 224, 412–420. [Google Scholar] [CrossRef]
- Ferraz, A.; Saatchi, S.; Mallet, C.; Meyer, V. Lidar detection of individual tree size in tropical forests. Remote Sens. Environ. 2016, 183, 318–333. [Google Scholar] [CrossRef]
- Ferraz, A.; Saatchi, S.S.; Longo, M.; Clark, D.B. Tropical tree size-frequency distributions from airborne lidar. Ecol. Appl. 2020, 30, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Rödig, E.; Knapp, N.; Fischer, R.; Bohn, F.J.; Dubayah, R.; Tang, H.; Huth, A. From small-scale forest structure to Amazon-wide carbon estimates. Nat. Commun. 2019, 10, 1–7. [Google Scholar] [CrossRef]
- Shugart, H.H.; Asner, G.P.; Fischer, R.; Huth, A.; Knapp, N.; Le Toan, T.; Shuman, J.K. Computer and remote-sensing infrastructure to enhance large-scale testing of individual-based forest models. Front. Ecol. Environ. 2015, 13, 503–511. [Google Scholar] [CrossRef]
- Fischer, R.; Knapp, N.; Bohn, F.; Shugart, H.H.; Huth, A. The relevance of forest structure for biomass and productivity in temperate forests: New perspectives for remote sensing. Surv. Geophys. 2019, 40, 709–734. [Google Scholar] [CrossRef]
- Fischer, F.J.; Maréchaux, I.; Chave, J. Improving plant allometry by fusing forest models and remote sensing. New Phytol. 2019, 223, 1159–1165. [Google Scholar] [CrossRef] [PubMed]

| Mean | Maximum | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Plot Side Length [m] | 10 | 20 | 50 | 100 | 10 | 20 | 50 | 100 | |
| CVin/CVp | 0.33 | 0.18 | 0.07 | 0.03 | 0.95 | 0.61 | 0.25 | 0.07 | |
| CVin/CVt | 0.64 | 0.21 | 0.07 | 0.04 | 16.23 | 1.44 | 0.31 | 0.07 | |
| CVout/CVt | 0.25 | 0.15 | 0.07 | 0.03 | 0.79 | 0.52 | 0.2 | 0.06 | |
| CSin/plot area | 0.3 | 0.16 | 0.07 | 0.03 | 1 | 0.66 | 0.26 | 0.06 | |
| CSout/plot area | 0.3 | 0.16 | 0.07 | 0.03 | 5.97 | 1.29 | 0.2 | 0.06 | |
| Mean | Maximum | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Plot Side Length [m] | 10 | 20 | 50 | 100 | 10 | 20 | 50 | 100 | |
| CVin/CVp | 0.37 | 0.2 | 0.08 | 0.04 | 0.98 | 0.77 | 0.29 | 0.09 | |
| CVin/CVt | 0.8 | 0.25 | 0.09 | 0.04 | 38.02 | 2.78 | 0.39 | 0.1 | |
| CVout/CVt | 0.27 | 0.17 | 0.08 | 0.04 | 0.85 | 0.56 | 0.27 | 0.07 | |
| CSin/plot area | 0.36 | 0.2 | 0.08 | 0.04 | 1 | 0.76 | 0.33 | 0.08 | |
| CSout/plot area | 0.36 | 0.2 | 0.08 | 0.04 | 10.25 | 1.86 | 0.32 | 0.07 | |
| Plot Size/Footprint | Perimeter-to-Area Ratio | With Border Effects | Without Border Effects | Contribution of Border Effects | |||||
|---|---|---|---|---|---|---|---|---|---|
| nRMSE | R² | nRMSE | R² | ΔnRMSE | Relative | ||||
| 10 m | 0.4 | 121% | 0.49 | 68% | 0.86 | 53% | 44% | ||
| 20 m | 0.2 | 48% | 0.66 | 29% | 0.87 | 19% | 40% | ||
| 50 m | 0.08 | 17% | 0.72 | 12% | 0.87 | 5% | 29% | ||
| 100 m | 0.04 | 7% | 0.86 | 7% | 0.85 | 0% | 0% | ||
| GEDI (23 m) | 0.174 | 52% | 0.54 | 40% | 0.72 | 12% | 23% | ||
| GLAS (65 m) | 0.062 | 15% | 0.7 | 14% | 0.74 | 1% | 6% | ||
| GEDI * (23 m) | 0.174 | 53% | 0.5 | 38% | 0.75 | 15% | 28% | ||
| GLAS * (65 m) | 0.062 | 12% | 0.8 | 10% | 0.87 | 2% | 17% | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knapp, N.; Huth, A.; Fischer, R. Tree Crowns Cause Border Effects in Area-Based Biomass Estimations from Remote Sensing. Remote Sens. 2021, 13, 1592. https://doi.org/10.3390/rs13081592
Knapp N, Huth A, Fischer R. Tree Crowns Cause Border Effects in Area-Based Biomass Estimations from Remote Sensing. Remote Sensing. 2021; 13(8):1592. https://doi.org/10.3390/rs13081592
Chicago/Turabian StyleKnapp, Nikolai, Andreas Huth, and Rico Fischer. 2021. "Tree Crowns Cause Border Effects in Area-Based Biomass Estimations from Remote Sensing" Remote Sensing 13, no. 8: 1592. https://doi.org/10.3390/rs13081592
APA StyleKnapp, N., Huth, A., & Fischer, R. (2021). Tree Crowns Cause Border Effects in Area-Based Biomass Estimations from Remote Sensing. Remote Sensing, 13(8), 1592. https://doi.org/10.3390/rs13081592