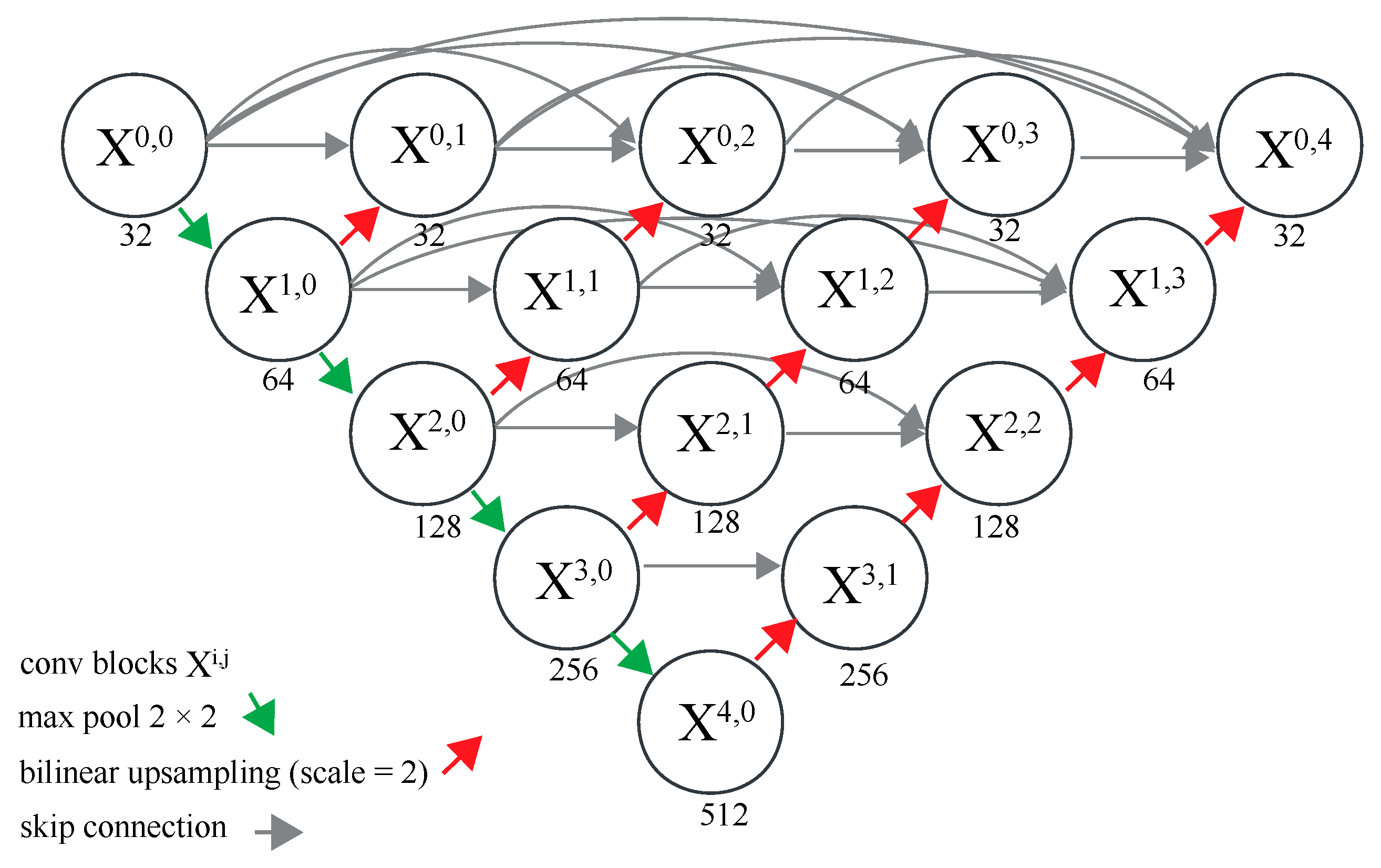
1. Introduction
Global caribou herd populations have been reportedly declining in recent decades because of multiple factors ranging from climate change to hunting and disease, according to “International Union for the Conservation of Nature” [
1]. Although the main driver of this population decline is still ambiguous, most factors thought to be contributing are directly or indirectly related to human activities [
2,
3], such as land-cover changes that can affect resource availability for caribou [
4] and can cause them to change their distribution, migration, and timing patterns when foraging for food [
5]. Lichen is an important source of food for caribou especially during winters [
6,
7].
Since the base data for caribou lichen mapping is generally UAV images covering small areas, one of the primary challenges is to extend the maps generated based on UAV images to much larger areas. Therefore, along with the type of data used, the method for lichen mapping is also of crucial importance to improve the results. This is due to the fact that the limited amount of data provided by UAV images may not help generalize results to larger scales. Given recent advances in the field of artificial intelligence (AI), this, therefore, demands for deploying advanced algorithmic approaches (such as semi-supervised learning (SSL)) that can partially compensate for the lack of sufficient labeled data.
Early research on lichen cover mapping using RS data was based on non-machine-learning (ML) approaches. In one of the earliest studies in this field, Petzold and Goward [
8] found that Normalized Difference Vegetation Index (NDVI) may not be a reliable measure for lichen cover estimation (especially where the surface is completely lichen covered) as the values of this index may be misinterpreted as sparse green vascular plants. The Normalized Difference Lichen Index (NDLI) proposed by Nordberg [
9] is an index specifically for lichen detection that is mainly used in conjunction with other vegetation indices. In a more recent study, Théau and Duguay [
10] improved the mapping of lichen abundance using Landsat TM data by employing spectral mixing analysis (SMA). The authors reported that the SMA overestimated (underestimated) lichen fractional coverage over sites with low (high) lichen presence.
Orthogonal to the aforementioned studies, in 2011, Gilichinsky et al. [
11] tested three classifiers for lichen mapping using SPOT-5 and Landsat ETM+ images. They found that
Mahalanobis distance classifier generated the highest accuracy (84.3%) for mapping lichen based on SPOT-5 images in their study. On the other hand, it turned out that a maximum likelihood classifier reached an overall accuracy (OA) of 76.8% on Landsat ETM+ imagery. However, the authors reported that the model did not perform well over areas where lichen cover was <50%. Later in 2014, Falldorf et al. [
12] developed a lichen volume estimator (LVE) based on Landsat TM images using a 2D Gaussian regression model. To train the model, the authors used the NDLI and Normalized Difference Moisture Index (NDMI) as the independent variables, and ground-based volumetric measurements of lichen cover as the dependent variable. Using a 10-fold cross-validation, the authors reported that the model achieved an R
2 of 0.67. In 2020, a study by Macander et al. [
13] employed different data sources, including plot data, UAV images, and airborne data, to map the fraction of lichen cover in Landsat imagery using a Random Forest (RF) regressor. However, the authors reported that they observed an overestimation for low lichen cover sites, and an underestimation for high lichen cover sites.
Although there have recently been seminal studies on lichen mapping using powerful ML models (e.g., deep neural networks (DNNs) [
14]), there are some gaps that have not yet been approached in this field. An important issue in this field is the lack of sufficient labeled, ground-truth lichen samples. As with any other environmental RS studies, lichen mapping suffers from “the curse of insufficient training data”. It is now clear that insufficient training data deteriorates the generalization power of a classifier, especially DNNs which can easily memorize training data [
15].
In case of limited labeled samples, a workaround is to use unlabeled samples that could be available in large volumes. In fact, learning from unlabeled samples could implicitly help improve the generalization performance of the model trained on limited labeled samples. There are a wide variety of methods for involving unlabeled data in a classification setup. Perhaps, one of the most commonly used approaches is unsupervised pre-training of a network and reusing its intermediate layers for training another network on the labeled set of interest [
16]. If unlabeled samples are not abundant, another method could be based on transfer learning [
17]. This approach is based on using a network trained on a large data set in another domain (e.g., natural images) or in the same domain but for a different application (e.g., urban mapping). This network can be then either fine-tuned on the limited labeled data of interest or used as a feature extractor for another classifier or other downstream tasks. The potential of transfer learning has also been proven in the RS community [
18].
Another class of methods that can take advantage of unlabeled data is semi-supervised learning (SSL). Some SSL methods, known as wrapper methods [
19], generally explicitly incorporate unlabeled samples into the training process by automatically labelling them to improve the performance of the model [
20]. The concept of
Teacher-Student SSL is an example of such a method that has shown to improve the generalization when limited labeled data are available [
20,
21]. In this regard,
Naïve-Student [
22] has been recently proposed as a potentially versatile SSL approach for the pixel-wise classification of street-level urban video sequences. This approach integrates SSL and self-training iteratively through a
Teacher and
Student network, where the
Student network can be the same as or larger than the
Teacher network. In contrast to a similar approach known as
Noisy-Student proposed earlier [
21],
Naïve-Student does not inject data-based noise (i.e., data augmentation) directly to the training process to improve the robustness of the
Student network. Instead, it uses test-time augmentation (TTA) functions when predicting pseudo-labels (i.e., labeling unlabeled samples) on which later the
Student network is trained. The premise of such an approach is to train a
Student network that outperforms the
Teacher network. Besides, the iterative nature of both
Noisy-Student and
Naive-Student approaches helps improve the
Student network further.
In environmental studies, there are cases where current maps were generated based on very high-spatial resolution aerial images covering small areas, but later the aim will be to reuse those maps to generate much larger-scale maps using another image source without conducting any further labeling. Scaling-up local maps is restricted by many factors (e.g., spatial/temporal misalignments, classification errors, etc.) that may prevent generating large-scale maps with appropriate accuracy. The accuracy of such a scale-up is further degraded in cases where the current high-resolution maps cover a very small, homogeneous area. This is due to differences in soil/substrate or climate conditions that could lead to variability in the spectral signature of the vegetation of interest [
23,
24]. Although multi-scale/sensor mapping in the field of RS is a common application, it has been mostly performed either using conventional ML approaches or using data-fusion frameworks [
25,
26,
27]. In this study, we approached this problem in a different way: To what degree is it possible to scale-up very-high-resolution lichen maps produced over small, homogenous areas to a much larger scale without labeling any new samples?
Taking into account the above question in this research, we aimed to scale-up a very high-resolution lichen map, generated from a small UAV scene, to a high-resolution satellite level, namely WorldView-2 (hereafter, WorldView). This scaling-up task posed several challenges. First, training a fully supervised approach in such a case would result in poor accuracy because training samples were not sufficient and diverse enough. Second, we expected the classifier performance to be negatively impacted by errors associated with image misalignments. These errors, caused by various spatial and temporal offsets, can lead to undesirable errors in the model training process. These sources of error caused noisy labels (i.e., wrong labels) in the WorldView image when the UAV-derived lichen map was scaled-up. We, therefore, investigated whether a small, noisy UAV-derived lichen map could be used to train a highly generalized model capable of producing accurate results over a much larger area. To improve classification performance, we adapted
Naive-Student [
22] to the lichen mapping task in this study to leverage unlabeled data in the training process.
3. Results
According to the experiments, the OA and F1-score of the classified UAV image were 97.26% and 96.84%, respectively.
Figure 5a illustrates the resampled (50-cm) classified UAV map superimposed onto the corresponding area of the WorldView scene. In this figure, it is also evident that there are some non-lichen pixels wrongly labeled as lichen or vice versa. The results of our assessment on the resampled map at the WorldView level showed an OA and F1-score of 83.61% and 90.74%, respectively, which indicates a tangible accuracy degradation compared to the original, non-scaled-up UAV map. No specific preprocessing or sampling was performed before training the SSL networks at the WorldView level.
The best result of training the
Teacher-
Student framework was achieved after three iterations (where the smallest labeled validation loss was achieved) namely that the
Student network was used as a
Teacher in two iterations.
Figure 6 shows a small portion of the WorldView image (also covering the UAV extent) that was used as an input to each of the three networks obtained after each iteration to predict lichen pixels. The
Student network improved gradually after each iteration. The most obvious performance improvement can be seen over the road pixels and some tree types that were differentiated more accurately from lichen patches as the network evolved. Essentially, the Type I error was reduced after each iteration without adding new training samples to the process. This confirms that an iterative
Teacher-Student SSL approach could help improve pixel-wise classification accuracy when only a small training set is available.
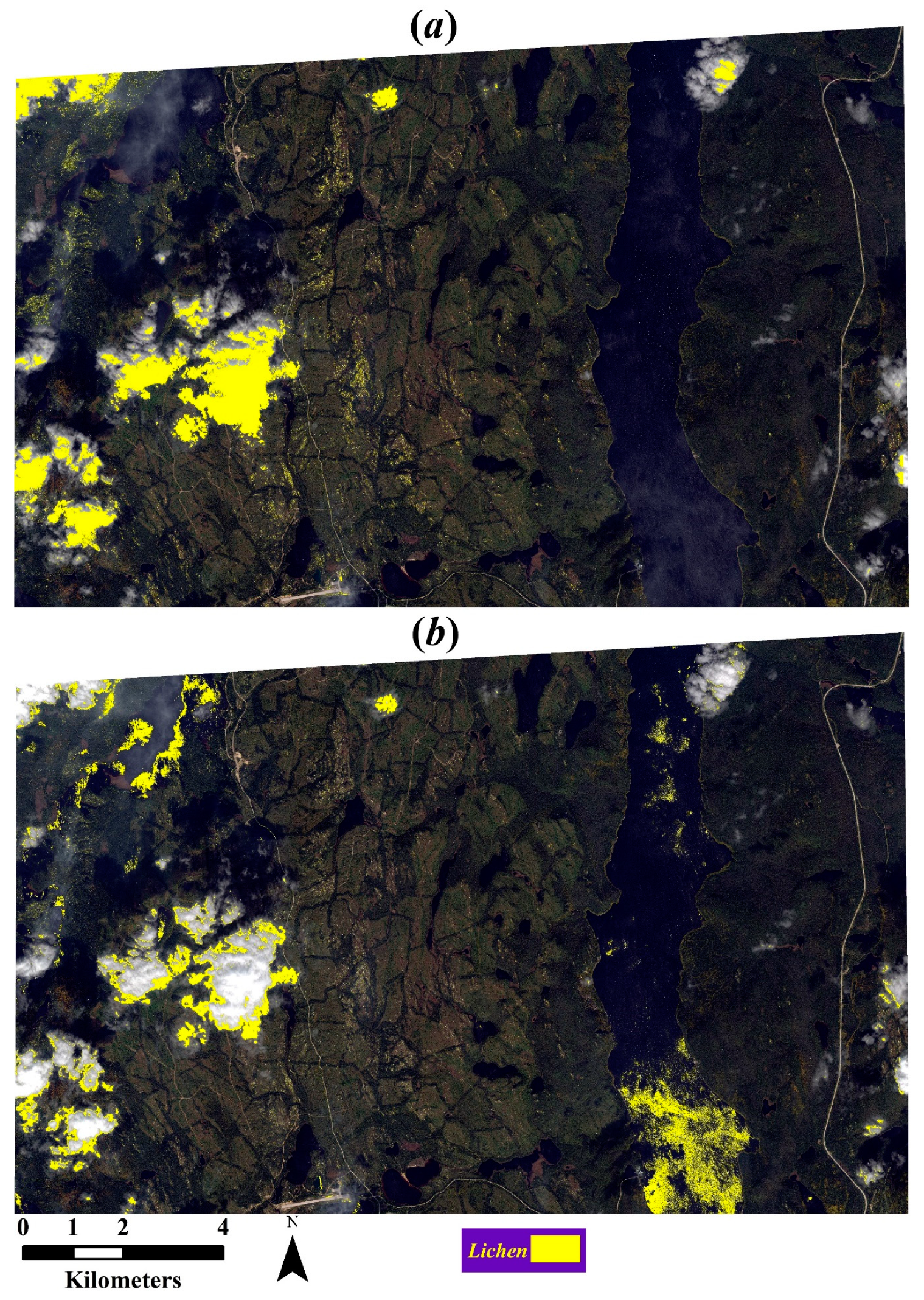
Figure 7 displays the map generated for the Northern part (i.e., test image) of the WorldView scene using the SSL approach. Unsurprisingly, some parts of the thick clouds were erroneously classified as lichen. However, most of the thin clouds and haze were correctly differentiated from lichen. We also observed that some rippling-water pixels were misclassified as lichen, although the number of misclassified water pixels was much less than that of misclassified cloud pixels. It should be noted that neither of these two classes were present in the labeled samples used for constructing the SSL models. Misclassification of some trees as lichen was caused by mislabeled pixels in the scaled-up UAV-derived lichen map. To handle misclassified pixels over cloud areas, we applied a simple cloud masking using the Fire Discrimination Index (FDI), which was reported to be useful for cloud-masking in WorldView imagery [
39]. Although applying this mask resulted in the removal of most of the lichen misclassifications over cloudy areas, there were still few cloud pixels that required manual removal as employing a larger threshold would remove some lichen pixels as well.
Based on our quantitative accuracy assessment, the SSL-based WorldView lichen map had an OA of 85.28% and F1-score of 84.38%. From the confusion matrix (
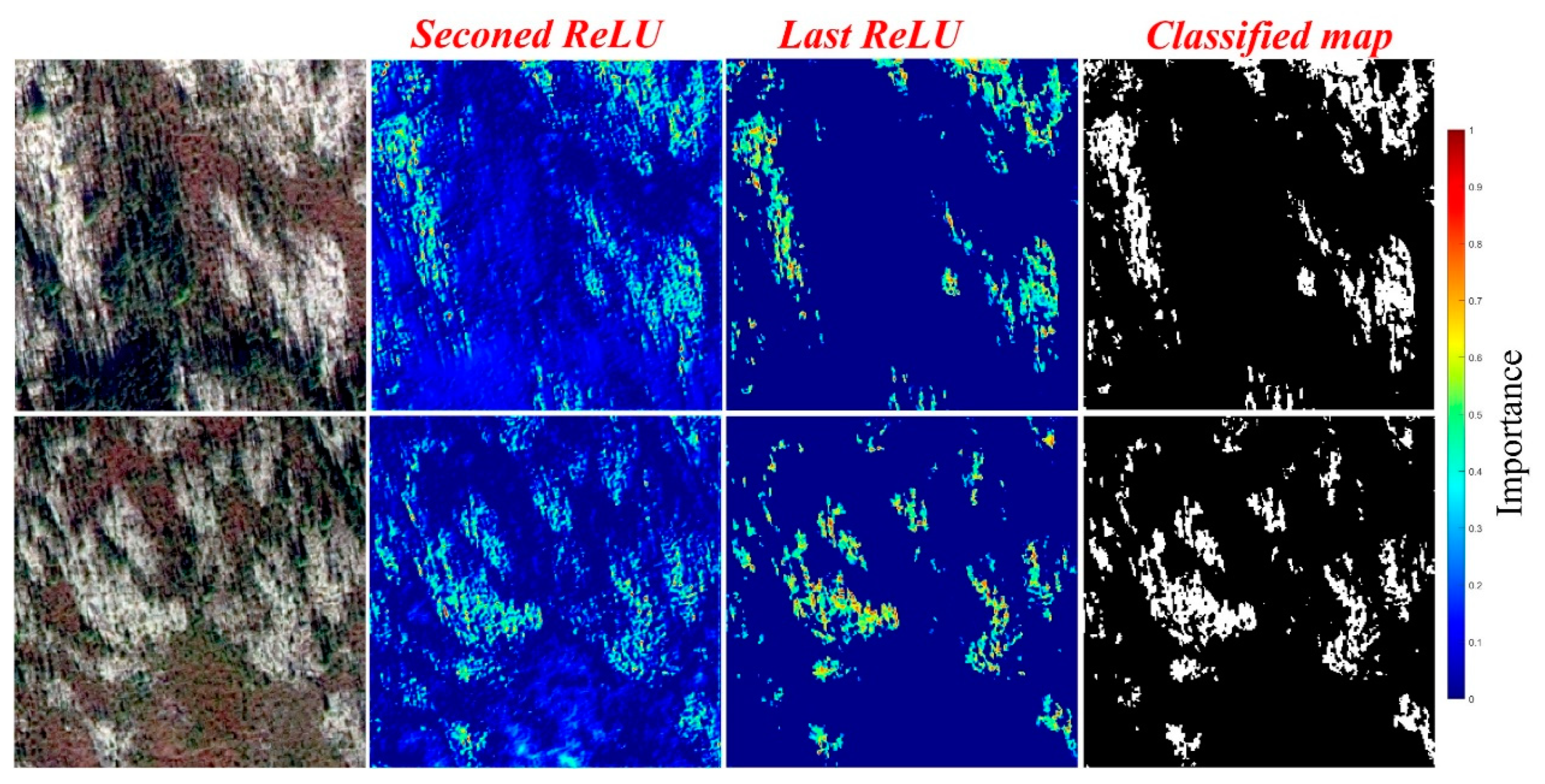
Table 2), it can be concluded that the rate of misclassification of background pixels as lichen was less than the rate of misclassification of lichen pixels as background (i.e., more lichen underestimation than lichen overestimation). The activation maps (Grad-CAM [
40]) of the SSL U-Net++ for two image titles containing lichen patches can be seen in
Figure 8. We can see that the network learned low-level features (like edges, and some basic relationships between lichens and neighboring areas) in the first few layers (first convolutional block). In the last layers, the network combined abstract and high-level features to perform final predictions, resulting in CAMs looking very similar to the prediction maps. Comparing the SSL U-Net++ with the supervised one showed a significant accuracy improvement (more than 15% and 22% improvement in OA and F1-score, respectively). The more important part of accuracy improvement was due to the higher true positive rate of the SSL U-Net++ (
Table 2), which is mainly because of the increase in the size of training data provided through the iterative pseudo-labeling and fine-tuning.
3.1. Noisy Labels vs. Cleaner Labels
We hypothesized that noise, specifically false positive detections, would have a deleterious effect on our results. In fact, since lichen maps generated based on high-resolution data are commonly used to map fractional lichen cover in coarser images (e.g., Landsat imagery), the priority may typically be to generate a map with a lower false positive rate (instead of a higher true positive rate). In this section, by rejecting the above-mentioned hypothesis, we discuss that not only these types of noise were not deleterious, but also they, to some degree, enhanced the robustness of the SSL along with the use of unlabeled data. To demonstrate this experimentally, rather than scaling-up the UAV-based lichen map to the WorldView image and then training the SSL baseline on it, we directly classified the same area in the WorldView image. For this purpose, we used the same 1825 points employed for training and testing the DL-GEOBIA model on the UAV image. However, this time we pre-screened the points to correct mislabeled samples and replace those falling inside the same segments (i.e., in the segmented image) with new randomly chosen ones. The samples were then used for training another DL-GEOBIA model capable of detecting lichen pixels over the area. According to the accuracy assessment, the OA and F1-score measures were 88.52% and 93.46%, respectively. As expected, this map had a higher accuracy than the resampled one. In other words, compared to the scaled-up UAV-based lichen map, this map visually appeared to have higher Precision (i.e., less background pixels were wrongly classified as lichen), especially over shadow and tree classes. Furthermore, a greater number of pure lichen pixels were detected correctly, which was at the expense of not detecting spectrally mixed lichen pixels that were more difficult for the classifier to identify in the WorldView image compared to the UAV image. As a result, this map provided a suitable case to analyze the effect of Type I error on the SSL training. This map was then used for training the SSL networks using the same procedure described earlier.
Figure 9a illustrates the map generated using the trained SSL framework on this cleaner training dataset (hereafter, cleaner model). We can clearly see that more cloud pixels were classified wrongly as lichen compared to the map generated by the main model in
Figure 7a. Accuracy assessment on this map showed an OA and F1-score 85.05% and 85.27%, respectively. According to the confusion matrix (
Table 2), the cleaner model classified less background pixels correctly compared to the main model trained on the noisy labels. However, it detected more lichen pixels correctly than the main model did.
3.2. Overfitted SSL
As discussed earlier, since DNNs generally first fit to clean samples and then overfit to noisy samples, preventing the network from extreme overfitting can significantly improve the performance of the model. To realize the effect of overfitting to our noisy labels for lichen mapping, we experimented a scenario where all the samples were used for training the SSL approach; that is, no validation set was used to perform early stopping. In fact, we let the networks overfit to the noise in our training data to realize the negative effect of natural noise in the case of overfitting in this study. After training the overfitted SSL, we applied it to our WorldView test image resulting in the map presented in
Figure 9b. Although clouds were better distinguished from lichen pixels in this map, the overfitted SSL produced a lichen map with significantly lower OA and F1-score (64.82% and 48.68%, respectively) than the main map did (
Figure 7). There is also a major increase in the number of non-lichen misclassifications, especially over water bodies. We expect that such misclassifications resulted from the lack of generalization in the model. Moreover, partly discriminating clouds from lichens was to some degree indicative of learning from noise. This, however, was at the expense of considering noisy labels as true labels (i.e., water pixels and thin clouds). The corresponding confusion matrix (
Table 2) shows that the network had a low performance for detecting lichen pixels correctly. This classifier had the worst accuracy compared to the other two SSL ones. The classifier detected lichen patches very sparsely as the network had a low lichen detection rate (
Figure 10). In fact, of 1214 lichen pixels, only 442 of them were correctly detected. Moreover, according to
Figure 9b, the unusual misclassifications of many background pixels (e.g., water bodies, thin clouds, etc.) proved the poor accuracy of the model overfitted to a noisy, limited labeled dataset.
A closer comparison among the maps generated by the three types of SSL models is provided in
Figure 10. According to this comparison, denser lichen patches were detected by the cleaner model. In other words, within a given lichen patch, more lichen pixels were detected correctly. This was less the case for the map generated by the main model as some lichen pixels within a given lichen patch were missed. The worst case occurred in the map derived from the overfitted model. These observations are in line with the previously described confusion matrices in
Table 2. As discussed earlier, the noisy model misclassified many cloudy pixels into lichen pixels. Although we were able to rule out many of those misclassifications using the FDI mask, there were still cloud pixels where the mask was not able to remove them. This was mainly the case in areas where there were thin clouds or haze. Overall, we observed that the SSL network trained on the naturally noisy labeled data was more consistent than the cleaner model was. In fact, it was found that there was a trade-off between false positive and false negative errors in this map.
3.3. Pixel-Based RF Model
In this section, we present and discuss the performance of the pixel-based RF applied for the scaling-up of the UAV-derived lichen map. As shown in
Figure 11, the map generated with the RF model was able to correctly classify many cloud pixels like the overfitted model did. However, it had a low lichen detection rate, even worse than the overfitted model, which is not surprising as conventional ML models are less generalizable than CNNs. As shown in
Figure 11, it is obvious that many cloud pixels were correctly classified as background, but it was at the expense of losing sufficient sensitivity for detecting many lichen pixels. According to the accuracy assessment results (
Table 2 and
Figure 12), the RF model had an OA of 60.89% and F1-score of 27.86%. Compared to the CNNs applied in this study, the much lower F1-score of the RF model is mainly due to its very low Recall performance. Considering the confusion matrix of the RF model (
Table 2), although the model was able to correctly classify most of the background test samples, it performed very poorly in detecting true lichen pixels in the test set.
4. Discussion
Scaling-up a small, very-high-resolution lichen map to a coarser, larger scale is a challenging task. In fact, given limited prior information, we expect an ML model to produce quality maps, which is a very hard problem for any model or training approach. Results presented in this study indicated that even using a small base map, we were able to carry out the scale-up process to obtain a reasonably accurate distribution map (i.e., OA of ~85% and F1-score of ~84%) for bright caribou lichens at a much larger scale. In addition to the small extent of the UAV image, another limiting factor was the lack of sufficient land-cover diversity over the UAV extent. The area surveyed with the UAV contained four primary land-cover types (i.e., soil, tree, bright caribou lichen, and grass). Beyond the extent of the area surveyed with UAV, we encountered several new land-cover types, some of which could mislead the classifier. Another potential issue for the scaling-up process was the presence of label noise in the training set prepared for the
Naïve-Student training. Although the behavior of DNNs against noise is not yet fully understood, past research has shown that DNNs have a tendency to first fit to clean samples and then overfit to noise after some epochs [
15,
41]. However, in this study, we had natural, not synthetic, noise in our label data. The resampled UAV-derived map had several mislabeled pixels caused by spatial and temporal data disagreements between the UAV and WorldView images. All these sources of label noise are different from data noise and classification-driven noise. Such noise can also affect the quality of the scaled-up maps that are generally difficult to avoid. The fourth problem was the presence of clouds and haze in the WorldView image. We depicted that the main model occasionally misclassified thick clouds as lichen and thus overestimated lichen cover. We addressed this issue by removing most of these misclassifications with an FDI mask. Conversely, the presence of haze over some areas with lichen led to misclassifications of lichen and therefore caused an underestimation of lichen cover. Given this, we found that clouds caused both false positive and false negative errors in cases where target lichens would be bright colored, as in this study.
The better performance of the cleaner model compared to the main model in detecting more lichen patches correctly (i.e., higher Recall) can be ascribed to the fact that more pure lichen training samples (i.e., less noisy labels) were used for training the network. This, however, degraded the Precision of the model compared to the main model.
Given the limited labeled samples and their lack of sufficient diversity, it appears that noisy labels helped prevent the model from memorizing homogenous patterns in the data. These findings are in line with those reported by Xie, Luong, Hovy, and Le [
21] where they found that injecting noise to the
Student network was one of the main reasons for improving the generalization power of their SSL approach. However, in our study, we had natural noise resulting from the scaling-up of the UAV-derived lichen map. This type of data noise affected both the
Teacher and
Student networks, as opposed to the
Noisy-Student approach in which data noise (i.e., image augmentation) is only injected to inputs of the
Student network, and as opposed to
Naïve-Student in which no explicit data noise is applied. This natural noise ultimately improved robustness related to Type I error in the
Naïve-Student approach applied in this research. Although the natural noise in this study partially improved the robustness of the model, overfitting to noise would be a critical problem if models are not trained appropriately. Considering this issue, we showed that overfitting to noise caused very poor accuracies, resulting in detecting many non-lichen pixels erroneously as lichen.
We also presented a mapping result generated with an RF model and a high-dimensional feature set. Although RF is a popular method in lichen mapping, accuracy assessment results showed that it did not lead to an accurate map. This was, however, to some degree expected as pixel-based models do not generalize well. Processing times of SSL and RF are presented in
Table 3. Training the SSL was more time consuming than RF and supervised U-Net++. However, the inference time of the RF was higher, although this is without considering the time spent on computing the GLCM features in the test image.
Given the above limitations, the use of unlabeled samples in a full-fledged SSL model was advantageous in scaling-up the UAV-derived lichen map to the WorldView scale. However, it is obvious that if lichens of interest are of different colors, and not all of them are present within the extent of the very-high-resolution maps, SSL will fail to detect those lichens in larger-scale images. The results also showed that although the UAV site was very homogenous, it was possible to obtain a map with a reasonable rate of false positive, which is important for lichen mapping tasks. The use of unlabeled data and noisy labels was found to improve the robustness of the network against samples that could have caused Type I error.
Although the main and cleaner SSL models led to better accuracies compared to the other approaches used in this study, one of the challenges with training these models was their computational complexity compared to supervised approaches. Not only were multiple models iteratively trained, but also the number of data used for training was also increased compared to the supervised models. This caused a significant increase in the execution time of the training process. In general, one of the main computational bottlenecks in the SSL approach was the dense prediction of pseudo-labels and then using them for training the Student models, which, as mentioned above, were then trained on a much larger data set. Another relevant factor affecting the computational time of the SSL approach was the TTA that was performed during pseudo-labeling. These two factors were the major computationally intensive components of the SSL in this study. Given the accuracy improvement provided by the main SSL model, and the expectations of the lichen mapping task under consideration, this inefficiency compared to the supervised models and to the very expensive labeling tasks is justifiable provided that sufficient computational resources are available.
5. Conclusions
In this study, we assessed the possibility of scaling-up a very fine lichen map acquired over a very small, homogenous area to a much larger area without collecting any new labeled samples. We used a Teacher-Student SSL approach (i.e., Naïve-Student) that trains Teacher and Student networks iteratively on both labeled and unlabeled data. This approach produced a reasonably accurate (OA of ~85% and F1-score of ~84%) lichen map at the WorldView scale. The main findings in this research are as follows:
A powerful SSL approach capable of taking advantage of abundant unlabeled data is beneficial for scaling-up small lichen maps to large scales using high-resolution satellite imagery, provided that there are samples for lichens of interest within the small lichen maps under consideration.
Different types of image misalignments can introduce noisy labels in the scaled-up training set. However, we found that if this noise is not massive, it may ultimately improve the model robustness without conducting any intensive data augmentation on training data.
The map generated by the cleaner model indicated a higher Recall but a lower Precision than the map generated by the main model.
Overfitting in the presence of noise significantly degraded the performance of the trained network when applied to the test image. That network failed to detect many lichen patches and resulted in low accuracy. In some cases, we observed that due to overfitting to noisy labels (especially dark areas), many lichen misdetections occurred over water bodies resembling noisy labels in the training set.
There is a need for more comprehensive studies on the effect of label noise on final classification results. This is an important gap in the context of lichen mapping although when using multi-source/sensor data, there will be several types of misalignments that cause label noise. In this regard, a potential future direction for RS-based lichen mapping could be to conduct more in-depth studies on the nature and amount of noise that can improve the robustness of a model. This can be performed based on a systematic study introducing synthetic and natural noise to the classification procedure. Similarly, it could be investigated whether an ensemble of fine-tuned Student networks is able to produce more accurate results than a single network. In fact, different networks have different prediction capabilities. Therefore, if they are ensembled together (e.g., based on an averaging/majority-voting output or a more complex aggregation strategy), they may improve lichen detection. This may be also achieved by distilling several interconnected Student networks to reach a single, lighter final Student network.
If sufficient labeled data are available, it is generally recommended to use the maximum input size fitting to the GPU memory when splitting a given image. However, due to the use of a small label map, one of the most important limitations in this study was the use of small image patches (i.e., 64 by 64 pixels) for training the Naïve-Student networks. Such an input size caused both of the Teacher and Student networks to be unable to extract more useful, representative contextual features in deeper encoding layers of the networks. We also found a shallower U-Net(++) less accurate than the one used in this study, although this improvement could have been more significant if larger image patches were used. If the likelihood of having more untraceable noise and uncertainty as well as training new, complex networks can be justified, one way of mitigating this problem could be to use Generative Adversarial Networks (GANs) to either synthesize more image patches from larger image patches or to improve the resolution of small image patches through a super-resolution setup.