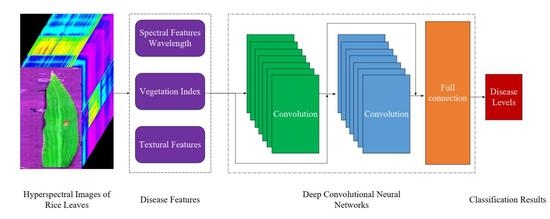
Rice Leaf Blast Classification Method Based on Fused Features and One-Dimensional Deep Convolutional Neural Network
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Data Acquisition and Processing
2.2.1. Sample Collection
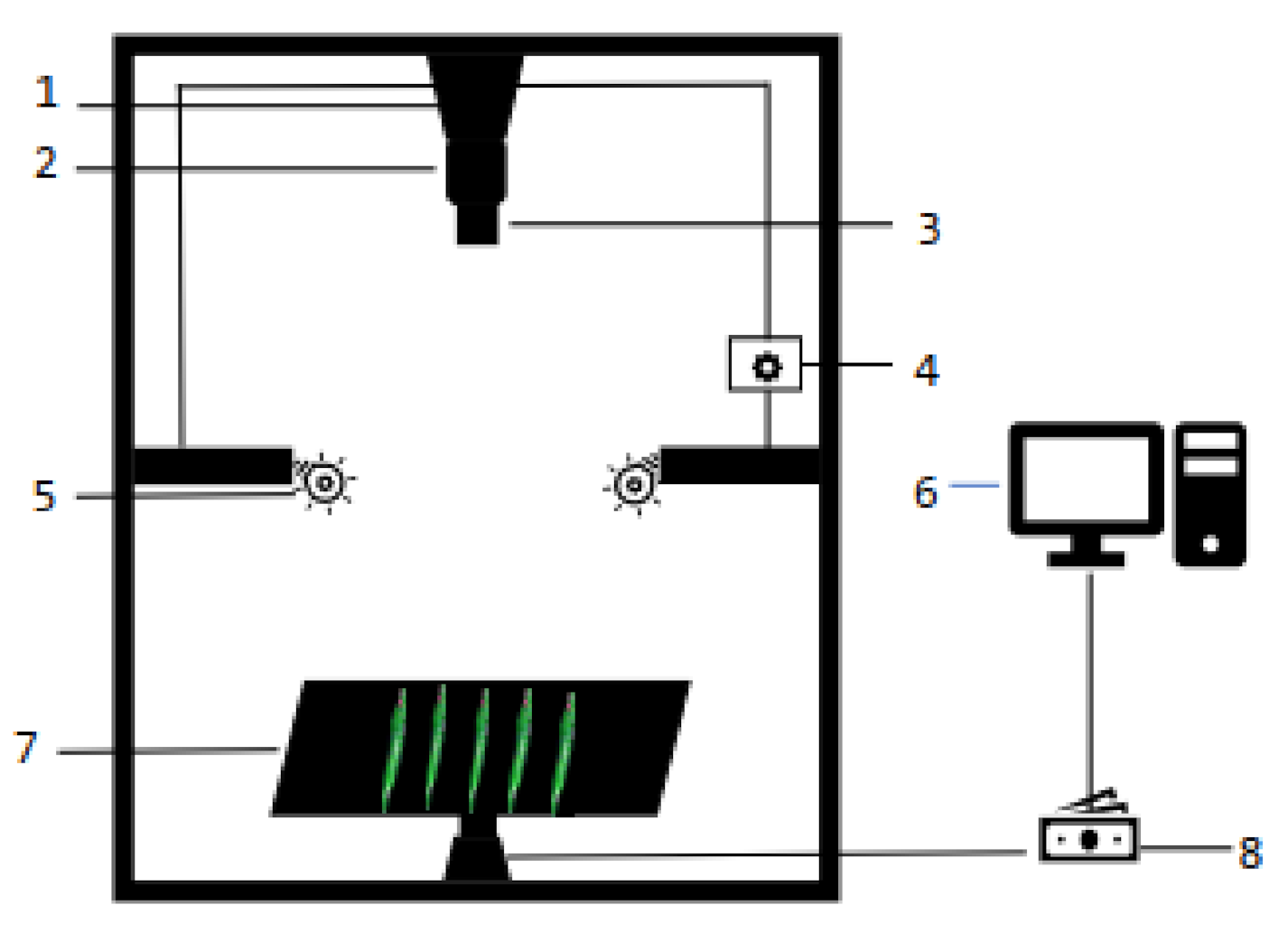
2.2.2. Hyperspectral Image Acquisition
2.2.3. Spectra Extraction and Processing
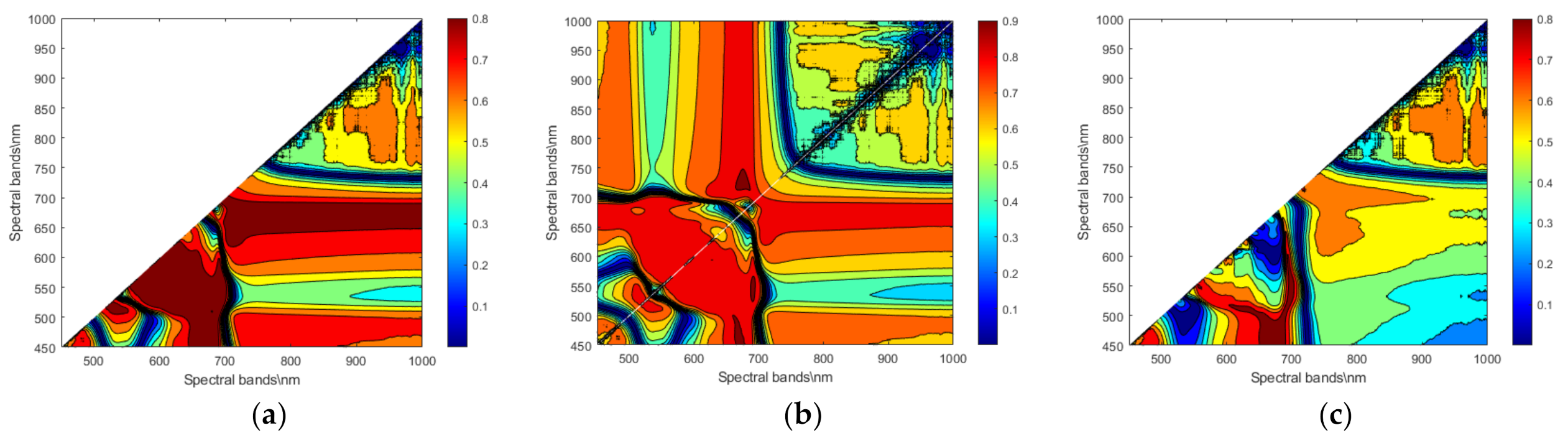
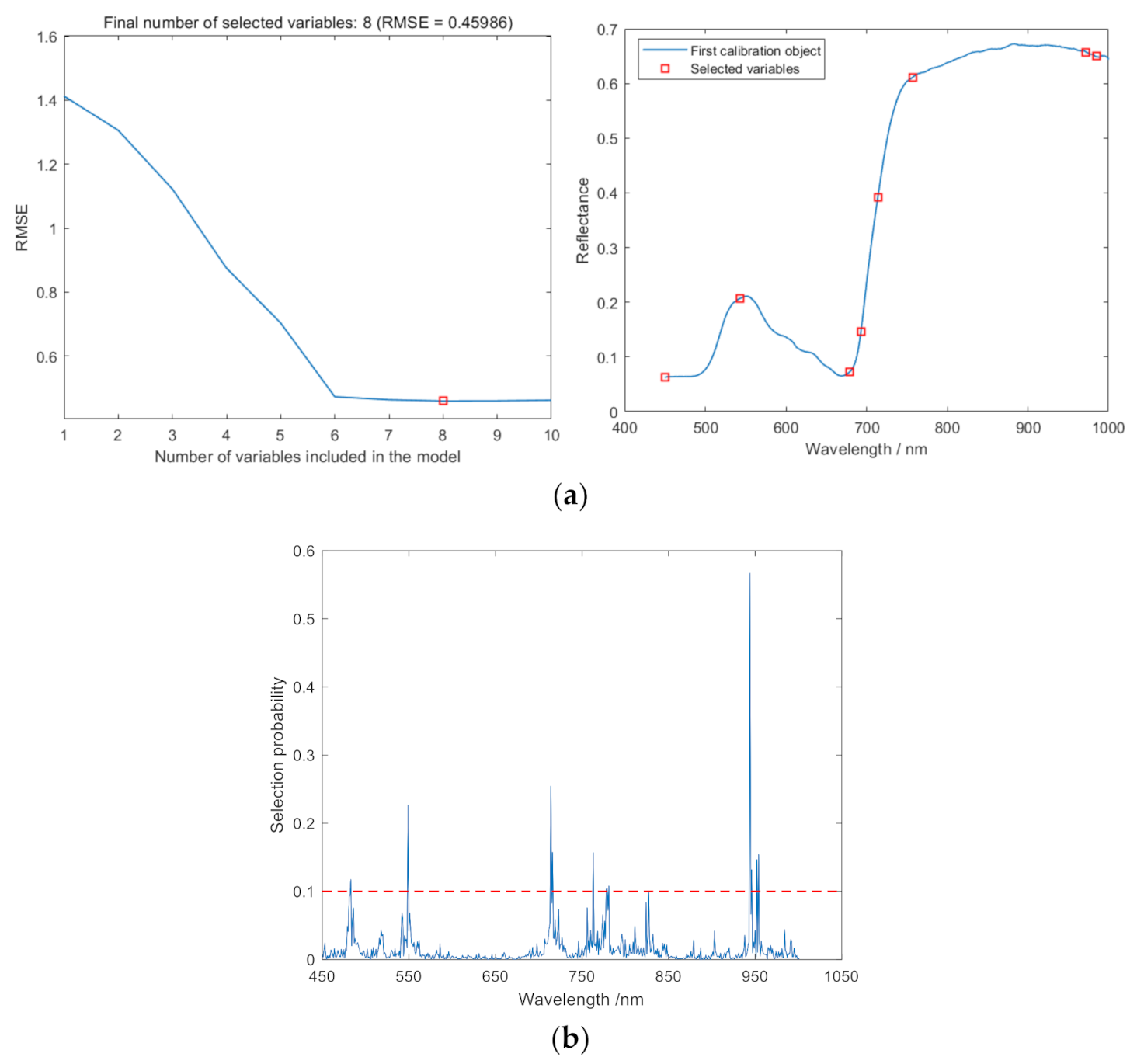
2.3. Optimal Spectral Feature Selection
2.4. Texture Features Extraction
2.5. Vegetation Index Extraction
2.6. Disease Classification Model
Deep Convolutional Neural Network
3. Results
3.1. Spectral Response Characteristics of Rice Leaves
3.2. Optimal Features
3.2.1. Vegetation Indices
3.2.2. Extraction of Hyperspectral Features
3.2.3. Extraction of Texture Features by GLCM
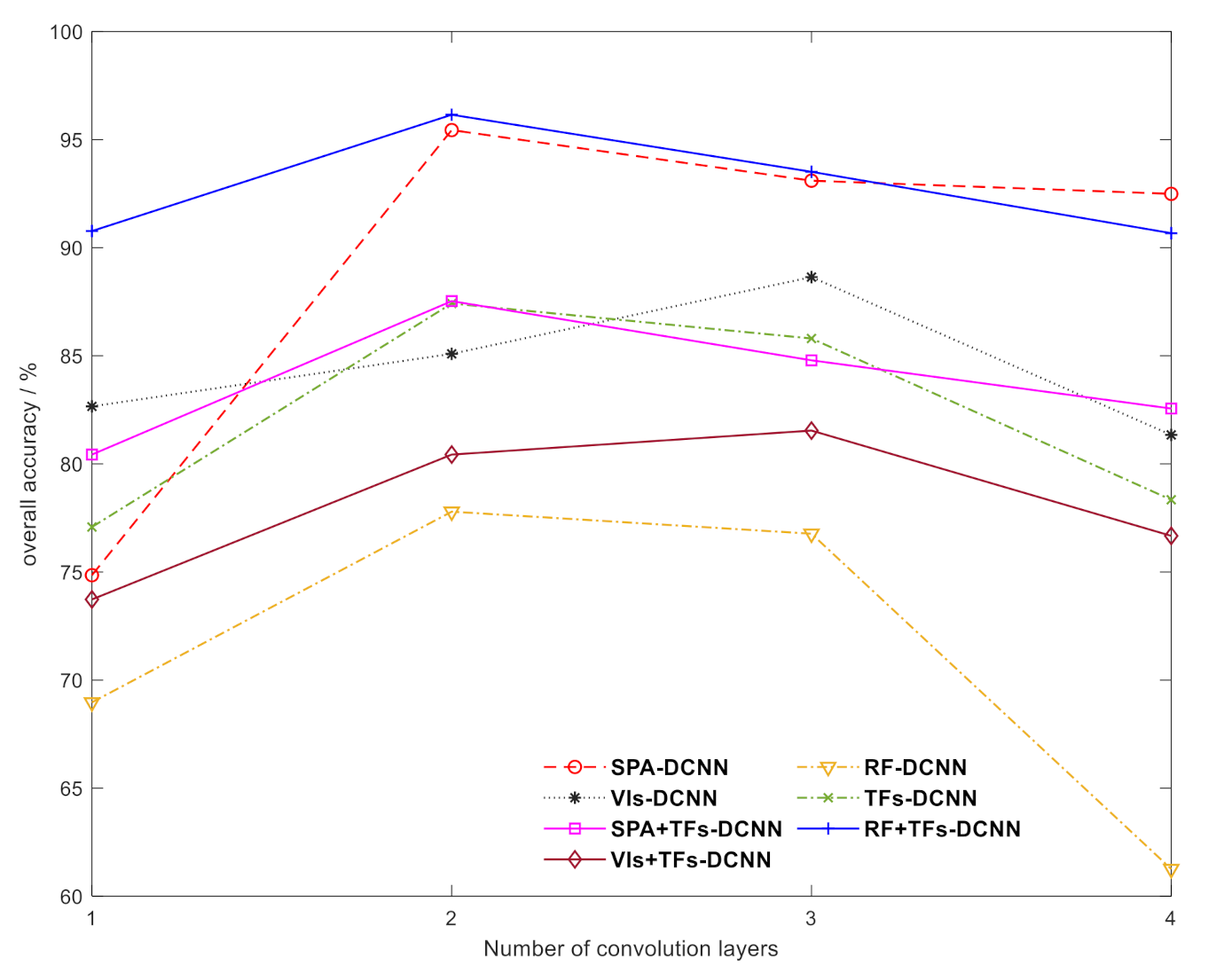
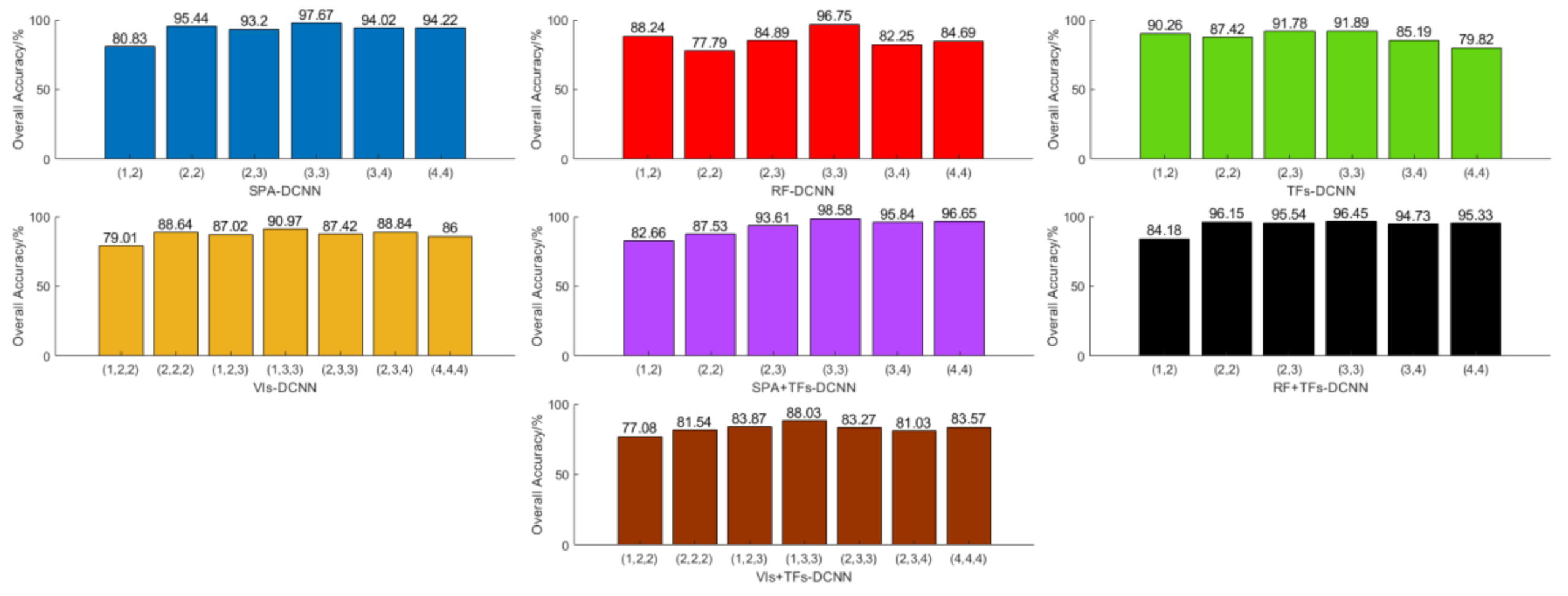
3.3. Sensitivity Analysis of the Number of Convolutional Layers and Convolutional Kernel Size for the DCNN
3.4. DCNN-Based Disease Classification of Rice Leaf Blast
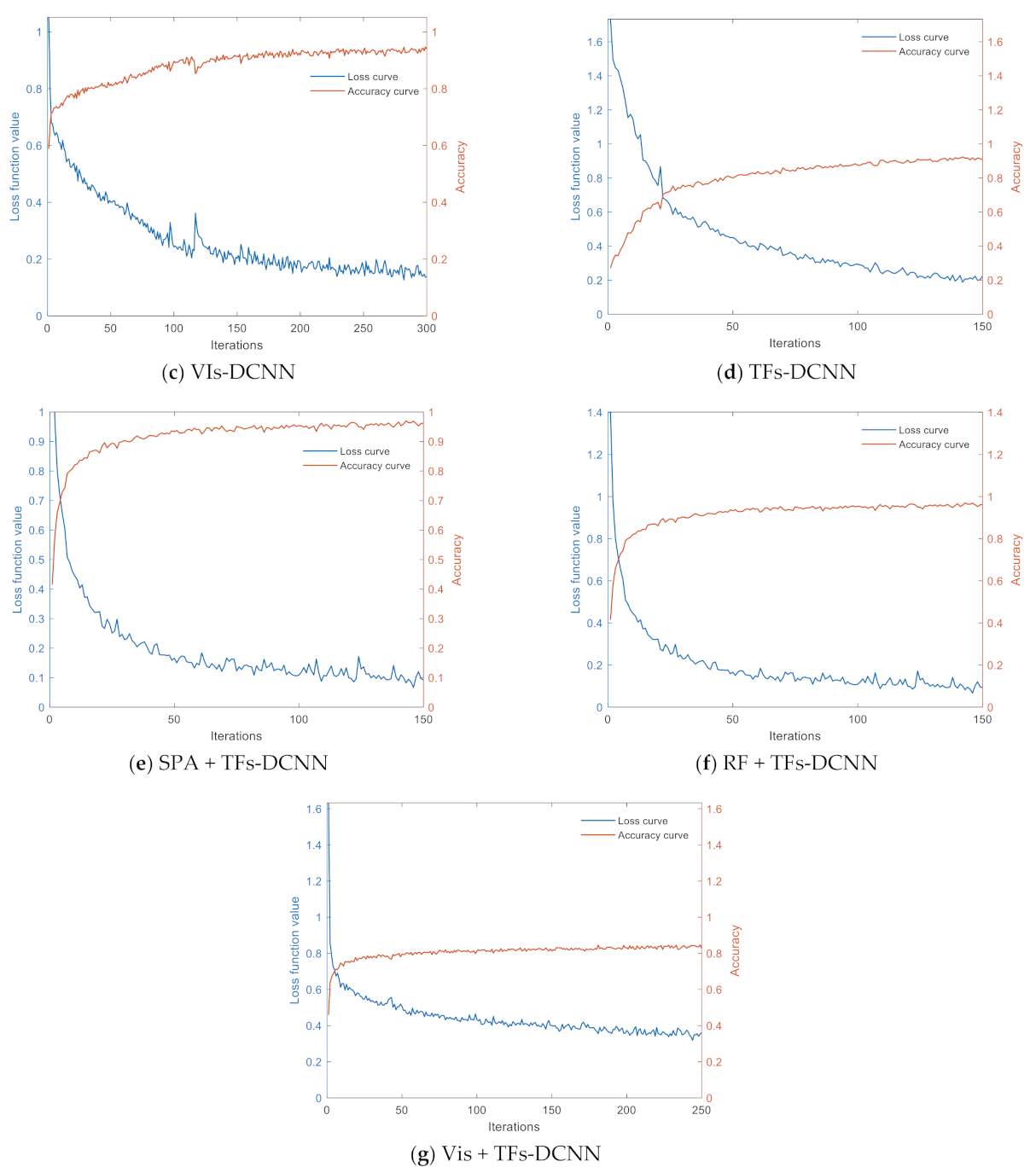
3.4.1. DCNN Model Training and Analysis
3.4.2. DCNN Model Testing and Analysis
3.4.3. Comparison with Other Classification Models
4. Discussion
5. Conclusions
Appendix A
| Methods | SVM - F1-Score /% | ELM - F1-Score /% | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Level 0 | Level 1 | Level 2 | Level 3 | Level 4 | Level 0 | Level 1 | Level 2 | Level 3 | Level 4 | |
| SPA | 100.00 | 92.93 | 90.41 | 87.89 | 95.81 | 97.53 | 88.64 | 87.31 | 83.96 | 92.46 |
| RF | 100.00 | 89.80 | 88.84 | 86.39 | 91.26 | 100 | 88.45 | 83.25 | 89.26 | 95.90 |
| VIs | 93.30 | 81.91 | 86.61 | 80.10 | 87.39 | 98.64 | 82.64 | 74.47 | 72.04 | 90.39 |
| TFs | 86.93 | 84.48 | 89.85 | 87.75 | 91.77 | 76.55 | 70.25 | 88.79 | 80.00 | 93.75 |
| SPA+TFs | 98.77 | 94.49 | 95.88 | 93.97 | 95.10 | 97.41 | 88.76 | 86.44 | 87.19 | 97.94 |
| RF+TFs | 97.03 | 90.03 | 93.56 | 93.64 | 96.90 | 97.94 | 88.69 | 86.55 | 86.17 | 95.77 |
| VIs+TFs | 95.93 | 79.58 | 78.99 | 64.09 | 83.25 | 95.81 | 66.09 | 67.29 | 65.35 | 80.98 |
| Methods | Inception V3 - F1-Score /% | ZF-Net - F1-Score /% | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Level 0 | Level 1 | Level 2 | Level 3 | Level 4 | Level 0 | Level 1 | Level 2 | Level 3 | Level 4 | |
| SPA | 100 | 94.43 | 94.54 | 93.01 | 94.39 | 100 | 96.61 | 89.74 | 87.61 | 97.04 |
| RF | 99.10 | 98.21 | 84.04 | 83.46 | 94.12 | 99.55 | 97.41 | 95.80 | 93.51 | 96.16 |
| VIs | 97.40 | 82.39 | 81.35 | 79.78 | 92.42 | 96.91 | 85.71 | 94.99 | 79.64 | 88.79 |
| TFs | 95.24 | 84.52 | 88.79 | 85.99 | 89.72 | 98.46 | 93.11 | 89.72 | 87.34 | 92.38 |
| SPA+TFs | 98.20 | 97.28 | 97.40 | 94.85 | 97.51 | 99.00 | 98.76 | 98.40 | 95.36 | 97.41 |
| RF+TFs | 97.16 | 94.64 | 96.88 | 91.91 | 95.77 | 98.06 | 96.59 | 96.28 | 92.71 | 96.07 |
| VIs+TFs | 96.06 | 73.19 | 76.14 | 73.94 | 90.75 | 96.52 | 83.12 | 80.57 | 77.68 | 91.13 |
| Methods | BiGRU - F1-Score /% | TextCNN - F1-Score /% | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Level 0 | Level 1 | Level 2 | Level 3 | Level 4 | Level 0 | Level 1 | Level 2 | Level 3 | Level 4 | |
| SPA | 100 | 94.94 | 93.43 | 95.82 | 98.50 | 100 | 94.22 | 92.02 | 93.82 | 97.99 |
| RF | 100 | 93.77 | 92.60 | 89.33 | 94.69 | 99.10 | 87.58 | 88.21 | 75.47 | 90.28 |
| VIs | 96.91 | 85.94 | 83.73 | 82.97 | 92.86 | 96.91 | 80.57 | 77.43 | 77.64 | 87.40 |
| TFs | 89.69 | 91.45 | 94.25 | 85.79 | 88.41 | 93.11 | 80.79 | 87.29 | 92.31 | 98.37 |
| SPA+TFs | 100 | 99.07 | 96.68 | 94.21 | 97.24 | 100 | 98.79 | 97.88 | 95.31 | 97.03 |
| RF+TFs | 96.06 | 95.58 | 98.13 | 94.95 | 96.73 | 97.22 | 94.74 | 96.31 | 93.26 | 95.85 |
| VIs+TFs | 95.51 | 73.90 | 74.44 | 73.04 | 89.45 | 96.61 | 8032 | 81.34 | 73.23 | 87.62 |
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Srivastava, D.; Shamim, M.; Kumar, M.; Mishra, A.; Pandey, P.; Kumar, D.; Yadav, P.; Siddiqui, M.H.; Singh, K.N. Current Status of Conventional and Molecular Interventions for Blast Resistance in Rice. Rice Sci. 2017, 24, 299–321. [Google Scholar] [CrossRef]
- Deutsch, C.A.; Tewksbury, J.J.; Tigchelaar, M.; Battisti, D.S.; Merrill, S.C.; Huey, R.B.; Naylor, R.L. Increase in crop losses to insect pests in a warming climate. Science 2018, 361, 916–919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, W.; Shi, Y.; Dong, Y.; Ye, H.; Wu, M.; Cui, B.; Liu, L. Progress and prospects of crop diseases and pests monitoring by remote sensing. Smart Agric. 2019, 1, 1–11. [Google Scholar]
- Akintayo, A.; Tylka, G.L.; Singh, A.K.; Ganapathysubramanian, B.; Singh, A.; Sarkar, S. A deep learning framework to discern and count microscopic nematode eggs. Sci. Rep. 2018, 8, 9145. [Google Scholar] [CrossRef] [PubMed]
- Bock, C.H.; Poole, G.H.; Parker, P.E.; Gottwald, T.R. Plant Disease Severity Estimated Visually, by Digital Photography and Image Analysis, and by Hyperspectral Imaging. Crit. Rev. Plant Sci. 2010, 29, 59–107. [Google Scholar] [CrossRef]
- Naik, H.S.; Zhang, J.; Lofquist, A.; Assefa, T.; Sarkar, S.; Ackerman, D.; Singh, A.; Singh, A.K.; Ganapathysubramanian, B. A real-time phenotyping framework using machine learning for plant stress severity rating in soybean. Plant Methods 2017, 13, 23. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Naik, H.S.; Assefa, T.; Sarkar, S.; Reddy, R.V.C.; Singh, A.; Ganapathysubramanian, B.; Singh, A.K. Computer vision and machine learning for robust phenotyping in genome-wide studies. Sci. Rep. 2017, 7, srep44048. [Google Scholar] [CrossRef]
- Zheng, Z.; Qi, L.; Ma, X.; Zhu, X.; Wang, W. Grading method of rice leaf blast using hyperspectral imaging technology. Trans. Chin. Soc. Agric. Eng. 2013, 29, 138–144. [Google Scholar]
- Asibi, A.E.; Chai, Q.; Coulter, J.A. Rice Blast: A Disease with Implications for Global Food Security. Agronomy 2019, 9, 451. [Google Scholar] [CrossRef] [Green Version]
- Larijani, M.R.; Asli-Ardeh, E.A.; Kozegar, E.; Loni, R. Evaluation of image processing technique in identifying rice blast disease in field conditions based on KNN algorithm improvement by K-means. Food Sci. Nutr. 2019, 7, 3922–3930. [Google Scholar] [CrossRef]
- Zarbafi, S.S.; Rabiei, B.; Ebadi, A.A.; Ham, J.H. Statistical analysis of phenotypic traits of rice (Oryza sativa L.) related to grain yield under neck blast disease. J. Plant Dis. Prot. 2019, 126, 293–306. [Google Scholar] [CrossRef]
- Bastiaans, L. Effects of leaf blast on photosynthesis of rice. 1. Leaf photosynthesis. Eur. J. Plant Pathol. 1993, 99, 197–203. [Google Scholar] [CrossRef] [Green Version]
- Nabina, N.; Kiran, B. A Review of Blast Disease of Rice in Nepal. J. Plant Pathol. Microbiol. 2021, 12, 1–5. [Google Scholar]
- Gowen, A.; O'Donnell, C.; Cullen, P.; Downey, G.; Frias, J. Hyperspectral imaging—An emerging process analytical tool for food quality and safety control. Trends Food Sci. Technol. 2007, 18, 590–598. [Google Scholar] [CrossRef]
- Feng, L.; Chai, R.-Y.; Sun, G.-M.; Wu, D.; Lou, B.-G.; He, Y. Identification and classification of rice leaf blast based on multi-spectral imaging sensor. Spectrosc. Spectr. Anal. 2009, 29, 2730–2733. [Google Scholar]
- Qi, L.; Ma, X.; Liao, X.-L. Rice bast resistance identification based on multi-spectral computer vision. J. Jilin Univ. Eng. Technol. Ed. 2009, 39, 356–359. [Google Scholar]
- Feng, L.; Wu, B.; Zhu, S.; Wang, J.; Su, Z.; Liu, F.; He, Y.; Zhang, C. Investigation on Data Fusion of Multisource Spectral Data for Rice Leaf Diseases Identification Using Machine Learning Methods. Front. Plant Sci. 2020, 11, 1664. [Google Scholar] [CrossRef]
- Wu, D.; Cao, F.; Zhang, H.; Sun, G.-M.; Feng, L.; He, Y. Study on disease level classification of rice panicle blast based on visible and near infrared spectroscopy. Spectrosc. Spectr. Anal. 2009, 29, 3295–3299. [Google Scholar]
- Barreto, A.; Paulus, S.; Varrelmann, M.; Mahlein, A.-K. Hyperspectral imaging of symptoms induced by Rhizoctonia solani in sugar beet: Comparison of input data and different machine learning algorithms. J. Plant Dis. Prot. 2020, 127, 441–451. [Google Scholar] [CrossRef]
- Abdulridha, J.; Ampatzidis, Y.; Roberts, P.; Kakarla, S.C. Detecting powdery mildew disease in squash at different stages using UAV-based hyperspectral imaging and artificial intelligence. Biosyst. Eng. 2020, 197, 135–148. [Google Scholar] [CrossRef]
- Fajardo, J.U.; Andrade, O.B.; Bonilla, R.C.; Cevallos-Cevallos, J.; Mariduena-Zavala, M.; Donoso, D.O.; Villardón, J.L.V. Early detection of black Sigatoka in banana leaves using hyperspectral images. Appl. Plant Sci. 2020, 8, e11383. [Google Scholar] [CrossRef]
- Bagheri, N.; Monavar, H.M.; Azizi, A.; Ghasemi, A. Detection of Fire Blight disease in pear trees by hyperspectral data. Eur. J. Remote Sens. 2017, 51, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.-Y.; Wu, H.-F.; Huang, J. Application of neural networks to discriminate fungal infection levels in rice panicles using hyperspectral reflectance and principal components analysis. Comput. Electron. Agric. 2010, 72, 99–106. [Google Scholar] [CrossRef]
- Zhang, G.; Xu, T.; Tian, Y.; Xu, H.; Song, J.; Lan, Y. Assessment of rice leaf blast severity using hyperspectral imaging during late vegetative growth. Australas. Plant Pathol. 2020, 49, 571–578. [Google Scholar] [CrossRef]
- Guo, A.; Huang, W.; Ye, H.; Dong, Y.; Ma, H.; Ren, Y.; Ruan, C. Identification of Wheat Yellow Rust Using Spectral and Texture Features of Hyperspectral Images. Remote Sens. 2020, 12, 1419. [Google Scholar] [CrossRef]
- Luo, Y.-H.; Jiang, P.; Xie, K.; Wang, F.-J. Research on optimal predicting model for the grading detection of rice blast. Opt. Rev. 2019, 26, 118–123. [Google Scholar] [CrossRef]
- Lu, B.; Jun, S.; Ning, Y.; Xiaohong, W.; Xin, Z. Identification of tea white star disease and anthrax based on hyperspectral image information. J. Food Process. Eng. 2020, 44, e13584. [Google Scholar] [CrossRef]
- Kang, L.; Yuan, J.Q.; Gao, R.; Kong, Q.M.; Jia, Y.J.; Su, Z.B. Early Identification of Rice Leaf Blast Based on Hyperspectral Imaging. Spectrosc. Spectr. Anal. 2021, 41, 898–902. [Google Scholar]
- Knauer, U.; Matros, A.; Petrovic, T.; Zanker, T.; Scott, E.S.; Seiffert, U. Improved classification accuracy of powdery mildew infection levels of wine grapes by spatial-spectral analysis of hyperspectral images. Plant Methods 2017, 13, 47. [Google Scholar] [CrossRef] [PubMed]
- Nagasubramanian, K.; Jones, S.; Sarkar, S.; Singh, A.K.; Singh, A.; Ganapathysubramanian, B. Hyperspectral band selection using genetic algorithm and support vector machines for early identification of charcoal rot disease in soybean stems. Plant Methods 2018, 14, 86. [Google Scholar] [CrossRef] [PubMed]
- Nettleton, D.F.; Katsantonis, D.; Kalaitzidis, A.; Sarafijanovic-Djukic, N.; Puigdollers, P.; Confalonieri, R. Predicting rice blast disease: Machine learning versus process-based models. BMC Bioinform. 2019, 20, 514–516. [Google Scholar] [CrossRef]
- Jia, D.; Chen, P. Effect of Low-altitude UAV Image Resolution on Inversion of Winter Wheat Nitrogen Concentration. Nongye Jixie Xuebao/Trans. Chin. Soc. Agric. Mach. 2020, 51, 164–169. [Google Scholar]
- Zhang, D.; Chen, G.; Zhang, H.; Jin, N.; Gu, C.; Weng, S.; Wang, Q.; Chen, Y. Integration of spectroscopy and image for identifying fusarium damage in wheat kernels. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 236, 118344. [Google Scholar] [CrossRef] [PubMed]
- Al-Saddik, H.; Laybros, A.; Billiot, B.; Cointault, F. Using Image Texture and Spectral Reflectance Analysis to Detect Yellowness and Esca in Grapevines at Leaf-Level. Remote Sens. 2018, 10, 618. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.-Y.; Chen, G.; Yin, X.; Hu, R.-J.; Gu, C.-Y.; Pan, Z.-G.; Zhou, X.-G.; Chen, Y. Integrating spectral and image data to detect Fusarium head blight of wheat. Comput. Electron. Agric. 2020, 175, 105588. [Google Scholar] [CrossRef]
- Zhu, H.; Chu, B.; Zhang, C.; Liu, F.; Jiang, L.; He, Y. Hyperspectral Imaging for Presymptomatic Detection of Tobacco Disease with Successive Projections Algorithm and Machine-learning Classifiers. Sci. Rep. 2017, 7, 4125. [Google Scholar] [CrossRef] [Green Version]
- Nagasubramanian, K.; Jones, S.; Singh, A.K.; Sarkar, S.; Singh, A.; Ganapathysubramanian, B. Plant disease identification using explainable 3D deep learning on hyperspectral images. Plant Methods 2019, 15, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Sun, C.; Qi, L.; Ma, X.; Wang, W. Rice panicle blast identification method based on deep convolution neural network. Trans. Chin. Soc. Agric. Eng. 2017, 33, 169–176. [Google Scholar]
- Zhang, X.; Han, L.; Dong, Y.; Shi, Y.; Huang, W.; Han, L.; González-Moreno, P.; Ma, H.; Ye, H.; Sobeih, T. A Deep Learning-Based Approach for Automated Yellow Rust Disease Detection from High-Resolution Hyperspectral UAV Images. Remote Sens. 2019, 11, 1554. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Jiang, H.; Li, C.; Jia, X.; Ghamisi, P. Deep Feature Extraction and Classification of Hyperspectral Images Based on Convolutional Neural Networks. IEEE Trans. Geosci. Remote Sens. 2016, 54, 6232–6251. [Google Scholar] [CrossRef] [Green Version]
- Araujo, M.; Saldanha, T.C.B.; Galvão, R.K.H.; Yoneyama, T.; Chame, H.C.; Visani, V. The successive projections algorithm for variable selection in spectroscopic multicomponent analysis. Chemom. Intell. Lab. Syst. 2001, 57, 65–73. [Google Scholar] [CrossRef]
- Li, H.-D.; Xu, Q.-S.; Liang, Y.-Z. Random frog: An efficient reversible jump Markov Chain Monte Carlo-like approach for variable selection with applications to gene selection and disease classification. Anal. Chim. Acta 2012, 740, 20–26. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the 29th IEEE Conference on Computer Vision and Pattern Recognition (CVPR 2016), Las Vegas, NV, USA, 26 June —1 July 2016; pp. 770–778. [Google Scholar]
- Xia, J.; Chanussot, J.; Du, P.; He, X. Rotation-Based Support Vector Machine Ensemble in Classification of Hyperspectral Data With Limited Training Samples. IEEE Trans. Geosci. Remote Sens. 2016, 54, 1519–1531. [Google Scholar] [CrossRef]
- Huang, G.-B.; Zhu, Q.-Y.; Siew, C.-K. Extreme learning machine: Theory and applications. Neurocomputing 2006, 70, 489–501. [Google Scholar] [CrossRef]
- Christian, S.; Vincent, V.; Sergey, L.; Jonathon, S.; Zbigniew, W. Rethinking the inception architecture for computer vision. arXiv 2015, arXiv:1512.00567. [Google Scholar]
- Matthew, D.Z.; Rob, F. Visualizing and understanding convolutional networks. arXiv 2013, arXiv:1311.2901. [Google Scholar]
- Chuang, J.Y.; Gulcehre, C.; Cho, K.; Bengio, Y. Empirical evaluation of gated recurrent neural networks on sequence modeling. arXiv 2014, arXiv:1412.3555. [Google Scholar]
- Kim, Y. Convolutional Neural Networks for Sentence Classification. Assoc. Comput. Linguist. 2014, 13, 1746–1751. [Google Scholar]
- Zhang, J.; Huang, Y.; Pu, R.; González-Moreno, P.; Yuan, L.; Wu, K.; Huang, W. Monitoring plant diseases and pests through remote sensing technology: A review. Comput. Electron. Agric. 2019, 165, 104943. [Google Scholar] [CrossRef]
- Le, T.T.H.; Kim, J.; Kim, H. An Effective Intrusion Detection Classifier Using Long Short-Term Memory with Gradient Descent Optimization. In Proceedings of the 2017 International Conference on Platform Technology and Service, IEEE 2017, Busan, Korea, 13–15 February 2017; pp. 155–160. [Google Scholar]
- Behmann, J.; Bohnenkamp, D.; Paulus, S.; Mahlein, A.-K. Spatial Referencing of Hyperspectral Images for Tracing of Plant Disease Symptoms. J. Imaging 2018, 4, 143. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Wang, G.; Liu, J.-D.; Ma, Z.-H.; Wang, J.; Li, T.-N. Extraction of Photosynthetic Parameters of Cotton Leaves under Disease Stress by Hyperspectral Remote Sensing. Spectrosc. Spectr. Anal. 2018, 38, 1834–1838. [Google Scholar]
- Ma, H.; Huang, W.; Jing, Y.; Pignatti, S.; Laneve, G.; Dong, Y.; Ye, H.; Liu, L.; Guo, A.; Jiang, J. Identification of Fusarium Head Blight in Winter Wheat Ears Using Continuous Wavelet Analysis. Sensors 2019, 20, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.C.; Xu, T.Y.; Zheng, W.; Deng, H.B. Classification and recognition approaches of tomato main organs based on DCNN. Trans. Chin. Soc. Agric. Eng. 2017, 33, 219–226. [Google Scholar]
- Huang, L.; Li, T.; Ding, C.; Zhao, J.; Zhang, D.; Yang, G. Diagnosis of the Severity of Fusarium Head Blight of Wheat Ears on the Basis of Image and Spectral Feature Fusion. Sensors 2020, 20, 2887. [Google Scholar] [CrossRef] [PubMed]
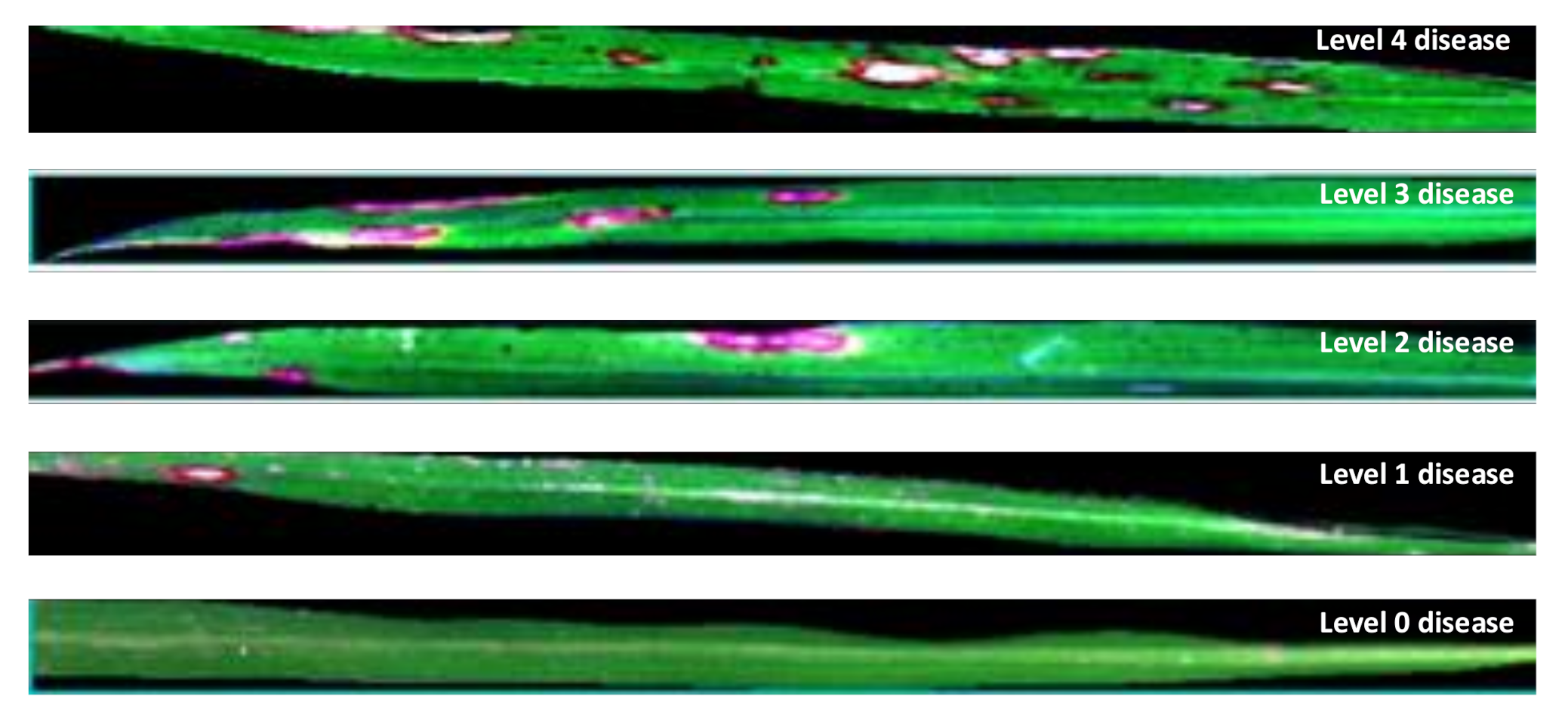
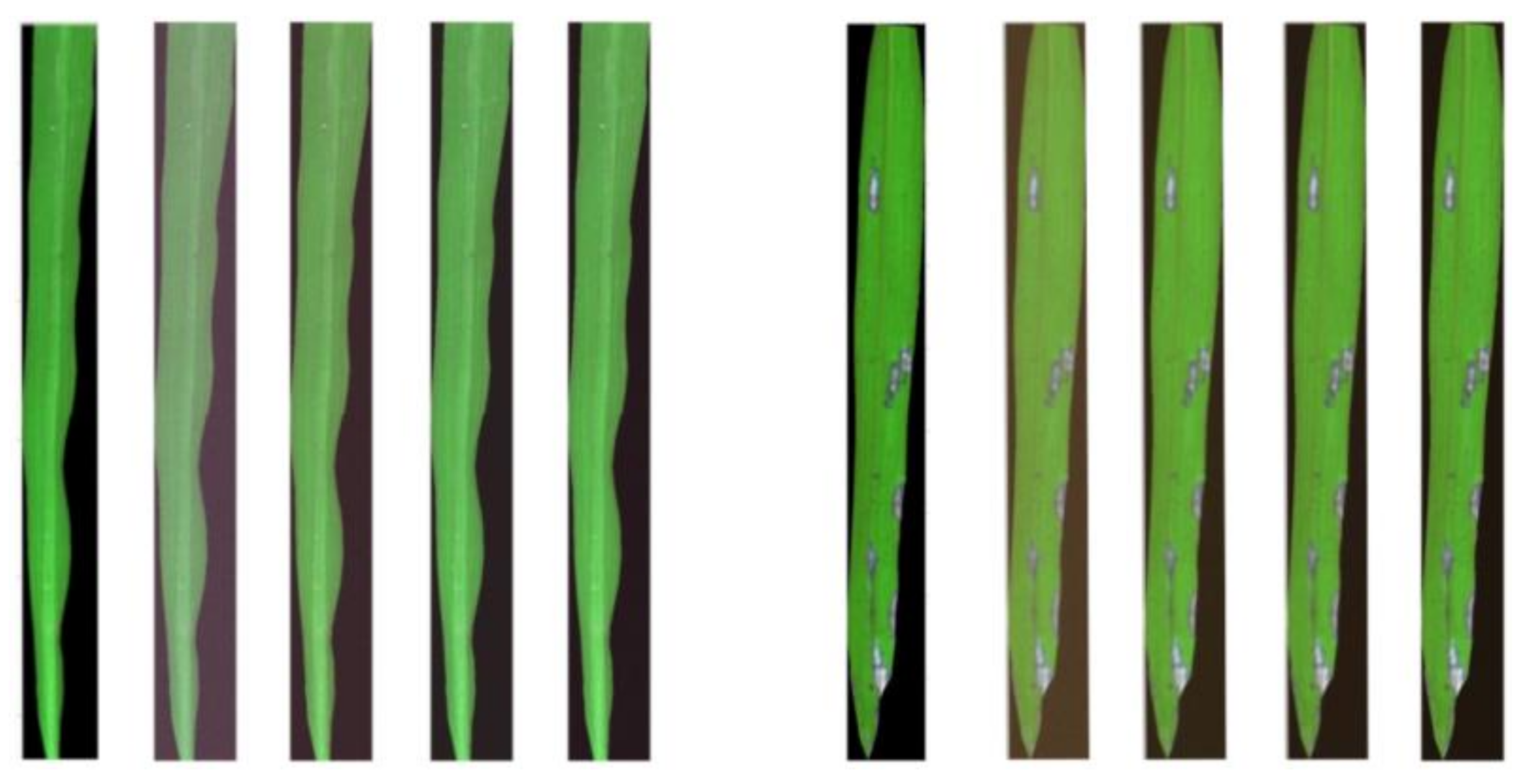





| Disease Level | Disease Level Determination Criteria | Sample Size |
|---|---|---|
| Level 0 | No disease spots. | 29 |
| Level 1 | Few and small spots, disease spot area less than 1% of leaf area. | 27 |
| Level 2 | Small and many spots or large and few disease spot area of 1~5% of leaf area. | 32 |
| Level 3 | Large and more spots, disease spot area of 5~10% of leaf area. | 27 |
| Level 4 | Large and more spots, disease spot area of 10~50% of leaf area. | 30 |
| Texture Features | Equation |
|---|---|
| Entropy | |
| Energy | |
| Correlation | |
| Contrast |
| Method | Variable Number | Wavelength/Nm |
|---|---|---|
| SPA | 8 | 450 543 679 693 714 757 972 985 |
| RF | 13 | 482 548 713 715 762 777 778 780 826 943 945 951 953 |
| Texture Features | Correlation Coefficient | p Value | Significance |
|---|---|---|---|
| MEne | 0.5618 | <0.001 | *** |
| SDEne | −0.2632 | <0.001 | *** |
| MEnt | −0.4914 | <0.001 | *** |
| SDEnt | −0.4263 | <0.001 | *** |
| MCon | −0.2308 | <0.001 | *** |
| SDCon | −0.2265 | <0.001 | *** |
| MCor | 0.1165 | <0.001 | *** |
| SDCor | −0.0365 | 0.0105 | ** |
| Descending Dimension Method | F1-Score (%) | OA (%) | Kappa (%) | ||||
|---|---|---|---|---|---|---|---|
| Level 0 | Level 1 | Level 2 | Level 3 | Level 4 | |||
| SPA | 100 | 97.44 | 95.74 | 96.15 | 98.54 | 97.67 | 97.08 |
| RF | 100 | 96.05 | 94.51 | 95.01 | 97.73 | 96.75 | 95.93 |
| VIs | 98.36 | 84.18 | 87.04 | 88.64 | 95.48 | 90.97 | 88.70 |
| TFs | 92.67 | 92.23 | 92.93 | 86.88 | 93.96 | 91.89 | 89.84 |
| SPA + TFs | 100.00 | 100.00 | 100.00 | 96.48 | 96.68 | 98.58 | 98.22 |
| RF + TFs | 100.00 | 100.00 | 97.93 | 91.36 | 93.66 | 96.45 | 95.55 |
| Vis + TFs | 97.17 | 83.66 | 85.79 | 80.72 | 92.13 | 88.03 | 85.04 |
| Methods | SVM | ELM | Inception V3 | ZF-Net | BiGRU | TextCNN | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OA (%) | Kappa (%) | OA (%) | Kappa (%) | OA (%) | Kappa (%) | OA (%) | Kappa (%) | OA (%) | Kappa (%) | OA (%) | Kappa (%) | |
| SPA | 93.41 | 91.74 | 90.19 | 87.82 | 95.44 | 94.28 | 94.42 | 93.01 | 96.65 | 95.81 | 95.74 | 94.66 |
| RF | 91.28 | 89.09 | 90.96 | 89.07 | 91.89 | 89.85 | 96.55 | 95.68 | 94.32 | 92.88 | 88.95 | 86.12 |
| VIs | 86.09 | 82.60 | 83.40 | 79.22 | 86.92 | 83.62 | 89.76 | 87.17 | 88.64 | 85.80 | 84.08 | 80.09 |
| TFs | 88.34 | 85.40 | 89.13 | 87.27 | 88.95 | 86.14 | 92.09 | 90.08 | 89.96 | 87.41 | 90.97 | 88.68 |
| SPA + TFs | 95.54 | 94.41 | 91.67 | 89.59 | 97.06 | 96.32 | 97.77 | 97.20 | 97.36 | 96.70 | 97.77 | 97.20 |
| RF + TFs | 94.42 | 93.01 | 91.02 | 88.82 | 95.33 | 94.16 | 96.04 | 95.05 | 96.35 | 95.43 | 95.54 | 94.41 |
| Vis + TFs | 80.61 | 75.69 | 74.94 | 68.79 | 83.47 | 79.30 | 86.00 | 82.49 | 81.14 | 76.40 | 83.77 | 79.73 |
| Method | OA (%) | Test Time (s) |
|---|---|---|
| SPA + TFs-SVM | 95.54 | 0.1058 |
| SPA + TFs-ELM | 91.67 | 0.0279 |
| SPA + TFs-Inception V3 | 97.06 | 0.5222 |
| SPA + TFs-ZF-Net | 97.77 | 0.4152 |
| SPA + TFs-BiGRU | 97.36 | 1.2086 |
| SPA + TFs-TextCNN | 97.77 | 0.3388 |
| SPA + TFs-DCNN (the model of this study) | 98.58 | 0.2200 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, S.; Cao, Y.; Xu, T.; Yu, F.; Zhao, D.; Zhang, G. Rice Leaf Blast Classification Method Based on Fused Features and One-Dimensional Deep Convolutional Neural Network. Remote Sens. 2021, 13, 3207. https://doi.org/10.3390/rs13163207
Feng S, Cao Y, Xu T, Yu F, Zhao D, Zhang G. Rice Leaf Blast Classification Method Based on Fused Features and One-Dimensional Deep Convolutional Neural Network. Remote Sensing. 2021; 13(16):3207. https://doi.org/10.3390/rs13163207
Chicago/Turabian StyleFeng, Shuai, Yingli Cao, Tongyu Xu, Fenghua Yu, Dongxue Zhao, and Guosheng Zhang. 2021. "Rice Leaf Blast Classification Method Based on Fused Features and One-Dimensional Deep Convolutional Neural Network" Remote Sensing 13, no. 16: 3207. https://doi.org/10.3390/rs13163207
APA StyleFeng, S., Cao, Y., Xu, T., Yu, F., Zhao, D., & Zhang, G. (2021). Rice Leaf Blast Classification Method Based on Fused Features and One-Dimensional Deep Convolutional Neural Network. Remote Sensing, 13(16), 3207. https://doi.org/10.3390/rs13163207