Abstract
Endmember estimation plays a key role in hyperspectral image unmixing, often requiring an estimation of the number of endmembers and extracting endmembers. However, most of the existing extraction algorithms require prior knowledge regarding the number of endmembers, being a critical process during unmixing. To bridge this, a new maximum distance analysis (MDA) method is proposed that simultaneously estimates the number and spectral signatures of endmembers without any prior information on the experimental data containing pure pixel spectral signatures and no noise, being based on the assumption that endmembers form a simplex with the greatest volume over all pixel combinations. The simplex includes the farthest pixel point from the coordinate origin in the spectral space, which implies that: (1) the farthest pixel point from any other pixel point must be an endmember, (2) the farthest pixel point from any line must be an endmember, and (3) the farthest pixel point from any plane (or affine hull) must be an endmember. Under this scenario, the farthest pixel point from the coordinate origin is the first endmember, being used to create the aforementioned point, line, plane, and affine hull. The remaining endmembers are extracted by repetitively searching for the pixel points that satisfy the above three assumptions. In addition to behaving as an endmember estimation algorithm by itself, the MDA method can co-operate with existing endmember extraction techniques without the pure pixel assumption via generalizing them into more effective schemes. The conducted experiments validate the effectiveness and efficiency of our method on synthetic and real data.
1. Introduction
Remotely sensed hyperspectral imaging (HSI) [1] provides a significant amount of information regarding different materials on the Earth’s surface, capturing their reflectance behavior in the presence of solar radiation (which depends on their chemical composition and physical structure) by measuring the degree of absorption along the wavelengths of the electromagnetic spectrum (usually focused on the visible, near infrared (NIR), and shortwave infrared (SWIR) spectrum [2]), in hundreds of narrow and continuous spectral bands [3]. As a result, each pixel contains the representation of a spectral signature, which is unique for each observed material. However, one typical problem that is associated with HSI data is its low spatial resolution [4], which inevitably leads to mixed pixels. A mixed pixel covers several types of materials, which are usually composed of macroscopic objects, such as water, soil, vegetation, or buildings [5,6], and the corresponding spectrum being a mixture of several ground cover spectra.
A challenge of the mixed pixel phenomenon is how to accurately identify the prime materials that compose it, determining the original spectral signatures and measuring the proportions in which they are present [7]. These principal constituent spectra are known as endmembers, and obtaining them is a hard and ill-posed problem, where the reflectance pattern is (in general) not known a priori. Additionally, the sensor inaccuracies and atmospheric/light conditions (which introduce an important amount of noise into the spectra), and the high spectral-dimensionality hinders the endmember extraction task [8], due to the high intra-class variability and inter-class similarity of the data [9]. Spectral unmixing is a significant technique for addressing this challenge [10,11,12,13,14]. In the available literature, spectral unmixing techniques [15,16,17] have been comprehensively investigated for the purpose of extracting endmembers and estimating their corresponding abundances, allowing for the processing of HSI scenes at the sub-pixel scale [18]. In fact, these techniques decompose each mixed pixel into a proportional composition of endmembers, where the constituent proportion with respect to different types of materials [19] for each pixel is defined as the abundance.
A significant challenge for existing spectral unmixing techniques is how to accurately extract endmembers [20,21,22,23,24] from remotely sensed HSI data. This is normally achieved by two seemingly independent, but, in fact, highly correlated procedures: (i) determining the number of endmembers, and (ii) extracting their spectral signatures. Because the accuracy of determining the number of endmembers has large influence on the endmember extraction step, it is natural to consider developing methods to essentially integrate both of the techniques seamlessly into an overall procedure of endmember estimation. This observation forms the motivation of our work.
2. Literature Review
In this paper, we propose the maximum distance analysis (MDA) method as a new unmixing framework that simultaneously counts and extracts endmembers without any prior knowledge. To introduce the proposed method in a detailed and comprehensive way, in Section 2.1 and Section 2.2, we review the literature regarding counting and extracting endmembers, respectively. In Section 2.3, we briefly describe the major contributions of our work in terms of presenting a new endmember estimation framework that simultaneously counts the number of endmembers and extracts their spectral signatures.
2.1. Endmember Counting
Estimating the number of endmembers is regarded as the first step of the overall endmember estimation task. The number of endmembers is often unavailable in an arbitrary HSI scene. In this scenario, most of the existing endmember extraction techniques cannot properly extract endmembers if the number of endmembers is not accurately determined. The limitation regarding an unknown number of endmembers hinders most existing endmember extraction techniques from an operational viewpoint. To address this limitation, several algorithms have been developed for counting the number of endmembers. Particularly, information theory-based algorithms, eigenvalue thresholding algorithms, and geometry characterization algorithms are three main families of techniques for estimating the number of endmembers [25]. A brief summary of each family of algorithms is provided below.
- The first family includes some kind of criteria information, such as theoretic criteria that are based on minimum description length [26,27], Akaike’s information criterion [28], and Bayesian information criterion [29]. Furthermore, different models have been developed for encoding a negative data log-likelihood term and a penalty term. An accurate estimation of the number of endmembers is expected to be obtained when the model achieves a global optimum. These strategies rely on the empirical configuration of specific mixed models or likelihood functions, and improper configurations will cause estimation errors of the number of endmembers.
- The second family is related to a thresholding scheme, in which a threshold is applied to the eigen-decomposition results from subspace analysis. Eigenvalue thresholding algorithms include principal component analysis (PCA) [30], hyperspectral signal subspace by minimum error (Hysime) [31], and the so-called Harsanyi-Farrand-Chang (HFC) method [32], coupled with its noise-whitened implementation (NWHFC) [33]. The PCA-based approaches aim to characterize a cutoff gap between the eigenvalues that are caused by signals and noise. However, these approaches will provide an incorrect estimation of the number of endmembers if the variation between the two eigenvalues is negligible. The Hysime approach conducts spectrum noise characterization and noise covariance estimation, and it requires high computational complexity. The HFC approach requires a fixed false alarm probability, which affects the estimated number of endmembers.
- The third family includes the geometry-based estimation of the number of endmembers (GENE), which includes the convex hull (GENE-CH) algorithm and affine hull (GENE-AH) algorithm [34]. Both GENE-CH and the GENE-AH utilize data geometry and exploit the fact that all the observed pixel vectors should lie in the convex hull (CH) and affine hull (AH) of the endmember signatures, respectively. Specifically, GENE algorithms operate along with an endmember extraction algorithm (EEA). In this scenario, a maximum hull volume would stop the EEA from extracting the next endmember signature. Therefore, the GENE algorithms depend on the effectiveness of the EEA used, and different endmember extraction algorithms (EEAs) cause different accuracies for counting endmembers.
2.2. Endmember Extraction
The subsequent step of endmember estimation is to extract endmembers (assuming that the number of endmembers has been previously determined). Linear unmixing assumes that all of the pixels are a linear combination of the endmember signatures present in the scene, weighted by their respective fractional abundances. The related techniques include two families, i.e., statistical algorithms and geometrical algorithms. Statistical algorithms address endmember extraction as an inference problem that is formulated under the Bayesian framework, whereas geometrical algorithms exploit the fact that the spectral vectors (under the linear mixing model) lie in a simplex whose vertices correspond to the endmembers. In our work, we focus on the geometrical approach to endmember extraction. Geometrical algorithms try to find a simplex set whose vertices correspond to the endmembers. As such, we extract endmembers by identifying the vertices of the aforementioned simplex. The main ideas that have been presented in recent research under this framework belong to two different groups. Pure pixel-based algorithms [9] presume that the image scene contains at least one pure pixel per endmember. Even more recently, some algorithms [35] dropped this assumption by assuming that no pure pixel points may be present in real image scenes. In the following, we outline these two types of algorithms.
- If there is at least one pure pixel for per endmember, then EEAs search for the spectral vectors in the data set that corresponds to the vertices of the data simplex. Typical algorithms that are based on the pure pixel assumption are pixel purity index (PPI) [36], N-FINDR [37,38], and vertex component analysis (VCA) [39], among others. The PPI algorithm projects all of the observed pixels onto randomly generated unit-norm vectors, and it records the number of times (i.e., scores) that the value of each projected pixel has an extreme value (either minimum or maximum projection value). Then, the endmembers are those pixels with the highest scores. The N-FINDR algorithm and its derivatives search for a simplex with the greatest volume over all pixel combinations, and the vertices of the simplex correspond to the endmembers. The VCA algorithm iteratively projects data onto a direction orthogonal to the subspace that is spanned by the endmembers already determined, and the new endmember is the pixel with the extreme value of the projected data. It is worth noting that PPI, N-FINDR, and VCA require the number of endmembers in advances to perform endmember extraction.
- If the pure pixel assumption is not satisfied (this is a more realistic scenario, since HSI data are often dominated by highly mixed pixel [9]), the endmember extraction process is a rather intractable task. The difficulty is that the HSI data set may not contain any endmembers or at least some of them. Some popular algorithms that are implemented under this assumption are the minimum volume constrained nonnegative matrix factorization (MVCNMF) [40], minimum-volume enclosing simplex (MVES) [41], minimum volume simplex analysis (MVSA) [42,43], etc. MVCNMF adopts constrained nonnegative matrix factorization, together with a volume-based constraint, to decompose mixed pixels in multispectral and hyperspectral remote sensing images. MVES finds a simplex containing all of the dimensionally-reduced pixels by minimizing the simplex volume subject to the constraint. MVSA fits a minimum volume simplex to the hyperspectral data, constraining the abundance fractions to belong to the probability simplex. Specifically, MVCNMF, MVES, and MVSA also require the number of endmembers when they implement endmember extraction.
2.3. From the Literature to Our Contributions
As reviewed in Section 2.1 and Section 2.2, most of the existing techniques for estimating the number of endmembers and extracting their signatures are performed as two independent procedures in the literature. Most existing EEAs require prior knowledge regarding the number of endmembers, but the algorithms for counting endmembers tend to be characterized by independent methods that may not seamlessly benefit the EEAs. To bridge these two seemingly independent (but, in fact, highly related) procedures, we establish a unifying endmember estimation framework. The contributions of our work are two-fold.
- The major contribution of our work is a novel endmember estimation method, referred to as maximum distance analysis (MDA), which accomplishes the overall endmember estimation task (that normally comprises of two independent steps, i.e., determining the number of endmembers and extracting their endmember signatures). Our newly proposed MDA method sequentially identifies pixel points as endmembers that are farthest from specific pixel point, line, plane, and affine hulls. The sequential endmember extraction terminates when the maximum distance between all the pixel points and the affine hull [41,44] formed by all extracted endmembers is zero. Our proposed MDA method does not require knowing the number of endmembers. It only involves straightforward vector-based computations, keeping its computational complexity very low. Moreover, our proposed MDA method does not use dimension reduction.
- A second major contribution of our work is that the proposed MDA method provides an effective way for generalizing most existing EEAs that require prior knowledge of the number of endmembers into more accurate ones without the requirement. We use the MVSA as a special case to demonstrate the generalization strategy that is based on the MDA. Specifically, MVSA requires the number of endmembers and uses the traditional VCA method as an initialization method for extracting endmembers. Therefore, our proposed MDA framework is not only capable of simultaneously counting and extracting endmembers by itself without any prior knowledge, but it also provides a general framework for developing new endmember extraction schemes that are based on arbitrary existing endmember extraction methods without the pure pixel assumption.
3. Maximum Distance Analysis
3.1. Extracting Endmember Signatures
Let denote a HSI dataset with L bands and N pixels, where the ith pixel represents an L-dimensional vector. From a geometric point of view, we know that endmembers are supposed to form a simplex with the greatest volume over all pixel combinations, and they are distributed at the outermost region of all pixels to enclose them. Therefore, if one pixel point is a candidate to be an endmember, then it must satisfy two conditions: (1) the pixel point is an essential component of the simplex with the greatest volume over all pixel combinations and (2) the pixel point is distributed in the outermost region of all pixels. Some of the EEAs utilize the idea of a simplex to search for some pixel points far away from each other as endmembers, and the operations inevitably render considerable computational overloads and make EEAs inefficient for tackling large-size remote sensing HSI scenes. To address this problem, we give some definitions regarding distances to effectively and efficiently extract endmembers, where the task of extracting endmembers does not require any prior knowledge regarding the number of endmembers.
Definition 1.
We define the farthest pixel point from the coordinate origin in the pixel space as the first endmember.
The Euclidean distance computes the distance between the ith and jth hyperspectral pixel points in the pixel space
We compute the distance between each pixel point and the coordinate origin by using Equation (1) to obtain the farthest pixel point from the coordinate origin, computing as a result of the first endmember. In this sense, this first point easily satisfies the two conditions to be an endmember. Specifically, the distance calculation in Equation (1) may need to be adapted if the hyperspectral data are nonlinearly mixed or includes noise.
Definition 2.
We define the farthest pixel point from the first extracted endmember as the second endmember.
We compute the distance between each pixel point and the first extracted endmember through Equation (1) to obtain the farthest pixel point from the first endmember, getting, as a result, the second endmember. We observe that the second extracted endmember also satisfies the two conditions to be considered as endmember.
Definition 3.
We define the farthest pixel point from the line that is defined by the first and second extracted endmembers as the third endmember.
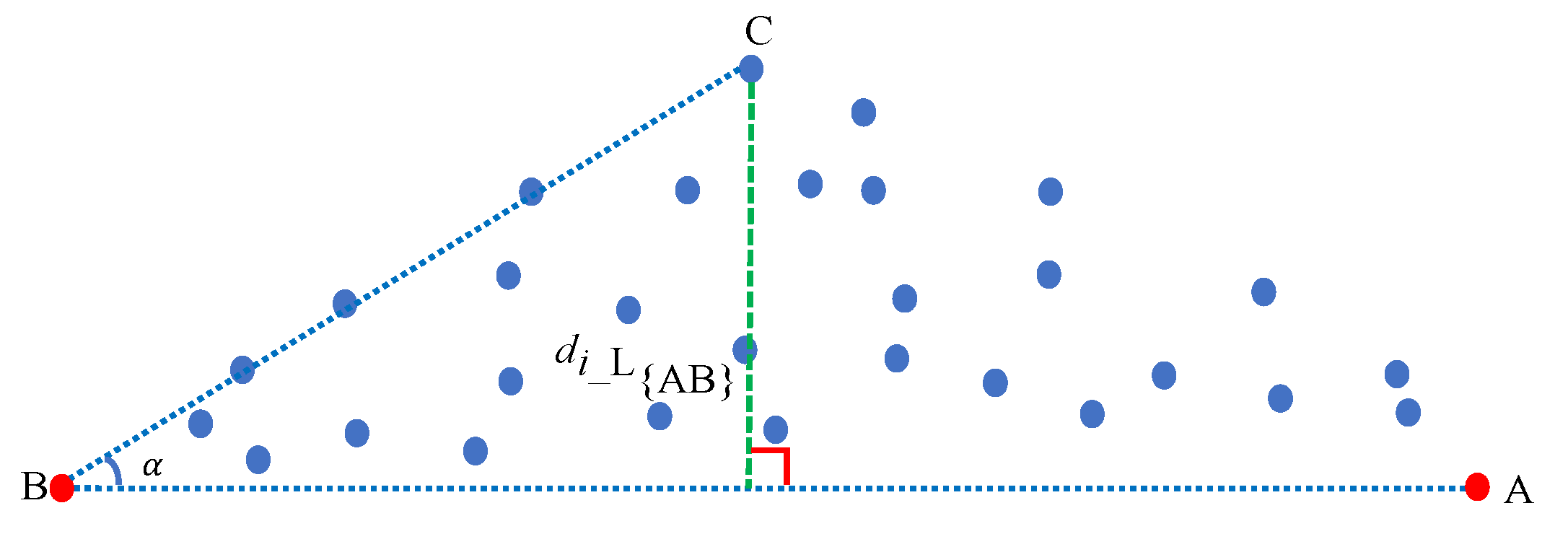
Assuming that we have acquired the first endmember A and the second endmember B, as shown in Figure 1, we compute the distance between any pixel point i and the line to obtain the third endmember. Subsequently, the angle between vectors and is trigonometrically computed by Equation (2)
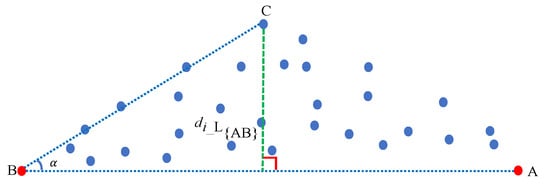
Figure 1.
Schematic diagram illustrating how to extract an endmember (from two previously available ones) by using our maximum distance analysis (MDA) method. The farthest pixel point from the line is the third endmember. The red dots represent endmembers that have been previously extracted, and the blue dots denote the pixel points in the pixel space.
In this sense, we obtain the index of the third endmember by maximizing the distance with
and we use the index to extract the third endmember C. Moreover, the third extracted endmember also satisfies the two conditions in order to be considered as an endmember.
Definition 4.
We define the farthest pixel point from the plane that is formed by the first, second, and third extracted endmembers as the fourth endmember.
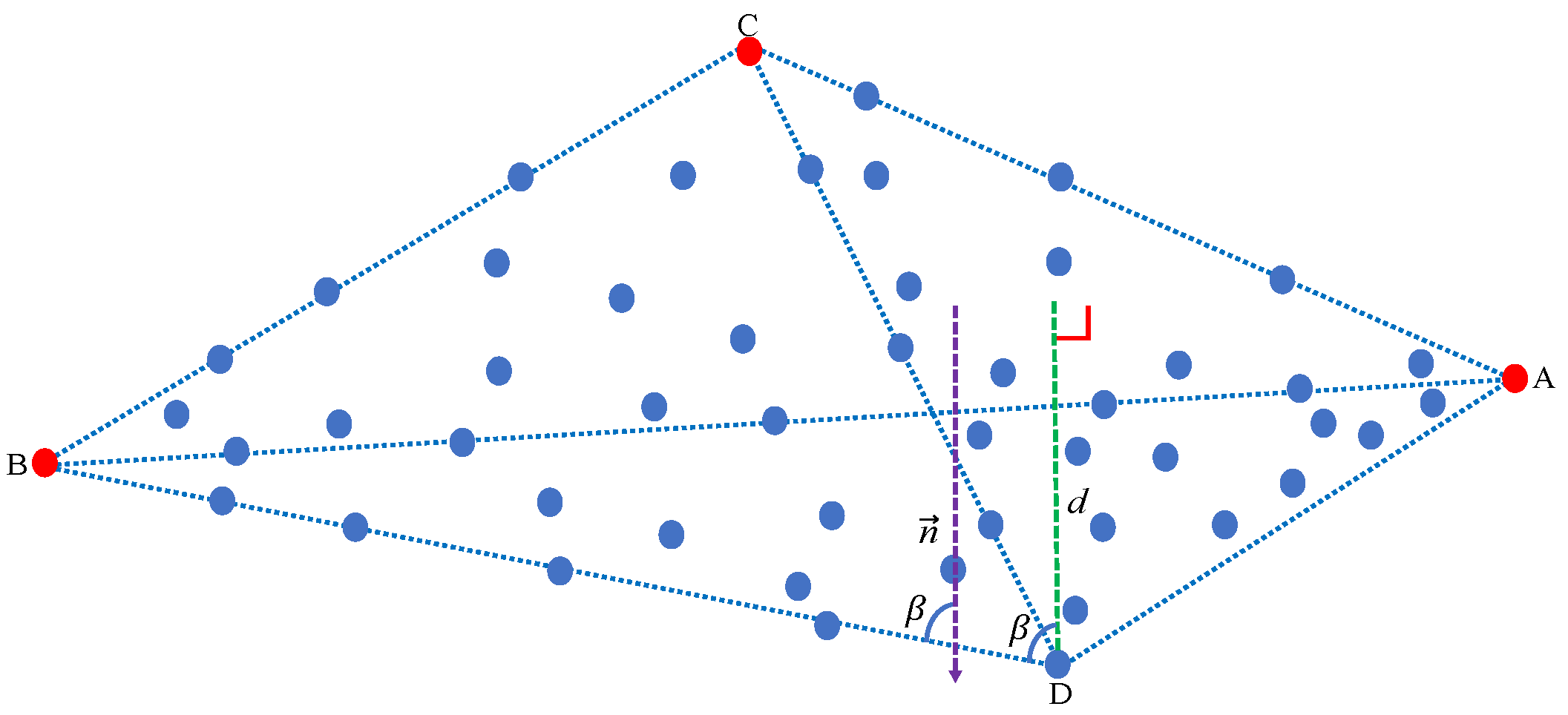
Assuming that the three extracted endmembers A, B, and C form a plane , as shown in Figure 2, we compute the distance between any pixel point i and the plane to obtain the fourth endmember. O is the coordinate origin in the pixel space and is the normal vector of the plane . However, in our work, we get the normal vector by a unique strategy. First, we get by
In addition, we know
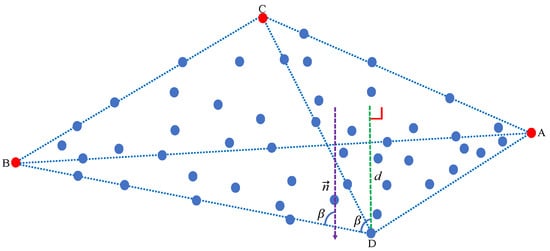
Figure 2.
Schematic diagram illustrating how to extract an endmember (from three previously available ones) by using our maximum distance analysis (MDA) method. The farthest pixel point from the plane is the fourth endmember.
Based on Equations (5) and (6), we have the expressions
We know that A, B, and C are three different endmembers, which mean that , , are not collinear with each other. Therefore, is not only the normal vector of the plane , but also orthogonal to , , . Therefore, we define matrix , where N is the number of pixels. We obtain the normal vector by addressing
The angle between vectors and is trigonometrically computed by
Therefore, the distance is computed by
where symbol denotes the absolute value of . With this, we obtain the index of the fourth endmember by
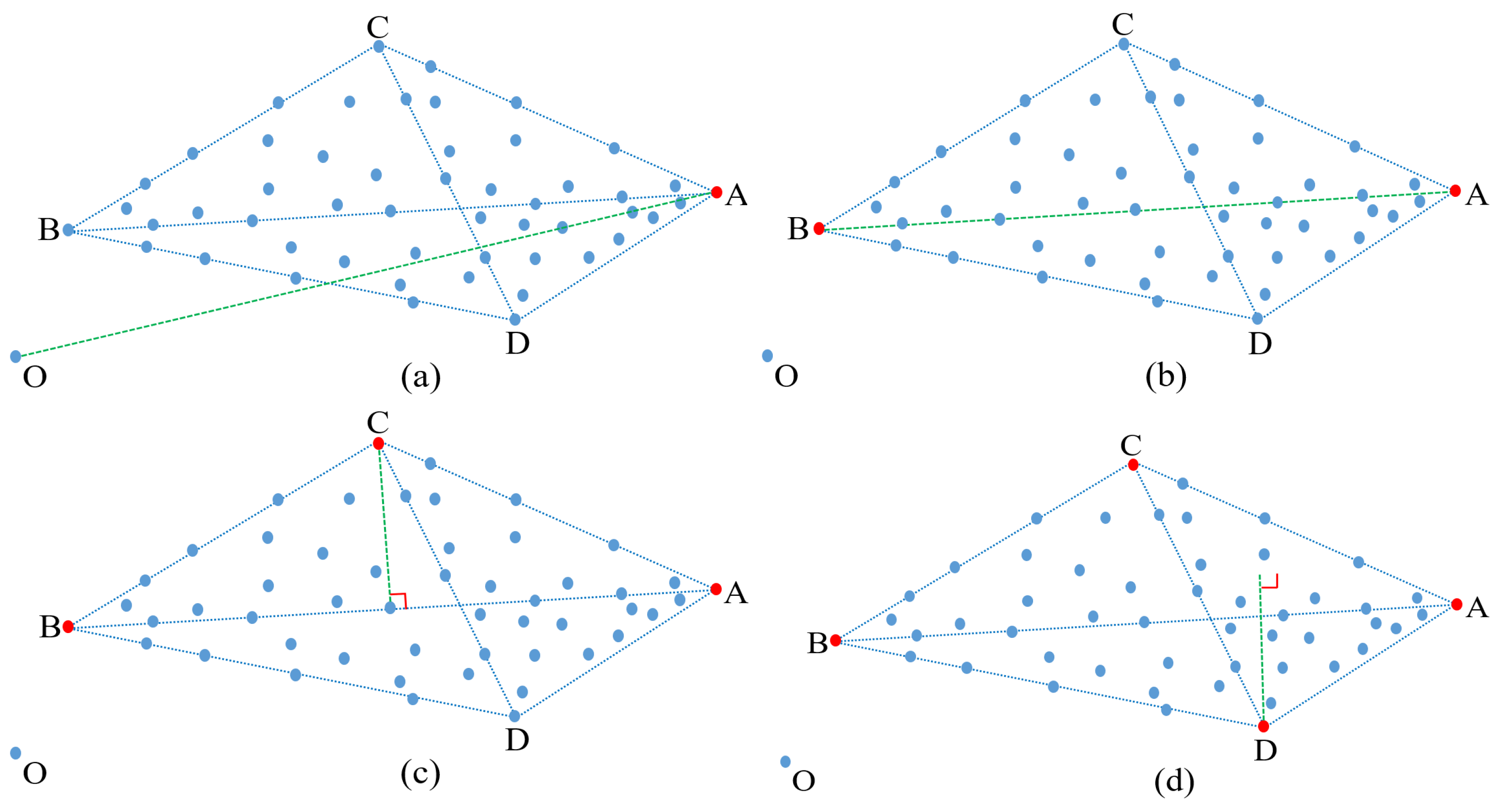
Finally, the index is used to extract the fourth endmember D, which also satisfies the two conditions to be considered as an endmember. Figure 3 graphically shows the process of extracting four endmembers.
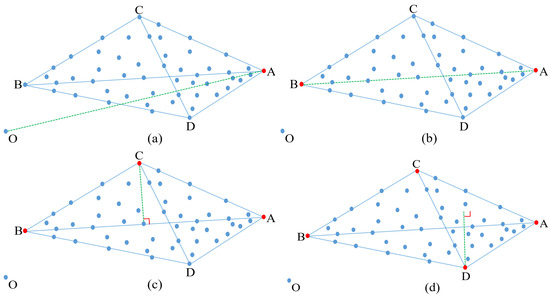
Figure 3.
(a–d) Step-by-step process illustrating how to extract four endmembers by using our maximum distance analysis (MDA) method.
Definition 5.
We define the farthest pixel point from the affine hull formed by all of the extracted endmembers as the next endmember.
Assuming that we have extracted endmembers , and these endmembers form a affine hull . We have the matrix , where N is the number of pixels and c is the number of endmembers that have been extracted. Assuming that is the normal vector of the affine hull and b is the offset or distance between the coordinate origin and affine hull, we compute the normal vector w by
is a pixel point of the affine hull , and the affine hull is written as Equation (12)
In this context, there is a pixel point , whose distance between it and the affine hull is denoted as d. Additionally, we consider point as the projection of onto the affine hull, where is the angle between vectors and , , and we have
and
and also
We know that from Equations (12) and (13) is represented as
Based on Equations (15) and (16), we have
Therefore, the distance between any pixel point and the considered affine hull is computed by
Specifically, support vector machines (SVMs) [45] provide the distance calculation that is given by Equation (18) between any pixel point and the affine hull. We obtain the index of the next endmember by
We then use the index to extract the next endmember, which also satisfies the aforementioned two conditions that any pixel point must meet in order to be considered an endmember.
3.2. Estimating the Number of Endmembers
From the mathematical discussions in the previous subsection, we know that: (1) if one pixel point belongs to one affine hull, the distance between it and the affine hull is zero, and (2) if one point does not belong to one affine hull, the distance between it and the affine hull is greater than zero. Based on the aforementioned concepts, we give the following definition as the stopping criterion of endmember extraction, also obtaining the final number of endmembers by counting those pixel points that have been extracted as endmembers.
Definition 6.
The stopping criterion of endmember extraction is that the maximum distance between all of the pixel points and affine hull formed by the extracted endmember is zero. Simultaneously, the number of endmembers is determined by counting the number of pixel points that have been extracted.
To conclude this section, we emphasize that our MDA method extracts endmembers without any previous knowledge regarding the number of endmembers, since it performs both endmember extraction and the estimation of the number of endmembers. It is worth noting that the maximum distance of the stopping criterion in Definition 6 is zero when the experimental data are linearly mixed and do not include noise; otherwise, the maximum distance is greater than zero. In addition, there is a clear difference between the distance between the non-endmember and endmember to the affine hull that formed by all extracted endmembers when the experimental data are non-linearly mixed and include noise. Therefore, we can observe the difference to obtain the maximum distance when the experimental data are non-linearly mixed and include noise, and then determine the number of endmembers.
4. Experiments
We use both synthetic and real hyperspectral data in order to evaluate the performance of our proposed MDA method in the tasks of estimating the number of endmembers and extracting their signatures. We perform the comparison tests using Matlab 2018b on an i5-8300 CPU (2.3 G). We conduct a series of experiments to demonstrate the effectiveness and efficiency of our newly proposed MDA method in counting and extracting endmembers. The endmember extraction capacity of our method is assessed with three endmember extraction algorithms, i.e., minimum-volume enclosing simplex (MVES) [41], vertex component analysis (VCA) [39], and minimum volume constrained nonnegative matrix factorization (MVCNMF) [40]. Additionally, the accuracy of our method in the task of estimating the number of endmembers is assessed with two endmember counting algorithms, i.e., hyperspectral signal subspace by minimum error (Hysime) [31] and Harsanyi–Farrand–Chang (HFC) [32]. We compare the estimated endmembers and abundances with the ground-truth ones that were used in our experiments by the following formulas
and
where stands for the Frobenius norm, is Euclidean norm, and are the estimated endmember signatures and abundances, and M and A denote the actual endmember signatures and abundances. The reconstruction error is another metric that is considered in our experiments, which is computed as
where and X are the estimated and actual spectral signatures, respectively. Additionally, we also use the spectral angle distance (SAD) to evaluate our method by
where c is the number of endmembers, and and are the ith estimated and actual spectral signatures, respectively.
EEAs get the most accurate results when the values of the above four metrics and SAD are zero.
4.1. Synthetic Data
The synthetic data are created based on the United States Geological Survey (USGS) library. The mineral signatures in USGS library both have 224 spectral bands. We generate synthetic data that are based on the linear mixture method [39,40]. Specifically, to improve the unmixing performance, we have removed the low SNR bands as well as the water vapor absorption bands (including bands 1-2, 104-113, 148-167, and 221-224) from the original 224-band data cube, and keep 188 spectral bands in our experiments. We consider four families of synthetic data to evaluate the effectiveness and efficiency of our proposed MDA method in counting and extracting endmembers: (1) synthetic data containing pure mineral signatures and no noise under different number of endmembers; (2) synthetic data containing pure mineral signatures and 2000 pixel points and five endmembers under different noise levles; (3) synthetic data containing nonpure mineral signatures and 2000 pixel points and five endmembers under different noise levels; and, (4) synthetic data, including nonpure spectral signatures with different number of endmembers and different sizes under specific noise (70 dB). We utilize Definitions 1 to 6 to extract the signatures and estimate the number of endmembers. For simplicity, we summarize the steps that were conducted for extracting and counting endmembers with our MDA method:
- We calculate the distance between every pixel point and coordinate origin in the pixel space and extract the pixel point with the maximum distance as the first endmember.
- We calculate the distance between every pixel point and the first extracted endmember in the pixel space and extract the pixel point with the maximum distance as the second endmember.
- We calculate the distance between every pixel point and the line defined by the first and second extracted endmembers, extracting the pixel point with the maximum distance as the third endmember.
- We calculate the distance between every pixel point and the plane formed by the first, second, and third extracted endmembers, extracting the pixel point with the maximum distance as the fourth endmember.
- We calculate the distance between every pixel point and the affine hull formed by all the previously extracted endmembers, extracting the pixel point with the maximum distance as the next endmember.
- We finish the overall endmember extraction procedure when the maximum distance between all pixel points and the affine hull formed by all the previously extracted endmembers is zero. Simultaneously, we determine the number of endmembers by counting the number of pixel points that have been extracted. In the following, we describe the conducted experiments.
4.1.1. Straightforward MDA Algorithm
This experiment uses the first family type of synthetic data to evaluate the proposed MDA algorithm in counting and extracting endmembers. The synthetic data are created by the procedure that was described in [40]. The synthetic data, with 3364 pixel points, have pure spectral signatures and no noise. We use the metrics and SAD to evaluate the endmember extraction performance of our proposed MDA method on synthetic data containing pure pixels and no noise under different number of endmembers. In addition, due to the fact that MDA can complete the two tasks of extracting endmembers and determining the number of endmembers, we employ VCA as one comparison method of extracting endmember, and Hysime and HFC as the two comparison methods of counting endmembers to evaluate the performance of our proposed MDA method. Table 1 illustrates the performance and efficiency of different methods in extracting their spectral signatures and determining the number of endmembers. Firstly, in terms of extracting endmembers, we observe and SAD from Table 1, and find that both VCA and our proposed MDA method acquire accurate results and they have high efficiency under different number of endmembers. Specifically, our method is slightly faster than VCA. Secondly, in terms of determining the number of endmembers, we find that both HFC and our proposed MDA method obtain accurate results, and Hysime has an error when the number of endmembers 20. In addition, we also observe that Hysime and HFC are faster than our proposed MDA method. However, VCA can only extract endmembers, Hysime and HFC can only determine the number of endmembers, but our proposed MDA method can simultaneously complete the two tasks (extracting endmembers and determining the number of endmembers). Specifically, due to the fact that MDA is faster than VCA, our proposed MDA method extracts endmembers more accurately and quickly as compared to Hysime (or HFC) +VCA. Therefore, our proposed MDA method is an effective and effcienet method in completing the overall endmember estimation task, i.e., determining the number of endmembers and extracting endmembers.

Table 1.
Performance and processing time (seconds) for our proposed MDA algorithm on synthetic data containing pure mineral signatures and no noise under different number of endmembers. c and are the actual and estimated number of endmembers, respectively. d denotes the maximum distance between all pixel points and the affine hull formed by all of the extracted endmembers.
4.1.2. MDA for Improving MVSA
As reviewed in Section 2.2, the original MVSA algorithm requires the number of endmembers before performing endmember extraction. Additionally, the original MVSA algorithm needs to utilize the VCA algorithm as an initialization method to extract endmembers. To this end, we embed our method into MVSA, which possibly extracts endmembers from nonpure pixels. Such composition (e.g., combining the MDA and MVSA) forms a new, overall unmixing framework that simultaneously and efficiently determines the number of endmembers and extracts their spectral signatures without any prior knowledge. We refer to this strategy as MDA-MVSA.
We conduct our experiments on synthetic data containing pure and nonpure spectral signatures with different noise levels, respectively. The two types of synthetic data both have 2000 pixel points and five endmembers. The two types of synthetic data are constructed according to the linear model that is given using the procedure described in [39]. Zero-mean white Gaussian noise has been added to the two types of synthetic data, and it is defined by
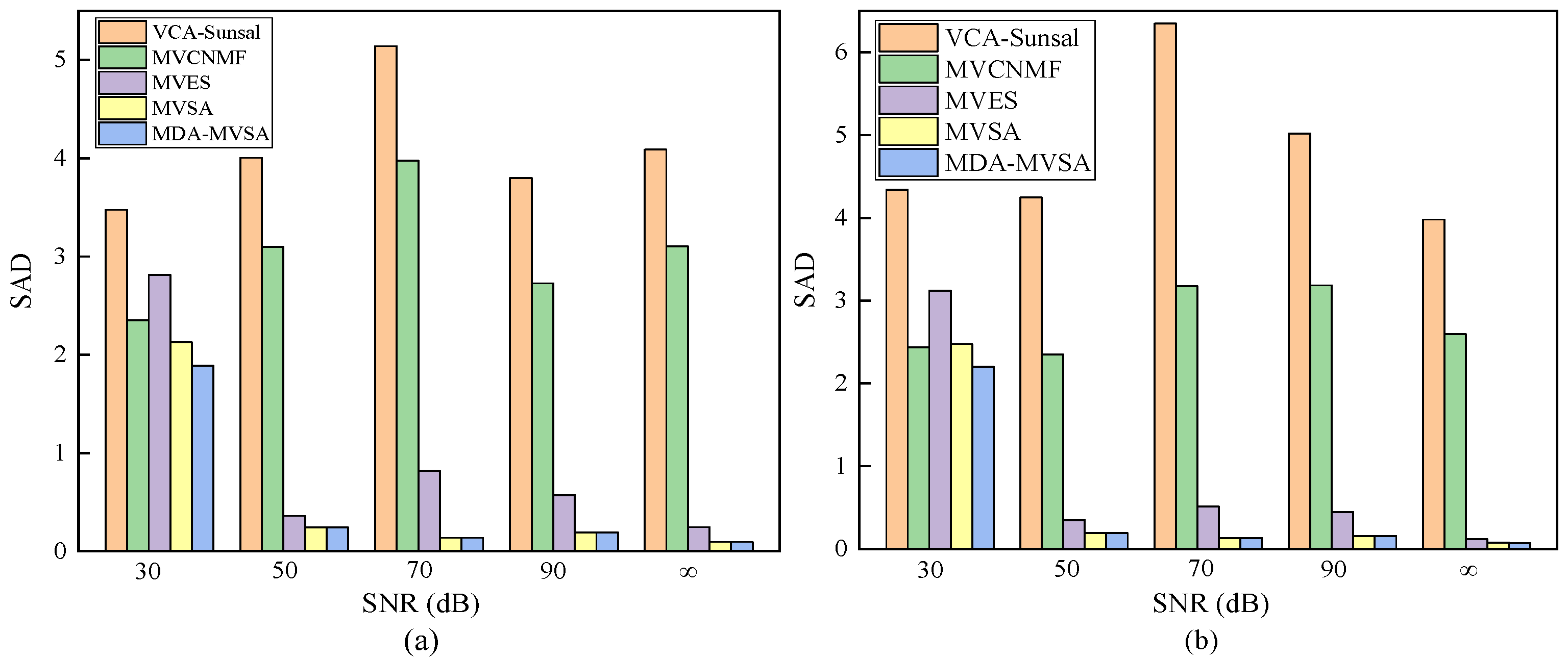
where X and N are the matrices of pixel points and noise, respectively. We continue to employ the metrics , and SAD to evaluate the performance of our proposed MDA-MVSA method in unmixing. Table 2 and Table 3 show the results that were obtained by different methods on synthetic data containing pure and non-pure spectral signatures under different noise levels. Specifically, VCA and sparse unmixing by variable splitting and augmented Lagrangian (Sunsal) [46] are the most typical endmember extraction method and abundance estimation method, respectively. Therefore, we combine VCA and Sunsal as VCA-Sunsal in order to evaluate the performance of different methods. From Table 2 and Table 3, we see that: (1) VCA-Sunsal is the fastest method when compared with other methods, but the time gap between it and our proposed MDA-MVSA method is very small. In addition, VCA-Sunsal requires the number of endmembers in endmember extraction compared to our proposed MDA-MVSA method. In this scenario, although VCA-Sunsal is faster than our proposed MDA-MVSA method, the processing of time regarding our proposed MDA-MVSA includes the processing time of determining the number of endmembers and VCA-Sunsal does not include; (2) our proposed MDA-MVSA method obtains better results than VCA-Sunsal, MVCNMF, and MVES in terms of the metrics , , and ; and, (3) our proposed MDA-MVSA method and original MVSA acquire almost same results in terms of the metrics , , and , but our proposed MDA-MVSA method completes endmember extraction without requiring the number of endmembers. In addition, the original MVSA uses VCA as initialization to obtain the final results of endmember extraction, and VCA may provide slightly different results in different runs due to its initialization. In this scenario, our proposed MDA-MVSA method is more stable than the original MVSA, and it extracts endmembers without requiring the number of endmembers. Therefore, our proposed MDA-MVSA method improves the performance of original MVSA method, and it is very effective and efficient in extracting endmembers. Figure 4 shows the results of SAD for different methods at various noise levels on the two considered synthetic data scenes. From Figure 4, we see that that our proposed MDA-MVSA method obtains more better results in terms of SAD compared to VCA-Sunsal, MVCNMF, and MVES. We also observe that our proposed MDA-MVSA method and original MVSA acquire the same results. Similarly, due to the fact that original MVSA uses VCA as initialization to complete endmember extraction and requires the number of endmembers when it extracts endmembers, our proposed MDA-MVSA method still improves theoriginal MVSA.

Table 2.
Performance and processing time (seconds) for different endmember extraction algorithms on synthetic data with 2000 pixel points and five endmembers and containing pure mineral signatures for the United States Geological Survey (USGS) library under different noise levels.

Table 3.
Performance and processing time (seconds) for different endmember extraction algorithms on synthetic data with 2000 pixel points and five endmembers and containing nonpure mineral signatures (maximum purity of 0.8) for the USGS library under different noise levels.
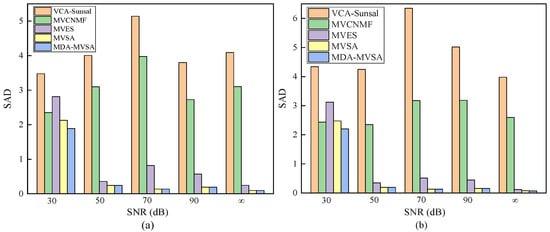
Figure 4.
Spectral angle distance (SAD) performance for different endmember extraction algorithms on (a) synthetic data with pure pixels under different noise levels and (b) synthetic data with nonpure pixels under different noise levels. The two types of synthetic data both have 3364 pixel points and five endmembers under different noise levels.
Table 4 reports the performance of our proposed MDA-MVSA method in counting the endmembers on synthetic data containing pure and non-pure spectral signatures under different noise levels. Specifically, the two types of synthetic data both have 2000 pixel points and five endmembers. From Table 4, we see that the gap between the maximum distance is very obvious when the estimated number of endmembers exceeds the actual number of endmembers. Therefore, our proposed MDA-MVSA method can accurately determine the number of endmembers. Table 5 shows the results (estimated number of endmembers + processing time) of different methods in the task of counting endmembers on the above two types of synthetic data, i.e., containing pure and non-pure spectral signatures under different noise levels. From Table 5, we see that Hysime and our proposed MDA-MVSA method obtain accurate results on synthetic data containing pure and non-pure spectral signatures with different noise levels, and HFC has errors when the actual number of endmembers is fibe on synthetic data containing pure and non-pure spectral signatures with different noise levels. Additionally, HFC is the most effective method in counting endmembers, but the time gap between it and our proposed MDA-MVSA method is very small. In addition, Hysime and HFC can only determine the number of endmembers and our proposed MDA-MVSA method can complete two tasks, i.e., determining the number of endmembers and extracting endmembers. In this scenario, the processing time of Hysime and HFC is the running time of determining the number of endmembers, while the processing time of our proposed MDA-MVSA method is the running time of determining the number of endmembers and extracting endmembers. Therefore, our proposed MDA-MVSA method is also very effective and efficient in determining the number of endmembers.

Table 4.
The relationship between the number of endmembers and the maximum distance on synthetic data containing pure and nonpure mineral signatures under different noise levels. The two types of synthetic data both have 2000 pixel points and five endmembers.

Table 5.
The results of estimated number of endmembers + the processing time by different methods on synthetic data containing pure and nonpure mineral signatures under different noise levels. The two types of synthetic data both have 2000 pixel points and five endmembers.
To take the experimental validation one step further, we conduct some experiments on synthetic data containing non-pure spectral signatures with different numbers of endmembers and sizes under a specific noise level (70 dB) to evaluate the performance of our proposed MDA-MVSA method in extracting and counting endmembers. Table 6 reports the performance and processing time for problems with N = 4000, 8000, and 12,000 pixel points while using different numbers of endmembers (c = 5, 10, 15 and 20) on synthetic data with non-pure spectral signatures. From Table 6, we see that our proposed MDA-MVSA method has good results in terms of the values of , SAD, and time over MVES. Specifically, this is a very difficult problem when the number of endmembers is 20. For instance, from Table 6, we see that our method takes about 12 s to perform the computation and in comparison MVES takes around 3.7 h on synthetic data with 12,000 pixel points and 20 endmembers. This fact reflects that our proposed MDA-MVSA method is very efficient for large-scale problems. From Table 6, we also observe that the gap of the maximum distance d is very obvious when the estimated number of endmembers exceeds the actual number of endmembers. Therefore, as compared to MVES, our algorithm is not only a high-speed algorithm, but it also does not require the number of endmembers when it implements endmember extraction.

Table 6.
Performance and processing time (seconds) obtained with our method and minimum-volume enclosing simplex (MVES) on synthetic data with N = 4000, 8000, 12,000, SNR = 70 dB, c = 5, 10, 15, and 20, and maximum purity of 0.8. is the estimated number of endmembers, and d is the maximum distance between all pixel points and the affine hull formed by all extracted endmembers.
4.2. Real Data
4.2.1. Cuprite Data
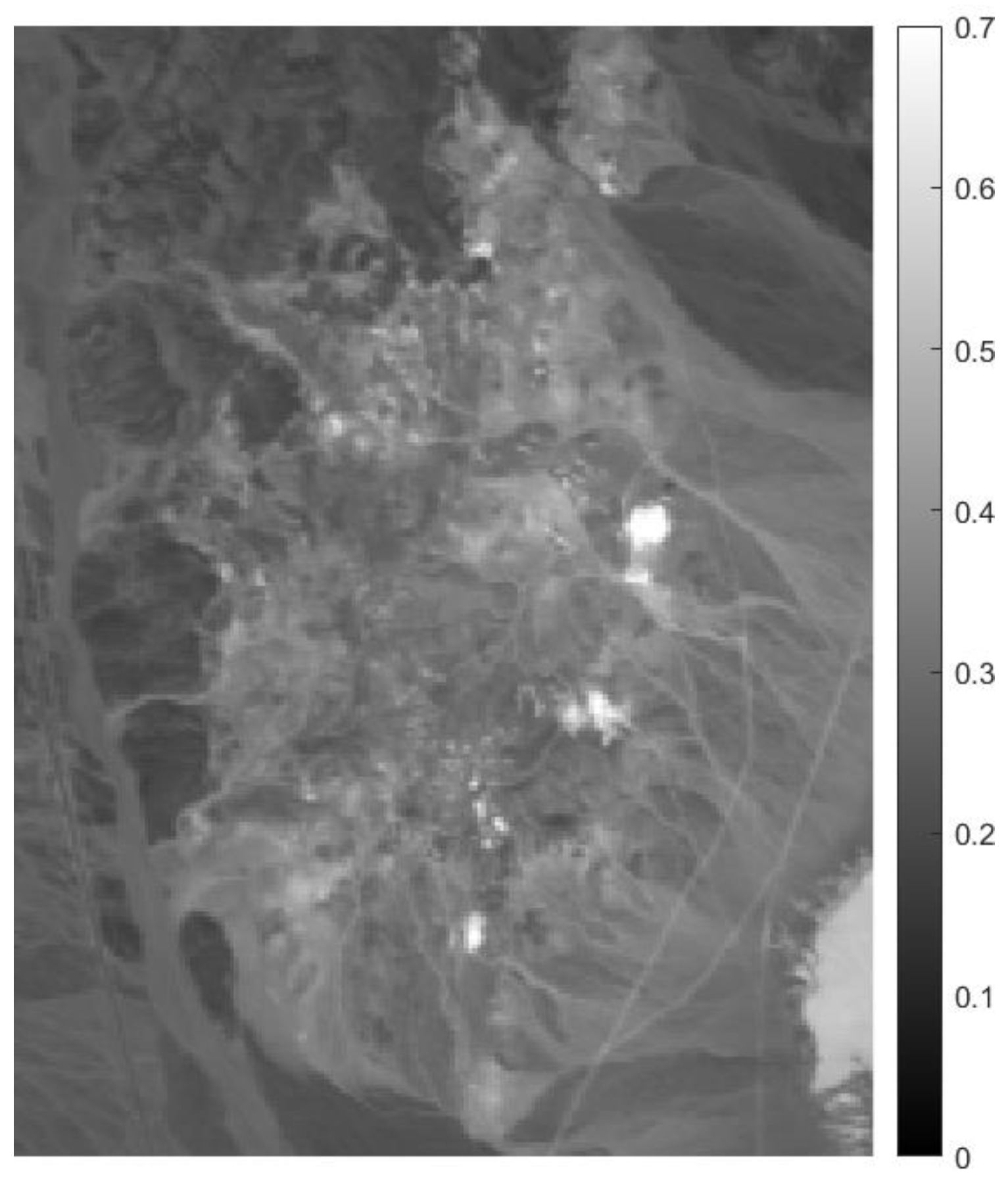
We first use the well-known AVIRIS Cuprite data (http://aviris.jpl.nasa.gov/html/aviris.freedata.html (accessed on 15 February 2021).) in order to evaluate the performance of the proposed approach in extracting endmembers. The scene has been widely used to evaluate the performance of endmember extraction algorithms. The portion used in experiments corresponds to a 250 × 191 pixel subset of the scene, with 224 spectral bands in the range 0.4–2.5 m and nominal spectral resolution of 10 nm. We remove the bands 1–6, 105–115, 150–170, and 222–224 (due to water absorption and low SNR), and retain 183 spectral bands in our experiments. Figure 5 presents the subimage of Cuprite data (displaying only the 30th band) for this experiment.
Figure 5.
30th band of the considered airborne visible/infrared imaging spectrometer (AVIRIS) Cuprite subimage.
We employ the metrics , and SAD to evaluate the endmember extraction performance of our method and MVCNMF on the Cuptite subimage. Table 7 shows the processing time and values of , and SAD for our method and MVCNMF. From Table 7, we see that our method gets more accurate results when compared with MVCNMF in terms of the values of the metrics , and SAD. This fact reflects that our method extracts endmembers effectively. From Table 7, we also find that our method takes less than 1 min. to perform the computation, while MVES spends around 20 min. in the considered environment. As a result, we conclude that our proposed method is very efficient for endmember extraction.

Table 7.
Performance and processing time (in seconds) obtained by our proposed MDA-minimum volume simplex analysis (MDA-MVSA) method and minimum volume constrained nonnegative matrix factorization (MVCNMF) on the considered Cuprite subimage.
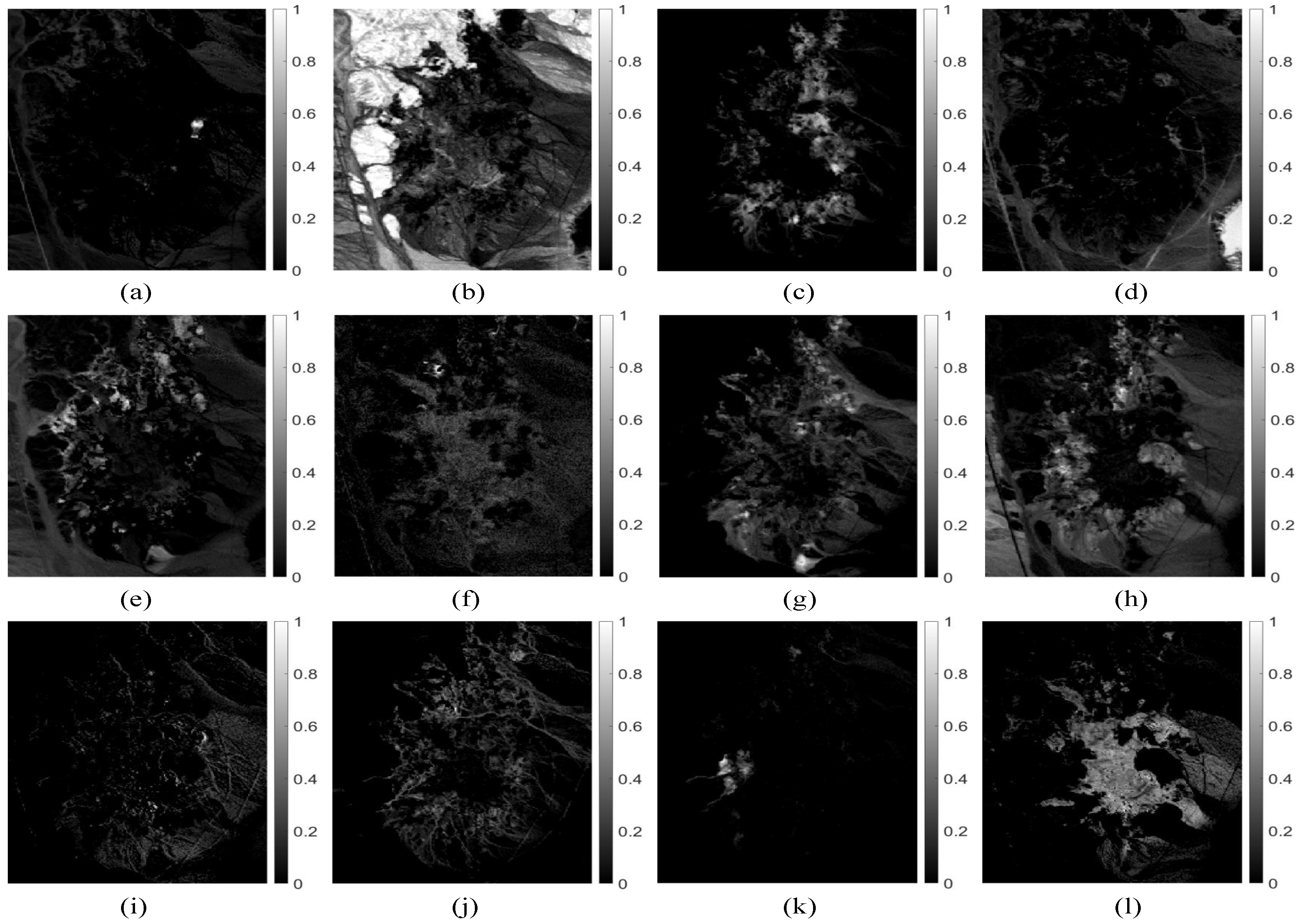
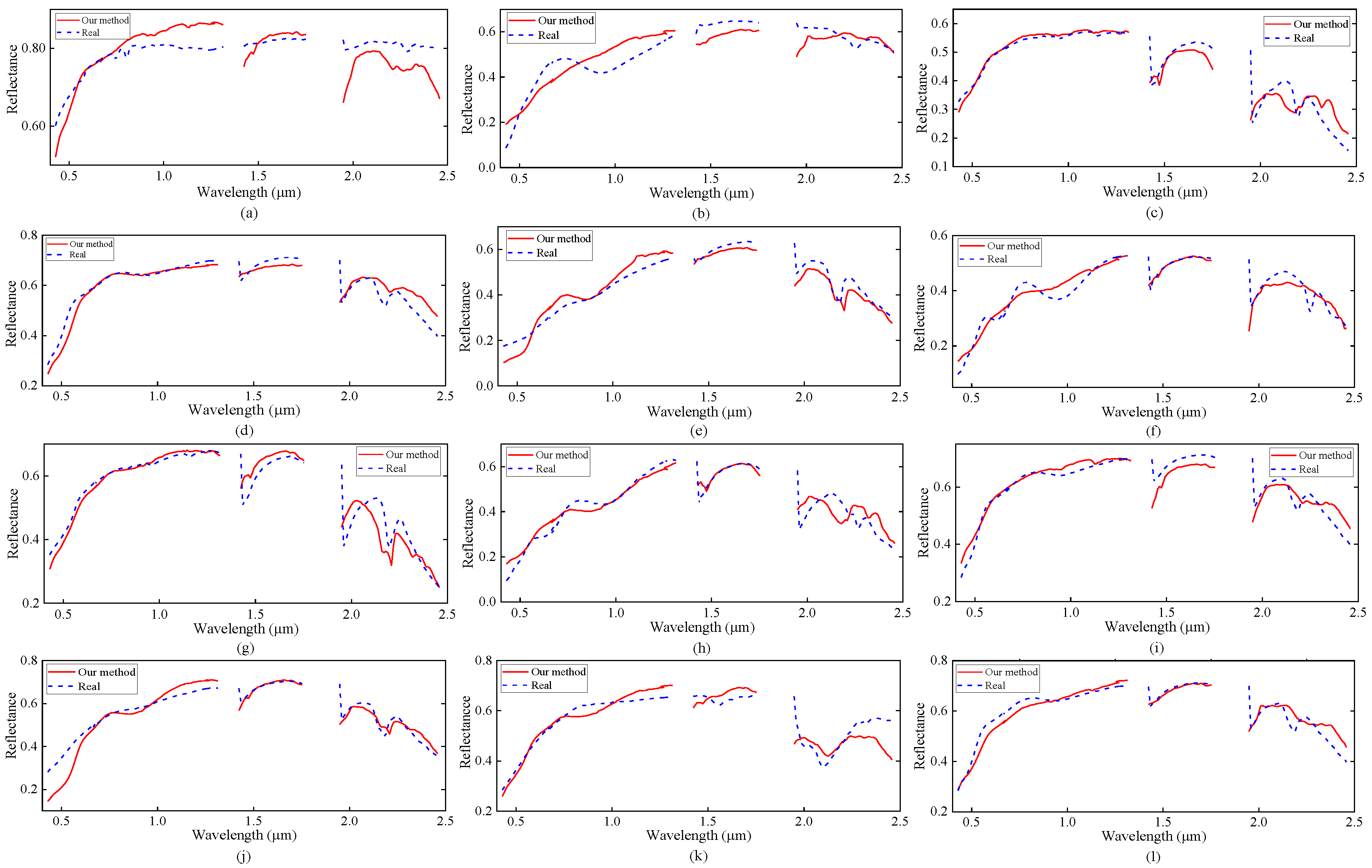
Figure 6 shows the abundance maps obtained by our MDA-MVSA method. The abundance maps that are obtained by MDA-MVSA are identified as mineral maps of Chlorapatite WS423, Jarosite WS368 Pb, Montmorillonite SCa-2.a, Kaolin/Smect KLF511 12%K #1, Kaolin/Smect KLF508 85%K, Nontronite NG-1.a, Montmorillonite SCa-2.b, Nontronite SWa-1.a, Kaolin/Smect KLF511 12%K #2, Kaolin/Smect H89-FR-5 30K, Buddingtonite GDS85 D-206, Kaolin/Smect KLF511 12%K #3, as shown in Figure 6a–n, respectively. In addition, Figure 7 presents the spectral signatures of the estimated endmembers. From Figure 7, we observe that the endmember signatures that are estimated by our method are in good agreement with the real spectral signatures. The results that are shown in Figure 7 reflect the high accuracy of our method in endmember extraction on the Cuprite subimage.
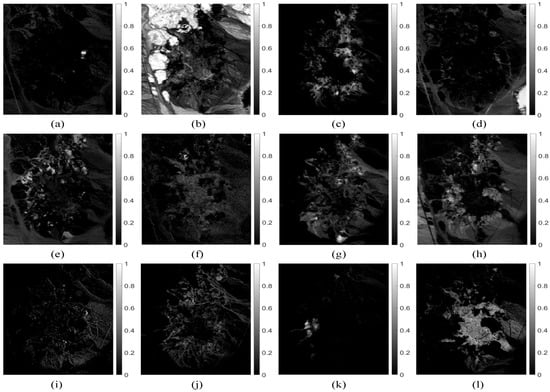
Figure 6.
Abundance fraction maps estimated by our MDA-MVSA method from the Cuprite subimage: (a) Chlorapatite WS423; (b) Jarosite WS368 Pb; (c) Montmorillonite SCa-2.a; (d) Kaolin/Smect KLF511 12%K #1; (e) Kaolin/Smect KLF508 85%K; (f) Nontronite NG-1.a; (g) Montmorillonite SCa-2.b; (h) Nontronite SWa-1.a; (i) Kaolin/Smect KLF511 12%K #2; (j) Kaolin/Smect H89-FR-5 30K; (k) Buddingtonite GDS85 D-206; and, (l) Kaolin/Smect KLF511 12%K #3.
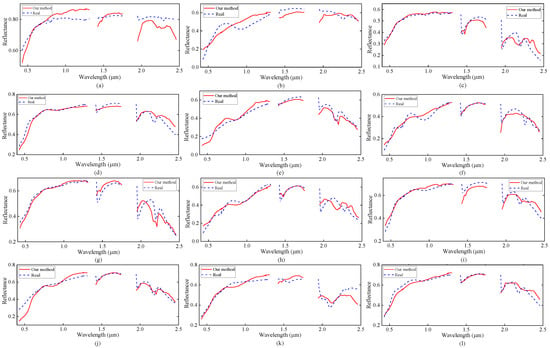
Figure 7.
Endmember signatures in the USGS library and the endmember estimates obtained by our MDA-MVSA method from the Cuprite subimage. The corresponding signatures are the following. (a) Chlorapatite WS423; (b) Jarosite WS368 Pb; (c) Montmorillonite SCa-2.a; (d) Kaolin/Smect KLF511 12%K #1; (e) Kaolin/Smect KLF508 85%K; (f) Nontronite NG-1.a; (g) Montmorillonite SCa-2.b; (h) Nontronite SWa-1.a; (i) Kaolin/Smect KLF511 12%K #2; (j) Kaolin/Smect H89-FR-5 30K; (k) Buddingtonite GDS85 D-206; and, (l) Kaolin/Smect KLF511 12%K #3.
4.2.2. Samson Data
We also utilize the Samson real hyperspectral image (http://lesun.weebly.com/hyperspectral-data-set.html (accessed on 15 February 2021).) to evaluate the performance of our method in extracting endmembers. The Samson image comprises 952 × 952 pixels, each of which has 156 bands that cover the wavelength range from 401 to 889 nm. The original image is quite large, leading to high computational cost for several of the considered algorithms. In this experiment, a subimage with 95 × 95 pixels is used. The spatial coordinate of the first pixel of the subimage in the full Samson image is (252,332). The Samson data contain three endmembers, i.e., tree, water, and soil (Figure 8a shows the false color composite of the considered subimage).
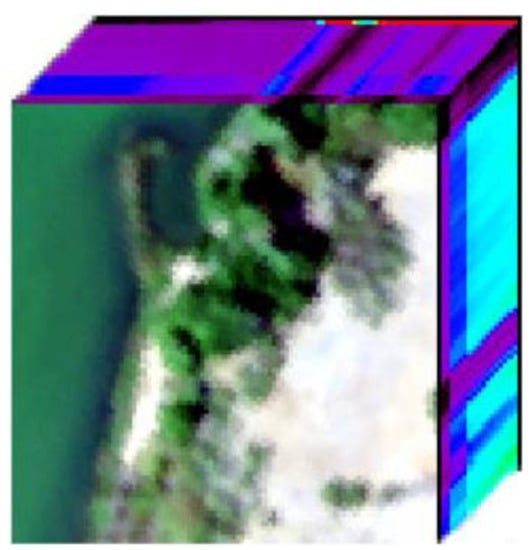
Figure 8.
False color composition of the considered Samson subimage.
We employ the metrics to evaluate the performance of our method and MVCNMF in extracting endmembers on the considered Samson data. Table 8 shows the performance metrics of different methods that are applied to the Samson subimage, together with the processing time (in seconds) for each method. From Table 8, we conclude that our method exhibits very low values of , while our method is faster than MVCNMF. In summary, this reveals that our method effectively and efficiently extracts endmembers from the considered Samson data.

Table 8.
Performance and processing time (seconds) obtained by our proposed MDA-MVSA method and MVCNMF on the considered Samson subimage.
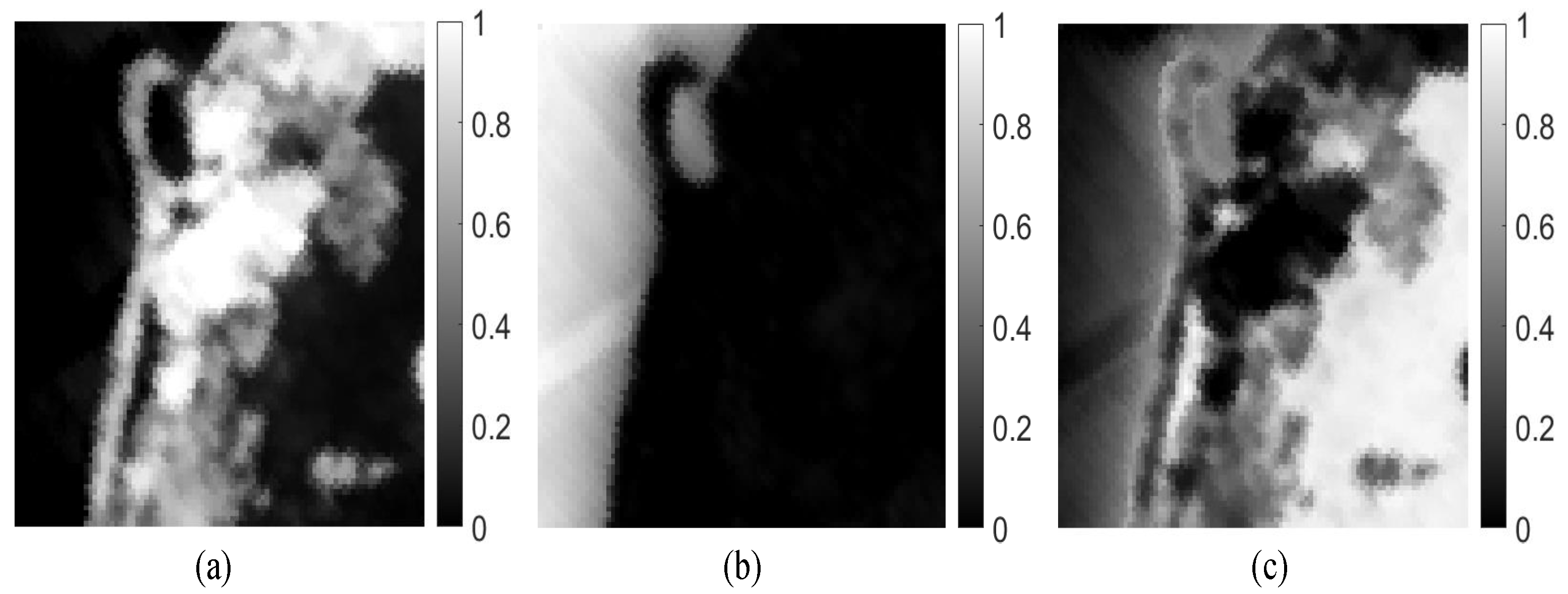
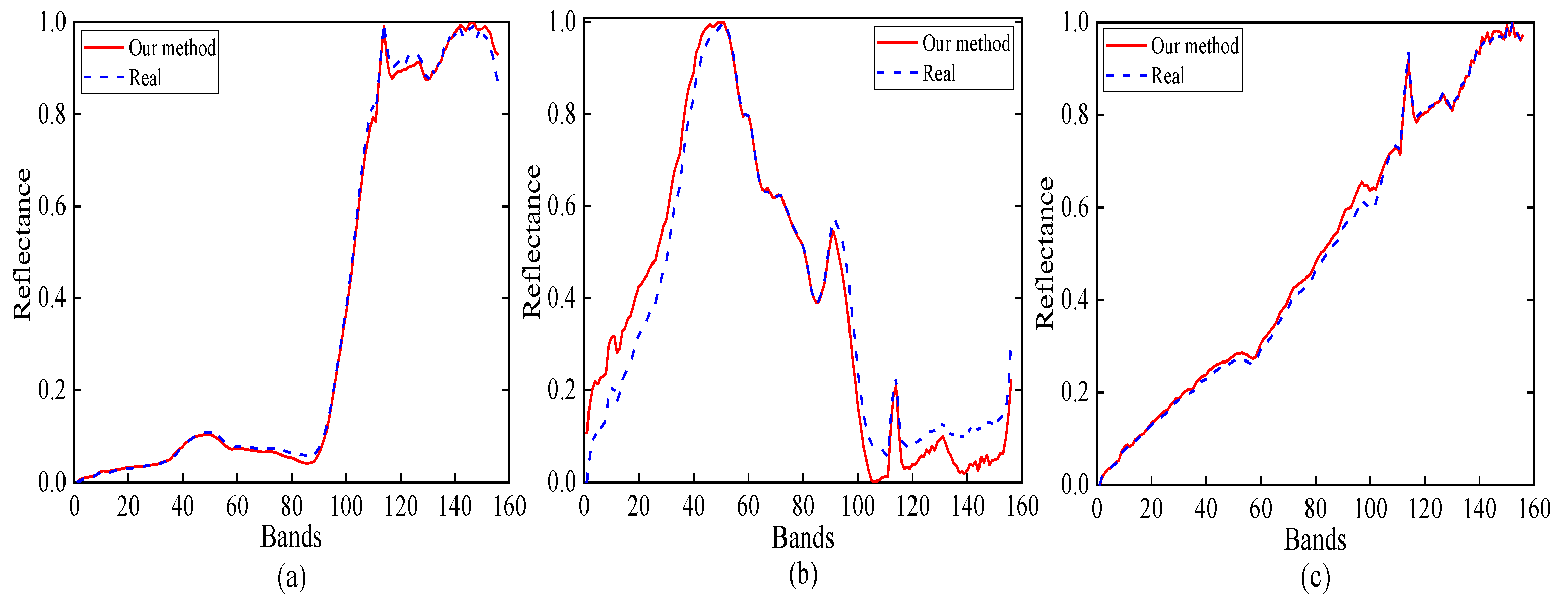
Figure 9 presents the estimated abundance maps by our method. Figure 10 shows a qualitative comparison between the endmembers that are extracted by our method from the Samson subimage and their corresponding reference (ground-truth) signatures. From Figure 10, we conclude that the endmember signatures estimated by our method are in very good spectral agreement with the reference signatures. The results that are shown in Figure 9 and Figure 10 suggest the high accuracy of our method in extracting endmembers from real data.
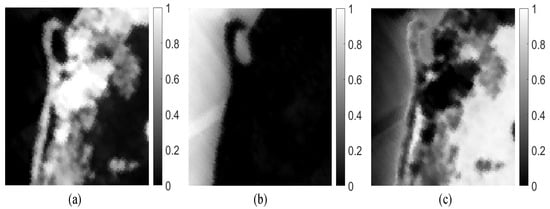
Figure 9.
Abundance fraction maps estimated by our MDA-MVSA method from the Samson subimage: (a) tree; (b) water and (c) soil.
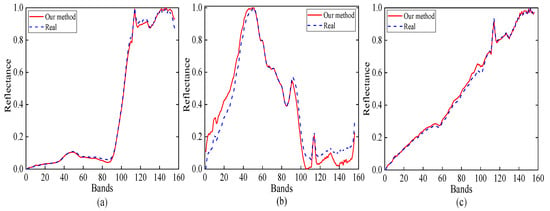
Figure 10.
Reference signatures and estimated endmember signatures by our MDA-MVSA method from the Samson subimage: (a) tree; (b) water; and, (c) soil.
5. Conclusions and Future Work
We have developed a new algorithm for estimating the number of endmembers and the corresponding endmember signatures, which is referred to as maximum distance analysis (MDA). Our proposed MDA method extracts endmember signatures without any prior knowledge about the number of endmembers. Moreover, our method cannot obtain the most accurate results of endmember estimation when the experimental data have noise due to the fact that the distance calculation that is shown in Equation (18) is given in the scenario of linear mixing. To address the issue, we embed our proposed MDA algorithm into the original MVSA algorithm to form a new and overall endmember estimation framework. The operation avoids the limitation that the original MVSA algorithm requires prior knowledge on the number of endmembers, and it has high efficiency for the overall endmember estimation task. We refer to this novel strategy as MDA-MVSA. The strategy has been validated while using synthetic data and real data, and it has shown to accurately perform both counting and extracting endmembers, exhibiting very high efficiency in comparison with other traditional endmember extraction methods. Specifically, MDA is usually used on simple scenes (i.e., experimental data containing pure pixels and no noise), and MDA-MVSA combines MDA and MVSA to form a new and effective unmixing framework for complex scenes (i.e., experimental data containing non-pure pixels and noise).
As with any new approach, there are still some unresolved issues that may present challenges over time. Specifically, the distance calculation in our proposed method may need to be adapted if the hyperspectral data are nonlinearly mixed or include noise. In our future work, we will investigate how to simultaneously determine the number of endmembers and extract their signatures from hyperspectral images when they are nonlinearly mixed.
Author Contributions
Methodology, X.T.; Software, J.M.H.; Writing—review & editing, M.E.P., P.R., J.P. and A.P. The authors have contributed as equally to this work. All authors have read and agreed to the published version of the manuscript.
Funding
This work has been funded by Spanish Ministry (FPU15/02090), Junta de Extremadura (GR18060), and European Union (734541-EXPOSURE).
Acknowledgments
We gratefully thank the Associate Editor and the three Anonymous Reviewers for theiroutstanding comments and suggestions, which greatly helped us to improve the technical quality and presentationof our work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Luo, F.; Du, B.; Zhang, L.; Zhang, L.; Tao, D. Feature learning using spatial-spectral hypergraph discriminant analysis for hyperspectral image. IEEE Trans. Cybern. 2018, 49, 2406–2419. [Google Scholar] [CrossRef] [PubMed]
- Vane, G.; Green, R.O.; Chrien, T.G.; Enmark, H.T.; Hansen, E.G.; Porter, W.M. The airborne visible/infrared imaging spectrometer (AVIRIS). Remote Sens. Environ. 1993, 44, 127–143. [Google Scholar] [CrossRef]
- Goetz, A.F.; Vane, G.; Solomon, J.E.; Rock, B.N. Imaging spectrometry for earth remote sensing. Science 1985, 228, 1147–1153. [Google Scholar] [CrossRef]
- Paoletti, M.; Haut, J.; Plaza, J.; Plaza, A. Deep learning classifiers for hyperspectral imaging: A review. ISPRS J. Photogramm. Remote Sens. 2019, 158, 279–317. [Google Scholar] [CrossRef]
- Keshava, N.; Mustard, J.F. Spectral unmixing. IEEE Signal Process. Mag. 2002, 19, 44–57. [Google Scholar] [CrossRef]
- Gao, F.; You, J.; Wang, J.; Sun, J.; Yang, E.; Zhou, H. A novel target detection method for SAR images based on shadow proposal and saliency analysis. Neurocomputing 2017, 267, 220–231. [Google Scholar] [CrossRef]
- Adams, J.B.; Smith, M.O.; Johnson, P.E. Spectral mixture modeling: A new analysis of rock and soil types at the Viking Lander 1 site. J. Geophys. Res. Solid Earth 1986, 91, 8098–8112. [Google Scholar] [CrossRef]
- Chan, T.H.; Ma, W.K.; Ambikapathi, A.; Chi, C.Y. An optimization perspective onwinter’s endmember extraction belief. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium, Vancouver, BC, Canada, 24–29 July 2011; pp. 1143–1146. [Google Scholar]
- Bioucas-Dias, J.M.; Plaza, A.; Dobigeon, N.; Parente, M.; Du, Q.; Gader, P.; Chanussot, J. Hyperspectral unmixing overview: Geometrical, statistical, and sparse regression-based approaches. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2012, 5, 354–379. [Google Scholar] [CrossRef]
- Li, J.; Bioucas-Dias, J.M.; Plaza, A.; Liu, L. Robust Collaborative Nonnegative Matrix Factorization for Hyperspectral Unmixing. IEEE Trans. Geosci. Remote Sens. 2016, 54, 6076–6090. [Google Scholar] [CrossRef]
- Tao, X.; Cui, T.; Plaza, A.; Ren, P. Simultaneously Counting and Extracting Endmembers in a Hyperspectral Image Based on Divergent Subsets. IEEE Trans. Geosci. Remote Sens. 2020, 58, 8952–8966. [Google Scholar] [CrossRef]
- Xu, X.; Pan, B.; Chen, Z.; Shi, Z.; Li, T. Simultaneously Multiobjective Sparse Unmixing and Library Pruning for Hyperspectral Imagery. IEEE Trans. Geosci. Remote Sens. 2020. [Google Scholar] [CrossRef]
- Xu, X.; Shi, Z.; Pan, B.; Li, X. A classification-based model for multi-objective hyperspectral sparse unmixing. IEEE Trans. Geosci. Remote Sens. 2019, 57, 9612–9625. [Google Scholar] [CrossRef]
- Geng, X. Study on Hyperspectral Target Detection and Classification. Ph.D. Thesis, Chinese Academy of Sciences, Beijing, China, January 2005. [Google Scholar]
- Wang, R.; Li, H.C.; Pizurica, A.; Li, J.; Plaza, A.; Emery, W.J. Hyperspectral unmixing using double reweighted sparse regression and total variation. IEEE Geosci. Remote Sens. Lett. 2017, 14, 1146–1150. [Google Scholar] [CrossRef]
- Fernandez-Beltran, R.; Plaza, A.; Plaza, J.; Pla, F. Hyperspectral unmixing based on dual-depth sparse probabilistic latent semantic analysis. IEEE Trans. Geosci. Remote Sens. 2018, 56, 6344–6360. [Google Scholar] [CrossRef]
- Delgado, J.; Martín, G.; Plaza, J.; Jiménez, L.I.; Plaza, A. Fast spatial preprocessing for spectral unmixing of hyperspectral data on graphics processing units. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 9, 952–961. [Google Scholar] [CrossRef]
- Zhang, L.; Du, B.; Zhong, Y. Hybrid detectors based on selective endmembers. IEEE Trans. Geosci. Remote Sens. 2010, 48, 2633–2646. [Google Scholar] [CrossRef]
- Kowkabi, F.; Ghassemian, H.; Keshavarz, A. A fast spatial–spectral preprocessing module for hyperspectral endmember extraction. IEEE Geosci. Remote Sens. Lett. 2016, 13, 782–786. [Google Scholar] [CrossRef]
- Kowkabi, F.; Keshavarz, A. Using spectral Geodesic and spatial Euclidean weights of neighbourhood pixels for hyperspectral Endmember Extraction preprocessing. ISPRS J. Photogramm. Remote Sens. 2019, 158, 201–218. [Google Scholar] [CrossRef]
- Zhang, C.; Qin, Q.; Zhang, T.; Sun, Y.; Chen, C. Endmember extraction from hyperspectral image based on discrete firefly algorithm (EE-DFA). ISPRS J. Photogramm. Remote Sens. 2017, 126, 108–119. [Google Scholar] [CrossRef]
- Sun, W.; Yang, G.; Wu, K.; Li, W.; Zhang, D. Pure endmember extraction using robust kernel archetypoid analysis for hyperspectral imagery. ISPRS J. Photogramm. Remote Sens. 2017, 131, 147–159. [Google Scholar] [CrossRef]
- Du, B.; Wei, Q.; Liu, R. An improved quantum-behaved particle swarm optimization for endmember extraction. IEEE Trans. Geosci. Remote Sens. 2019, 57, 6003–6017. [Google Scholar] [CrossRef]
- Tong, L.; Du, B.; Liu, R.; Zhang, L. An improved multiobjective discrete particle swarm optimization for hyperspectral endmember extraction. IEEE Trans. Geosci. Remote Sens. 2019, 57, 7872–7882. [Google Scholar] [CrossRef]
- Lin, C.H.; Chi, C.Y.; Chen, L.; Miller, D.J.; Wang, Y. Detection of sources in non-negative blind source separation by minimum description length criterion. IEEE Trans. Neural Networks Learn. Syst. 2017, 29, 4022–4037. [Google Scholar] [CrossRef]
- Rissanen, J. Modeling by shortest data description. Automatica 1978, 14, 465–471. [Google Scholar] [CrossRef]
- Schwarz, G. Estimating the dimension of a model. Ann. Stat. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Akaike, H. Akaike, H. A new look at the statistical model identification. In Selected Papers of Hirotugu Akaike; Springer: Berlin/Heidelberg, Germany, 1974; pp. 716–723. [Google Scholar]
- Graham, M.W.; Miller, D.J. Unsupervised learning of parsimonious mixtures on large spaces with integrated feature and component selection. IEEE Trans. Signal Process. 2006, 54, 1289–1303. [Google Scholar] [CrossRef]
- Peres-Neto, P.R.; Jackson, D.A.; Somers, K.M. How many principal components? Stopping rules for determining the number of non-trivial axes revisited. Comput. Stat. Data Anal. 2005, 49, 974–997. [Google Scholar] [CrossRef]
- Bioucas-Dias, J.M.; Nascimento, J.M. Hyperspectral subspace identification. IEEE Trans. Geosci. Remote Sens. 2008, 46, 2435–2445. [Google Scholar] [CrossRef]
- Chang, C.I.; Du, Q. Estimation of number of spectrally distinct signal sources in hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2004, 42, 608–619. [Google Scholar] [CrossRef]
- Halimi, A.; Honeine, P.; Kharouf, M.; Richard, C.; Tourneret, J.Y. Estimating the intrinsic dimension of hyperspectral images using a noise-whitened eigengap approach. IEEE Trans. Geosci. Remote Sens. 2016, 54, 3811–3821. [Google Scholar] [CrossRef]
- Ambikapathi, A.; Chan, T.H.; Chi, C.Y.; Keizer, K. Hyperspectral data geometry-based estimation of number of endmembers using p-norm-based pure pixel identification algorithm. IEEE Trans. Geosci. Remote Sens. 2012, 51, 2753–2769. [Google Scholar] [CrossRef]
- Parente, M.; Plaza, A. Survey of geometric and statistical unmixing algorithms for hyperspectral images. In Proceedings of the 2010 2nd Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing, Reykjavik, Iceland, 14–16 June 2010; pp. 1–4. [Google Scholar]
- Boardman, J.W.; Kruse, F.A.; Green, R.O. Mapping target signatures via partial unmixing of AVIRIS data. In Proceedings of the Summaries 5th JPL Airborne Earth Science Workshop, Pasadena, CA, USA, 23–26 January 1995; Volume 1, pp. 23–26. [Google Scholar]
- Tao, X.; Cui, T.; Yu, Z.; Ren, P. Locality Preserving Endmember Extraction for Estimating Green Algae Area. In Proceedings of the 2018 OCEANS-MTS/IEEE Kobe Techno-Oceans (OTO), Kobe, Japan, 28–31 May 2018; pp. 1–4. [Google Scholar]
- Tao, X.; Cui, T.; Ren, P. Cofactor-Based Efficient Endmember Extraction for Green Algae Area Estimation. IEEE Geosci. Remote Sens. Lett. 2019, 16, 849–853. [Google Scholar] [CrossRef]
- Nascimento, J.M.; Dias, J.M. Vertex component analysis: A fast algorithm to unmix hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2005, 43, 898–910. [Google Scholar] [CrossRef]
- Miao, L.; Qi, H. Endmember extraction from highly mixed data using minimum volume constrained nonnegative matrix factorization. IEEE Trans. Geosci. Remote Sens. 2007, 45, 765–777. [Google Scholar] [CrossRef]
- Chan, T.H.; Chi, C.Y.; Huang, Y.M.; Ma, W.K. A convex analysis-based minimum-volume enclosing simplex algorithm for hyperspectral unmixing. IEEE Trans. Signal Process. 2009, 57, 4418–4432. [Google Scholar] [CrossRef]
- Li, J.; Bioucas-Dias, J.M. Minimum volume simplex analysis: A fast algorithm to unmix hyperspectral data. In Proceedings of the IGARSS 2008-2008 IEEE International Geoscience and Remote Sensing Symposium, Boston, MA, USA, 7–11 July 2008; Volume 3, p. III–250. [Google Scholar]
- Li, J.; Agathos, A.; Zaharie, D.; Bioucas-Dias, J.M.; Plaza, A.; Li, X. Minimum volume simplex analysis: A fast algorithm for linear hyperspectral unmixing. IEEE Trans. Geosci. Remote Sens. 2015, 53, 5067–5082. [Google Scholar]
- Lin, C.H.; Chi, C.Y.; Wang, Y.H.; Chan, T.H. A fast hyperplane-based minimum-volume enclosing simplex algorithm for blind hyperspectral unmixing. IEEE Trans. Signal Process. 2015, 64, 1946–1961. [Google Scholar] [CrossRef]
- Hearst, M.A.; Dumais, S.T.; Osuna, E.; Platt, J.; Scholkopf, B. Support vector machines. IEEE Intell. Syst. Their Appl. 1998, 13, 18–28. [Google Scholar] [CrossRef]
- Bioucas-Dias, J.M.; Figueiredo, M.A. Alternating direction algorithms for constrained sparse regression: Application to hyperspectral unmixing. In Proceedings of the 2010 2nd Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing, Reykjavik, Iceland, 14–16 June 2010; pp. 1–4. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).