Inversion of Phytoplankton Pigment Vertical Profiles from Satellite Data Using Machine Learning
Abstract
1. Introduction
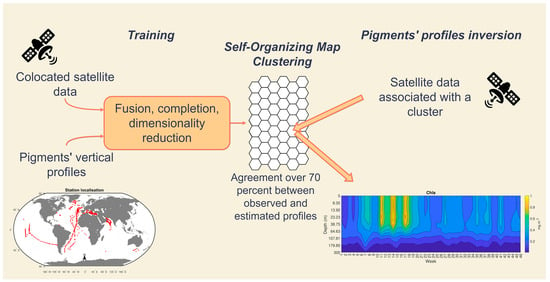
2. Materials and Methods
2.1. Data
2.1.1. Pigment Observations
2.1.2. Satellite Observations
2.1.3. Combined Dataset
2.2. Inverse Method: From Satellite Data to Vertical Profiles
2.2.1. Algorithms
2.2.2. Sat2profile Methodology
- Selecting an initial set of explanatory variables proposed by an expert.
- Completing the missing data occurring on the pigment observations using ITCOMPSOM.
- Applying a PCA to filter and compress the vertical profiles to be retrieved by Sat2Profile. During this phase, two hyper parameters are determined: the number of PCA () and the size of the map.
2.2.3. Methodological Workflow
Training Phase
Retrieval Phase
Cross-Validation of the Model
2.2.4. Test of Spatial and Temporal Coherence
3. Results
3.1. Parameters of the Method
3.2. Cross Validation Performance
3.3. Test Performance
3.4. Spatial and Temporal Coherence
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AVHRR | Advanced Very High-Resolution Radiometer |
| Chla | Chlorophyll-A |
| Chla_sat | Chlorophylle-A Satellite measured |
| DVChla | Divinyl Chlorophyll-A |
| ESA | European Space Agency |
| fucox | fucoxanthin |
| HPLC | High Performance Liquid Chromatography |
| ITCOMP-SOM | Iterative Completion Self Organizing Map |
| KDPAR | coefficient of attenuation of photosynthesis available radiance |
| KD490 | light coefficient of attenuation at 490 nm |
| MERIS | Medium Resolution Imaging Spectrometer |
| MODIS | Moderate Resolution Imaging Spectroradiometer |
| NOAA | National Oceanic and Atmospheric Administration |
| OLCI | Ocean and Land Colour Instrument |
| PCA | Principal Component Analysis |
| PAR | Photosynthesis available radiance |
| perid | peridinin |
| PFTs | Phytoplankton Functional Types |
| PSC | Phytoplankton Size Classes |
| RRS412 | Remote Sensing Reflectance at 412 nm |
| RRS443 | Remote Sensing Reflectance at 443 nm |
| RRS490 | Remote Sensing Reflectance at 490 nm |
| RRS555 | Remote Sensing Reflectance at 555 nm |
| SOM | Self Organizing Maps |
| SST | Sea Surface Temperature |
| VIIRS | Visible Infrared Imaging Radiometer Suite |
| zeax | zeaxanthin |
| ZEU | Depth of the euphotic layer |
| ZHL | Depth of the warmed layer |
| 19hex | 19’hexanoyloxyfucoxanthin |
References
- Turley, C.; Gattuso, J.P. Future biological and ecosystem impacts of ocean acidification and their socioeconomic-policy implications. Curr. Opin. Environ. Sustain. 2012, 4, 278–286. [Google Scholar] [CrossRef]
- Roessig, J.M.; Woodley, C.M.; Cech, J.J.; Hansen, L.J. Effects of global climate change on marine and estuarine fishes and fisheries. Rev. Fish Biol. Fish. 2004, 14, 251–275. [Google Scholar] [CrossRef]
- Harley, C.D.; Randall Hughes, A.; Hultgren, K.M.; Miner, B.G.; Sorte, C.J.; Thornber, C.S.; Rodriguez, L.F.; Tomanek, L.; Williams, S.L. The impacts of climate change in coastal marine systems. Ecol. Lett. 2006, 9, 228–241. [Google Scholar] [CrossRef] [PubMed]
- Macías, D.; Castilla-Espino, D.; García-del Hoyo, J.; Navarro, G.; Catalán, I.A.; Renault, L.; Ruiz, J. Consequences of a future climatic scenario for the anchovy fishery in the Alboran Sea (SW Mediterranean): A modeling study. J. Mar. Syst. 2014, 135, 150–159. [Google Scholar] [CrossRef]
- Gregg, W.W.; Rousseaux, C.S. Decadal trends in global pelagic ocean chlorophyll: A new assessment integrating multiple satellites, in situ data, and models. J. Geophys. Res. Ocean. 2014, 119, 5921–5933. [Google Scholar] [CrossRef]
- Kwiatkowski, L.; Torres, O.; Bopp, L.; Aumont, O.; Chamberlain, M.; Christian, J.R.; Dunne, J.P.; Gehlen, M.; Ilyina, T.; John, J.G.; et al. Twenty-first century ocean warming, acidification, deoxygenation, and upper-ocean nutrient and primary production decline from CMIP6 model projections. Biogeosciences 2020, 17, 3439–3470. [Google Scholar] [CrossRef]
- Meredith, M.; Sommerkorn, M.; Cassotta, S.; Derksen, C. Polar Regions. IPCC Special Report on the Ocean and Cryosphere in a Changing Climate; Pörtner, H.-O., Roberts, D.C., Masson-Delmotte, V., Zhai, P., Tignor, M., Poloczanska, E., Mintenbeck, K., Eds.; IPCC, WMO, UNEP: Geneva, Switzerland, 2019; pp. 1–173. [Google Scholar]
- Cavicchioli, R.; Ripple, W.J.; Timmis, K.N.; Azam, F.; Bakken, L.R.; Baylis, M.; Behrenfeld, M.J.; Boetius, A.; Boyd, P.W.; Classen, A.T.; et al. Scientists’ warning to humanity: Microorganisms and climate change. Nat. Rev. Microbiol. 2019, 17, 569–586. [Google Scholar] [CrossRef]
- Alvain, S.; Moulin, C.; Dandonneau, Y.; Bréon, F.M. Remote sensing of phytoplankton groups in case 1 waters from global SeaWiFS imagery. Deep Sea Res. Part I Oceanogr. Res. Pap. 2005, 52, 1989–2004. [Google Scholar] [CrossRef]
- Sathyendranath, S.; Aiken, J.; Alvain, S.; Barlow, R.; Bouman, H.; Bracher, A.; Brewin, R.; Bricaud, A.; Brown, C.; Ciotti, A.; et al. Phytoplankton Functional Types from Space; (Reports of the International Ocean-Colour Coordinating Group (IOCCG), 15); International Ocean-Colour Coordinating Group: Dartmouth, NS, Canada, 2014; pp. 1–156. [Google Scholar]
- El Hourany, R.; Abboud-Abi Saab, M.; Faour, G.; Aumont, O.; Crépon, M.; Thiria, S. Estimation of Secondary Phytoplankton Pigments From Satellite Observations Using Self-Organizing Maps (SOMs). J. Geophys. Res. Ocean. 2019, 124, 1357–1378. [Google Scholar] [CrossRef]
- El Hourany, R.; Abboud-abi Saab, M.; Faour, G.; Mejia, C.; Crépon, M.; Thiria, S. Phytoplankton diversity in the Mediterranean Sea from satellite data using self-organizing maps. J. Geophys. Res. Ocean. 2019, 124, 5827–5843. [Google Scholar] [CrossRef]
- Morel, A.; Berthon, J.F. Surface pigments, algal biomass profiles, and potential production of the euphotic layer: Relationships reinvestigated in view of remote-sensing applications. Limnol. Oceanogr. 1989, 34, 1545–1562. [Google Scholar] [CrossRef]
- Uitz, J.; Claustre, H.; Morel, A.; Hooker, S.B. Vertical distribution of phytoplankton communities in open ocean: An assessment based on surface chlorophyll. J. Geophys. Res. Ocean. 2006, 111. [Google Scholar] [CrossRef]
- Bracher, A.; Bouman, H.A.; Brewin, R.J.; Bricaud, A.; Brotas, V.; Ciotti, A.M.; Clementson, L.; Devred, E.; Di Cicco, A.; Dutkiewicz, S.; et al. Obtaining phytoplankton diversity from ocean color: A scientific roadmap for future development. Front. Mar. Sci. 2017, 4, 55. [Google Scholar] [CrossRef]
- Charantonis, A.; Badran, F.; Thiria, S. Retrieving the evolution of vertical profiles of Chlorophyll-a from satellite observations using Hidden Markov Models and Self-Organizing Topological Maps. Remote Sens. Environ. 2015, 163, 229–239. [Google Scholar] [CrossRef]
- Cortivo, F.D.; Chalhoub, E.S.; Velho, H.F.C.; Kampel, M. Chlorophyll profile estimation in ocean waters by a set of artificial neural networks. Comput. Assist. Methods Eng. Sci. 2017, 22, 63–88. [Google Scholar]
- Sauzède, R.; Claustre, H.; Jamet, C.; Uitz, J.; Ras, J.; Mignot, A.; D’Ortenzio, F. Retrieving the vertical distribution of chlorophyll a concentration and phytoplankton community composition from in situ fluorescence profiles: A method based on a neural network with potential for global-scale applications. J. Geophys. Res. Ocean. 2015, 120, 451–470. [Google Scholar] [CrossRef]
- Sammartino, M.; Marullo, S.; Santoleri, R.; Scardi, M. Modelling the vertical distribution of phytoplankton biomass in the Mediterranean Sea from satellite data: A neural network approach. Remote Sens. 2018, 10, 1666. [Google Scholar] [CrossRef]
- Sammartino, M.; Buongiorno Nardelli, B.; Marullo, S.; Santoleri, R. An Artificial Neural Network to Infer the Mediterranean 3D Chlorophyll-a and Temperature Fields from Remote Sensing Observations. Remote Sens. 2020, 12, 4123. [Google Scholar] [CrossRef]
- Farikou, O.; Sawadogo, S.; Niang, A.; Diouf, D.; Brajard, J.; Mejia, C.; Dandonneau, Y.; Gasc, G.; Crépon, M.; Thiria, S. Inferring the seasonal evolution of phytoplankton groups in the Senegalo-Mauritanian upwelling region from satellite ocean-color spectral measurements. J. Geophys. Res. Ocean. 2015, 120, 6581–6601. [Google Scholar] [CrossRef]
- Jouini, M.; Béranger, K.; Arsouze, T.; Beuvier, J.; Thiria, S.; Crépon, M.; Taupier-Letage, I. The Sicily Channel surface circulation revisited using a neural clustering analysis of a high-resolution simulation. J. Geophys. Res. Ocean. 2016, 121, 4545–4567. [Google Scholar] [CrossRef]
- Chapman, C.; Charantonis, A.A. Reconstruction of subsurface velocities from satellite observations using iterative self-organizing maps. IEEE Geosci. Remote Sens. Lett. 2017, 14, 617–620. [Google Scholar] [CrossRef]
- Reichstein, M.; Camps-Valls, G.; Stevens, B.; Jung, M.; Denzler, J.; Carvalhais, N. Deep learning and process understanding for data-driven Earth system science. Nature 2019, 566, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Peloquin, J.; Smith, W.O., Jr. The MAREDAT global database of high performance liquid chromatography marine pigment measurements. Earth Syst. Sci. Data 2013, 5, 109. [Google Scholar] [CrossRef]
- Pesant, S.; Not, F.; Picheral, M.; Kandels-Lewis, S.; Le Bescot, N.; Gorsky, G.; Iudicone, D.; Karsenti, E.; Speich, S.; Troublé, R.; et al. Open science resources for the discovery and analysis of Tara Oceans data. Sci. Data 2015, 2, 1–16. [Google Scholar] [CrossRef]
- Vidussi, F.; Claustre, H.; Manca, B.B.; Luchetta, A.; Marty, J.C. Phytoplankton pigment distribution in relation to upper thermocline circulation in the eastern Mediterranean Sea during winter. J. Geophys. Res. Ocean. 2001, 106, 19939–19956. [Google Scholar] [CrossRef]
- Hirata, T.; Aiken, J.; Hardman-Mountford, N.; Smyth, T.; Barlow, R. An absorption model to determine phytoplankton size classes from satellite ocean colour. Remote Sens. Environ. 2008, 112, 3153–3159. [Google Scholar] [CrossRef]
- Hirata, T.; Hardman-Mountford, N.; Brewin, R.; Aiken, J.; Barlow, R.; Suzuki, K.; Isada, T.; Howell, E.; Hashioka, T.; Noguchi-Aita, M.; et al. Synoptic relationships between surface Chlorophyll-a and diagnostic pigments specific to phytoplankton functional types. Biogeosciences 2011, 8, 311–327. [Google Scholar] [CrossRef]
- Jeffrey, S. Algal pigment systems. In Primary Productivity in the Sea; Springer: Berlin/Heidelberg, Germany, 1980; pp. 33–58. [Google Scholar]
- Jeffrey, S.; Hallegraeff, G. Chlorophyllase distribution in ten classes of phytoplankton: A problem for chlorophyll analysis. Mar. Ecol. Prog. Ser. 1987, 35, 293–304. [Google Scholar] [CrossRef]
- Wright, S.W.; Jeffrey, S. Fucoxanthin pigment markers of marine phytoplankton analysed by HPLC and HPTLC. Mar. Ecol. Prog. Ser. 1987, 38, 259–266. [Google Scholar] [CrossRef]
- Guillard, R.; Murphy, L.; Foss, P.; Liaaen-Jensen, S. Synechococcus spp. as likely zeaxanthin-dominant ultraphytoplankton in the North Atlantic 1. Limnol. Oceanogr. 1985, 30, 412–414. [Google Scholar] [CrossRef]
- Dandonneau, Y.; Deschamps, P.Y.; Nicolas, J.M.; Loisel, H.; Blanchot, J.; Montel, Y.; Thieuleux, F.; Bécu, G. Seasonal and interannual variability of ocean color and composition of phytoplankton communities in the North Atlantic, equatorial Pacific and South Pacific. Deep Sea Res. Part II Top. Stud. Oceanogr. 2004, 51, 303–318. [Google Scholar] [CrossRef]
- Mitchell, B.G.; Brody, E.A.; Holm-Hansen, O.; McClain, C.; Bishop, J. Light limitation of phytoplankton biomass and macronutrient utilization in the Southern Ocean. Limnol. Oceanogr. 1991, 36, 1662–1677. [Google Scholar] [CrossRef]
- Fenton, N.; Priddle, J.; Tett, P. Regional variations in bio-optical properties of the surface waters in the Southern Ocean. Antarct. Sci. 1994, 6, 443–448. [Google Scholar] [CrossRef]
- Arrigo, K.R.; Worthen, D.; Schnell, A.; Lizotte, M.P. Primary production in Southern Ocean waters. J. Geophys. Res. Ocean. 1998, 103, 15587–15600. [Google Scholar] [CrossRef]
- Mitchell, B.G.; Holm-Hansen, O. Bio-optical properties of Antarctic Peninsula waters: Differentiation from temperate ocean models. Deep Sea Res. Part A Oceanogr. Res. Pap. 1991, 38, 1009–1028. [Google Scholar] [CrossRef]
- Mitchell, B.G.; Holm-Hansen, O. Observations of modeling of the Antartic phytoplankton crop in relation to mixing depth. Deep Sea Res. Part A Oceanogr. Res. Pap. 1991, 38, 981–1007. [Google Scholar] [CrossRef]
- Mitchell, B.G. Predictive bio-optical relationships for polar oceans and marginal ice zones. J. Mar. Syst. 1992, 3, 91–105. [Google Scholar] [CrossRef]
- Korb, R.E.; Whitehouse, M.J.; Ward, P. SeaWiFS in the southern ocean: Spatial and temporal variability in phytoplankton biomass around South Georgia. Deep Sea Res. Part II Top. Stud. Oceanogr. 2004, 51, 99–116. [Google Scholar] [CrossRef]
- Hirawake, T.; Takao, S.; Horimoto, N.; Ishimaru, T.; Yamaguchi, Y.; Fukuchi, M. A phytoplankton absorption-based primary productivity model for remote sensing in the Southern Ocean. Polar Biol. 2011, 34, 291–302. [Google Scholar] [CrossRef]
- Dierssen, H.M.; Smith, R.C. Bio-optical properties and remote sensing ocean color algorithms for Antarctic Peninsula waters. J. Geophys. Res. Ocean. 2000, 105, 26301–26312. [Google Scholar] [CrossRef]
- Reynolds, R.A.; Stramski, D.; Mitchell, B.G. A chlorophyll-dependent semianalytical reflectance model derived from field measurements of absorption and backscattering coefficients within the Southern Ocean. J. Geophys. Res. Ocean. 2001, 106, 7125–7138. [Google Scholar] [CrossRef]
- Kahru, M.; Mitchell, B.G. Blending of ocean colour algorithms applied to the Southern Ocean. Remote Sens. Lett. 2010, 1, 119–124. [Google Scholar] [CrossRef]
- Casey, K.S.; Brandon, T.B.; Cornillon, P.; Evans, R. The past, present, and future of the AVHRR Pathfinder SST program. In Oceanography from Space; Springer: Berlin/Heidelberg, Germany, 2010; pp. 273–287. [Google Scholar]
- Saha, K.; Zhao, X.; Zhang, H.; Casey, K.; Zhang, D.; Baker-Yeboah, S.; Kilpatrick, K.; Evans, R.; Ryan, T.; Relph, J. AVHRR Pathfinder Version 5.3 Level 3 Collated (L3C) Global 4km Sea Surface Temperature for 1981-Present; NOAA National Centers for Environmental Information: Asheville, NC, USA, 2018. [Google Scholar]
- Belgrano, A.; Lima, M.; Stenseth, N.C. Nonlinear dynamics in marine-phytoplankton population systems. Mar. Ecol. Prog. Ser. 2004, 273, 281–289. [Google Scholar] [CrossRef]
- Kohonen, T. Essentials of the self-organizing map. Neural Netw. 2013, 37, 52–65. [Google Scholar] [CrossRef]
- Mwasiagi, J.I. Self Organizing Maps: Applications and Novel Algorithm Design; BoD–Books on Demand: Norderstedt, Germany, 2011. [Google Scholar]
- Meza-Padilla, R.; Enriquez, C.; Liu, Y.; Appendini, C.M. Ocean circulation in the western Gulf of Mexico using self-organizing maps. J. Geophys. Res. Ocean. 2019, 124, 4152–4167. [Google Scholar] [CrossRef]
- Jouini, M.; Lévy, M.; Crépon, M.; Thiria, S. Reconstruction of satellite chlorophyll images under heavy cloud coverage using a neural classification method. Remote Sens. Environ. 2013, 131, 232–246. [Google Scholar] [CrossRef]
- Charantonis, A.A.; Testor, P.; Mortier, L.; D’ortenzio, F.; Thiria, S. Completion of a sparse GLIDER database using multi-iterative Self-Organizing Maps (ITCOMP SOM). Procedia Comput. Sci. 2015, 51, 2198–2206. [Google Scholar] [CrossRef]
- Ilin, A.; Raiko, T. Practical approaches to principal component analysis in the presence of missing values. J. Mach. Learn. Res. 2010, 11, 1957–2000. [Google Scholar]
- Bracher, A.; Xi, H.; Dinter, T.; Mangin, A.; Strass, V.; Von Appen, W.J.; Wiegmann, S. High resolution water column phytoplankton composition across the Atlantic Ocean from ship-towed vertical undulating radiometry. Front. Mar. Sci. 2020, 7, 235. [Google Scholar] [CrossRef]
- Letelier, R.M.; Bidigare, R.R.; Hebel, D.V.; Ondrusek, M.; Winn, C.; Karl, D.M. Temporal variability of phytoplankton community structure based on pigment analysis. Limnol. Oceanogr. 1993, 38, 1420–1437. [Google Scholar] [CrossRef]
- Siokou-Frangou, I.; Christaki, U.; Mazzocchi, M.G.; Montresor, M.; Ribera d’Alcalá, M.; Vaqué, D.; Zingone, A. Plankton in the open Mediterranean Sea: A review. Biogeosciences 2010, 7, 1543–1586. [Google Scholar] [CrossRef]
- Quere, C.L.; Harrison, S.P.; Colin Prentice, I.; Buitenhuis, E.T.; Aumont, O.; Bopp, L.; Claustre, H.; Cotrim Da Cunha, L.; Geider, R.; Giraud, X.; et al. Ecosystem dynamics based on plankton functional types for global ocean biogeochemistry models. Glob. Chang. Biol. 2005, 11, 2016–2040. [Google Scholar] [CrossRef]
- Rumyantseva, A.; Henson, S.; Martin, A.; Thompson, A.F.; Damerell, G.M.; Kaiser, J.; Heywood, K.J. Phytoplankton spring bloom initiation: The impact of atmospheric forcing and light in the temperate North Atlantic Ocean. Prog. Oceanogr. 2019, 178, 102202. [Google Scholar] [CrossRef]
- Barton, A.D.; Lozier, M.S.; Williams, R.G. Physical controls of variability in N orth A tlantic phytoplankton communities. Limnol. Oceanogr. 2015, 60, 181–197. [Google Scholar] [CrossRef]
- Kheireddine, M.; Ouhssain, M.; Claustre, H.; Uitz, J.; Gentili, B.; Jones, B.H. Assessing pigment-based phytoplankton community distributions in the Red Sea. Front. Mar. Sci. 2017, 4, 132. [Google Scholar] [CrossRef]
- Pearman, J.K.; Ellis, J.; Irigoien, X.; Sarma, Y.; Jones, B.H.; Carvalho, S. Microbial planktonic communities in the Red Sea: High levels of spatial and temporal variability shaped by nutrient availability and turbulence. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef]








| Pigment | Missing Data (%) |
|---|---|
| Chla | 30 |
| DVChla | 48 |
| 19hex | 32 |
| fucox | 30 |
| perid | 32 |
| zeax | 40 |
| Satellite data | 70 |
| Pigment | RMSE (mg m) | |
|---|---|---|
| Chl-A | 0.70 | 0.181 |
| DVChl-A | 0.78 | 0.016 |
| 19-Hex | 0.64 | 0.032 |
| Fucox | 0.74 | 0.035 |
| Perid | 0.53 | 0.005 |
| Zeax | 0.73 | 0.014 |
| Mean Spearman Correlation | Mean | Mean RMSE (mg m) | Mean RMSE (% of Mean Concentration) | Mean Concentration (mg m) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Without PCA | With PCA | Without PCA | With PCA | Without PCA | With PCA | Without PCA | With PCA | ||
| Chla | 0.65 | 0.81 | 0.56 | 0.81 | 0.083 | 0.036 | 36.4 | 15.8 | 0.2280 |
| DVChla | 0.475 | 0.79 | 0.42 | 0.68 | 0.011 | 0.006 | 43.5 | 23.7 | 0.0253 |
| 19hex | 0.62 | 0.82 | 0.53 | 0.81 | 0.02 | 0.008 | 35.4 | 14.2 | 0.0565 |
| fucox | 0.52 | 0.84 | 0.4 | 0.83 | 0.012 | 0.005 | 40.3 | 16.8 | 0.0298 |
| perid | 0.42 | 0.78 | 0.34 | 0.76 | 0.002 | 0.001 | 45.5 | 22.7 | 0.0044 |
| zeax | 0.59 | 0.77 | 0.57 | 0.81 | 0.01 | 0.005 | 30.2 | 15.1 | 0.0331 |
| PCA | SOM | |||
|---|---|---|---|---|
| Mean RMSE (mg m) | Mean RMSE (% of the Mean Concentration) | Mean RMSE (mg m) | Mean RMSE (% of the mean Concentration) | |
| Chla | 0.046 | 20.2 | 0.036 | 15.8 |
| DVChla | 0.006 | 23.7 | 0.006 | 23.7 |
| 19hex | 0.011 | 19.5 | 0.008 | 14.2 |
| Fucox | 0.005 | 16.8 | 0.005 | 16.8 |
| Perid | 0.001 | 22.7 | 0.001 | 22.7 |
| Zeax | 0.005 | 15.1 | 0.005 | 15.1 |
| Mean Spearman Coefficient | Mean | Mean RMSE (mg m) | Mean RMSE (% of Mean Concentration) | |
|---|---|---|---|---|
| Chla | 0.75 | 0.74 | 0.042 | 18.4 |
| DVChla | 0.74 | 0.65 | 0.012 | 47.4 |
| 19hex | 0.78 | 0.74 | 0.008 | 14.2 |
| fucox | 0.82 | 0.79 | 0.003 | 10.1 |
| perid | 0.72 | 0.72 | 0.001 | 22.7 |
| zeax | 0.80 | 0.86 | 0.007 | 21.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puissant, A.; El Hourany, R.; Charantonis, A.A.; Bowler, C.; Thiria, S. Inversion of Phytoplankton Pigment Vertical Profiles from Satellite Data Using Machine Learning. Remote Sens. 2021, 13, 1445. https://doi.org/10.3390/rs13081445
Puissant A, El Hourany R, Charantonis AA, Bowler C, Thiria S. Inversion of Phytoplankton Pigment Vertical Profiles from Satellite Data Using Machine Learning. Remote Sensing. 2021; 13(8):1445. https://doi.org/10.3390/rs13081445
Chicago/Turabian StylePuissant, Agathe, Roy El Hourany, Anastase Alexandre Charantonis, Chris Bowler, and Sylvie Thiria. 2021. "Inversion of Phytoplankton Pigment Vertical Profiles from Satellite Data Using Machine Learning" Remote Sensing 13, no. 8: 1445. https://doi.org/10.3390/rs13081445
APA StylePuissant, A., El Hourany, R., Charantonis, A. A., Bowler, C., & Thiria, S. (2021). Inversion of Phytoplankton Pigment Vertical Profiles from Satellite Data Using Machine Learning. Remote Sensing, 13(8), 1445. https://doi.org/10.3390/rs13081445