Towards Amazon Forest Restoration: Automatic Detection of Species from UAV Imagery
Abstract
:1. Introduction
2. Materials and Methods
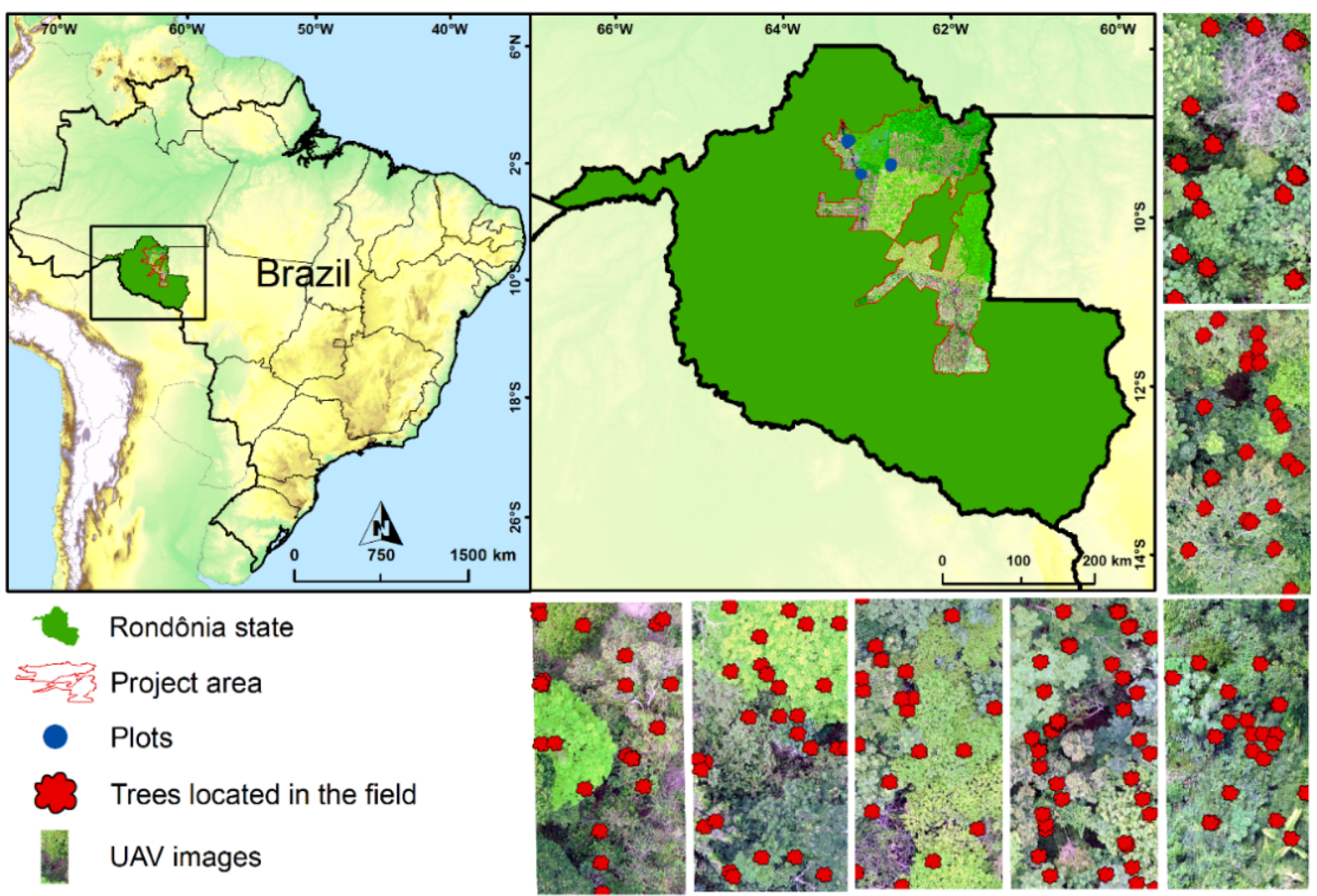
2.1. Study Area
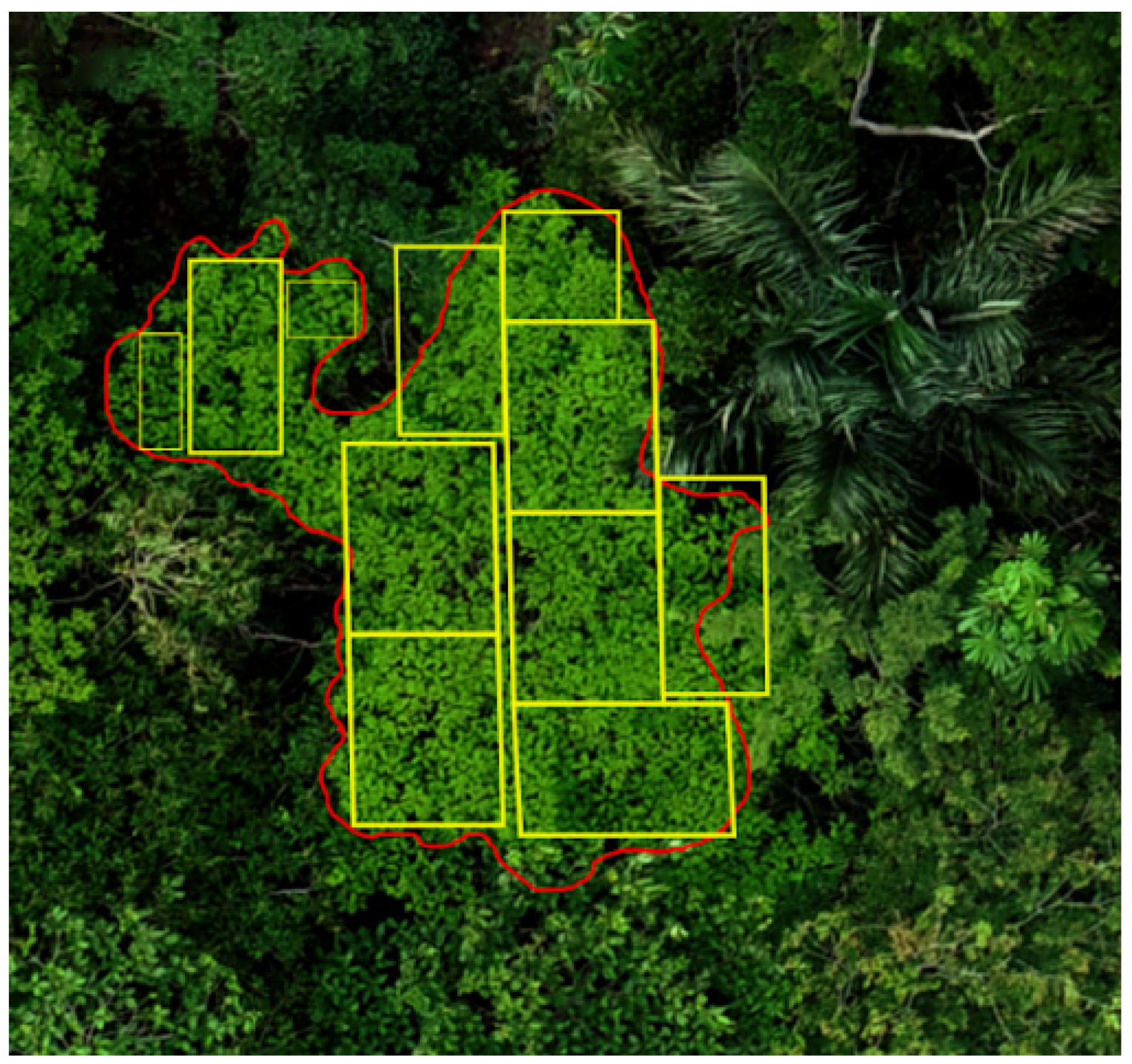
2.2. Image Acquisition and Pre-Processing
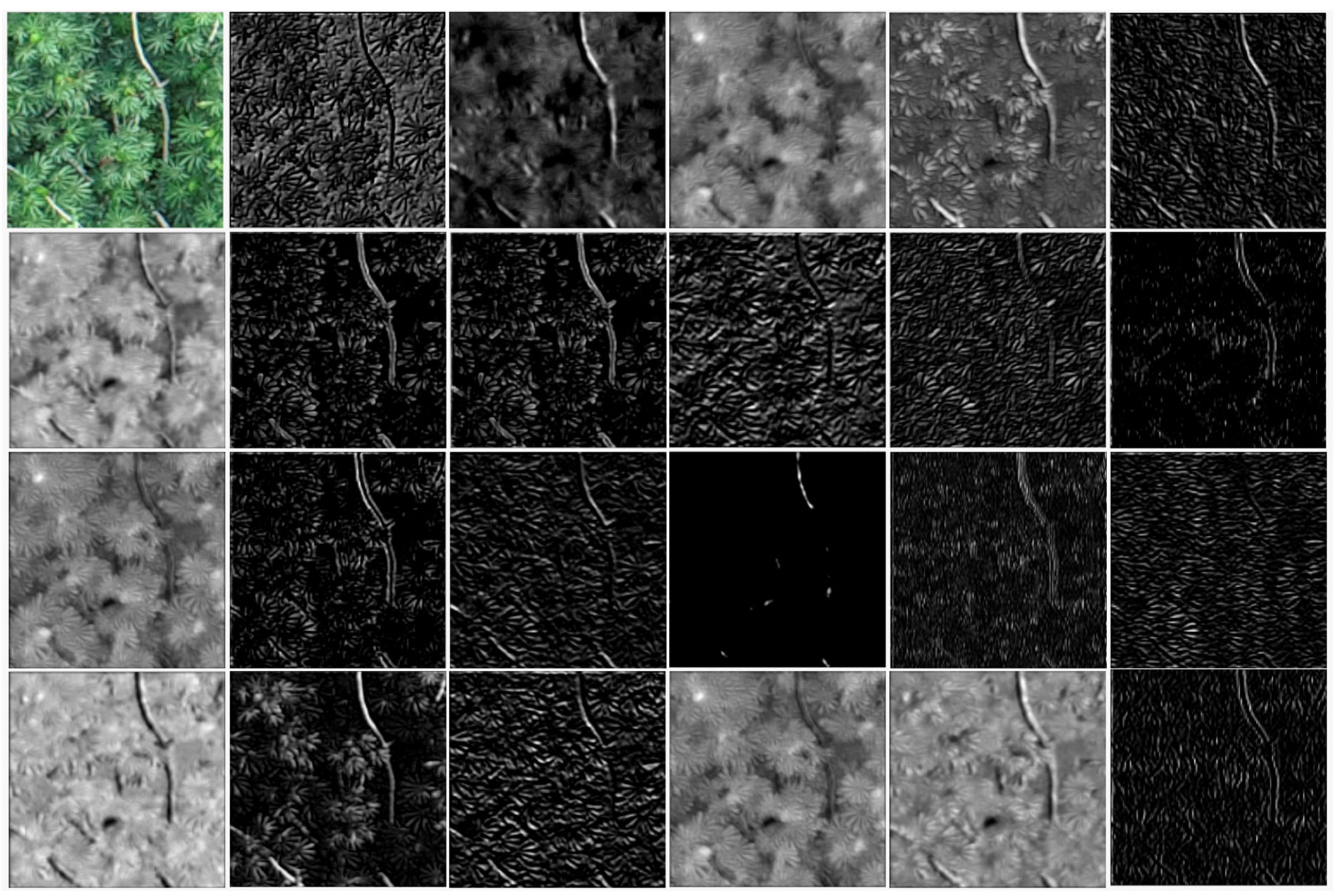
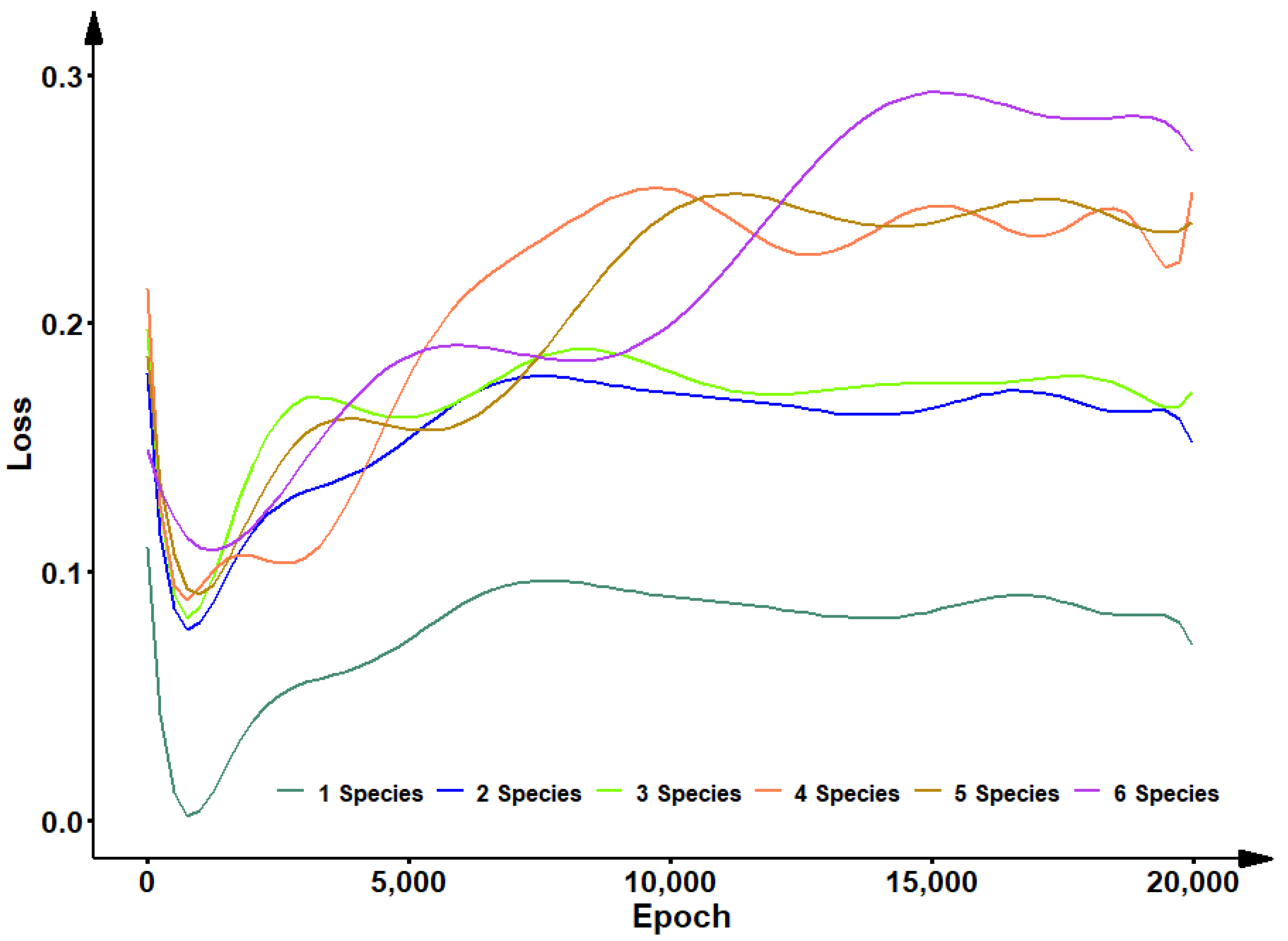
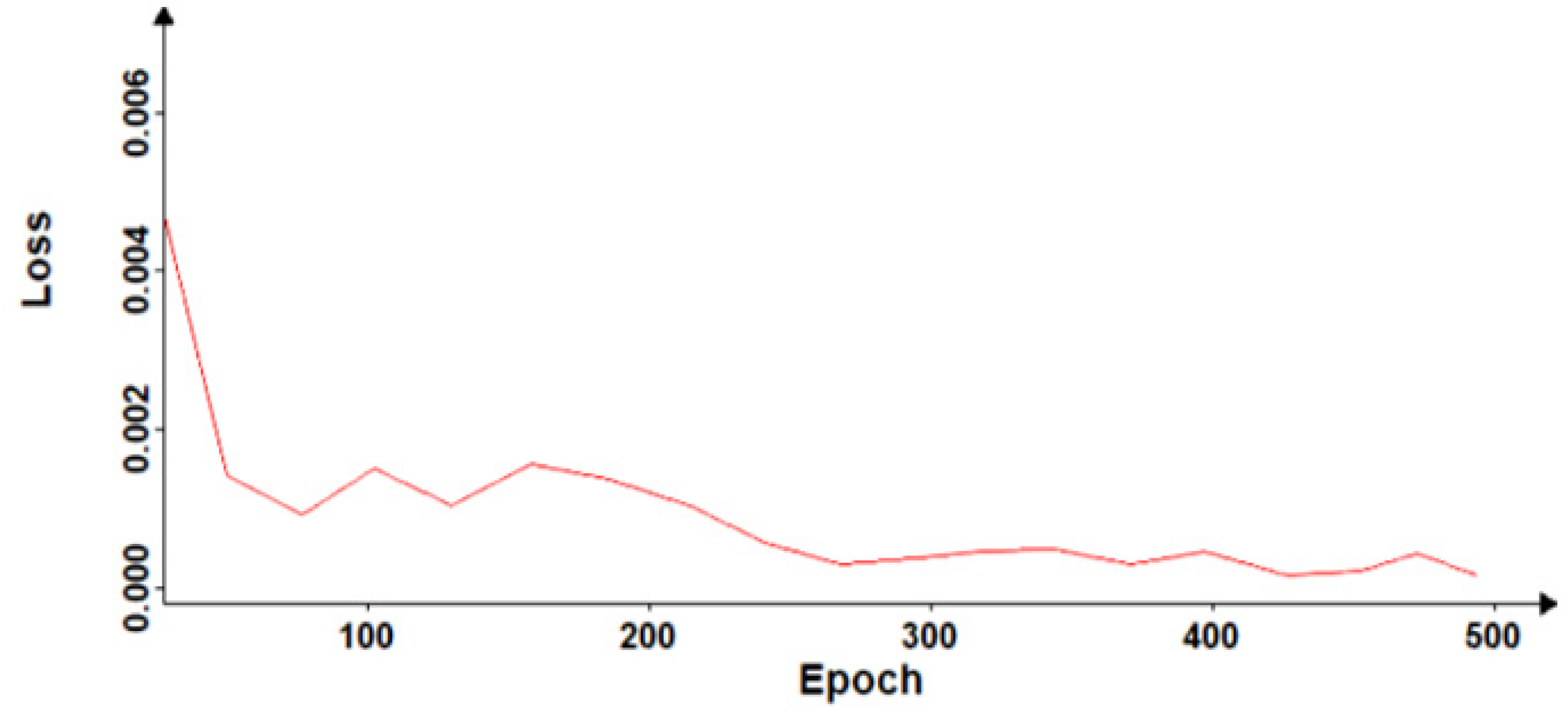
2.3. Convolutional Neural Networks Training
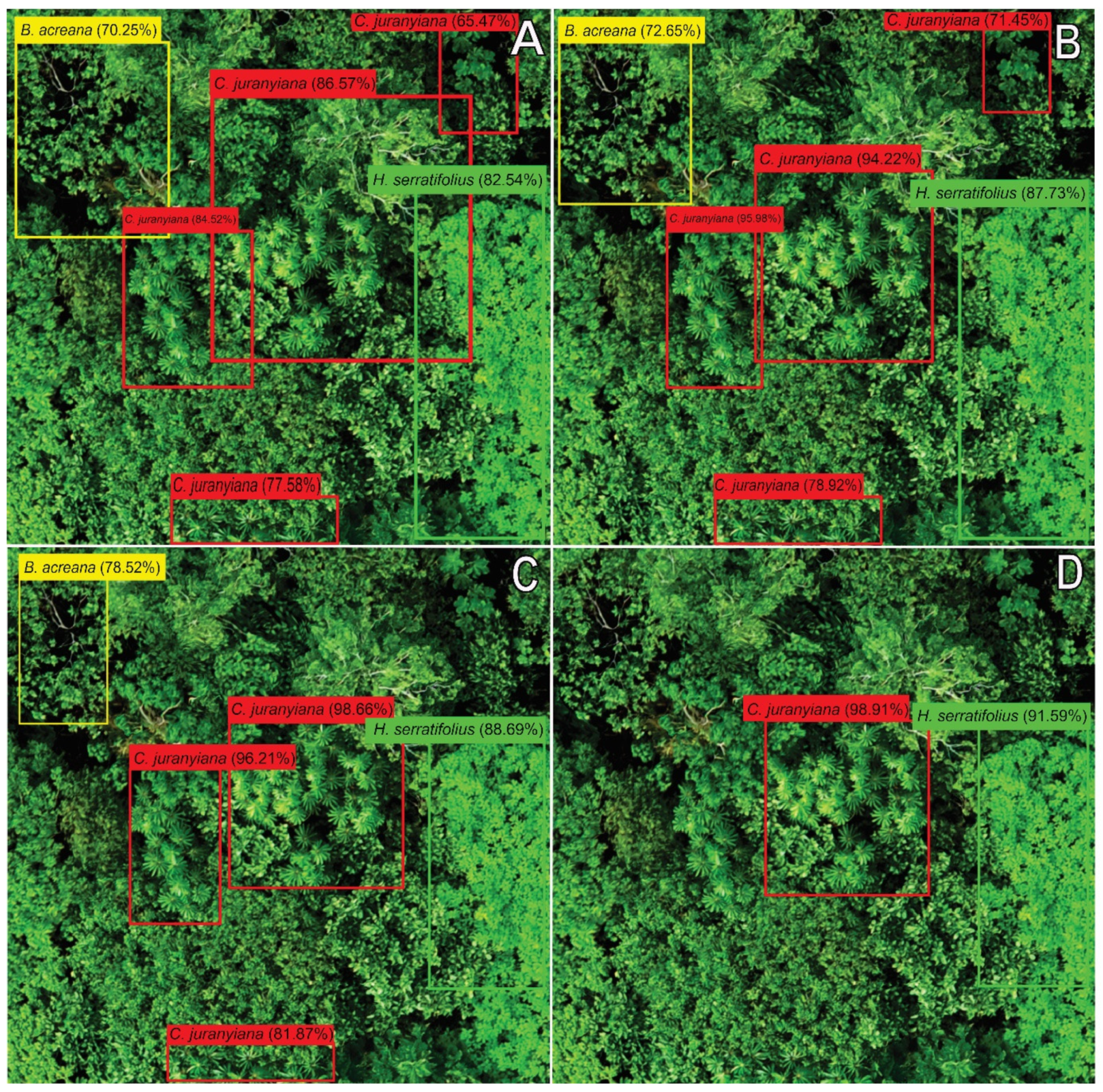
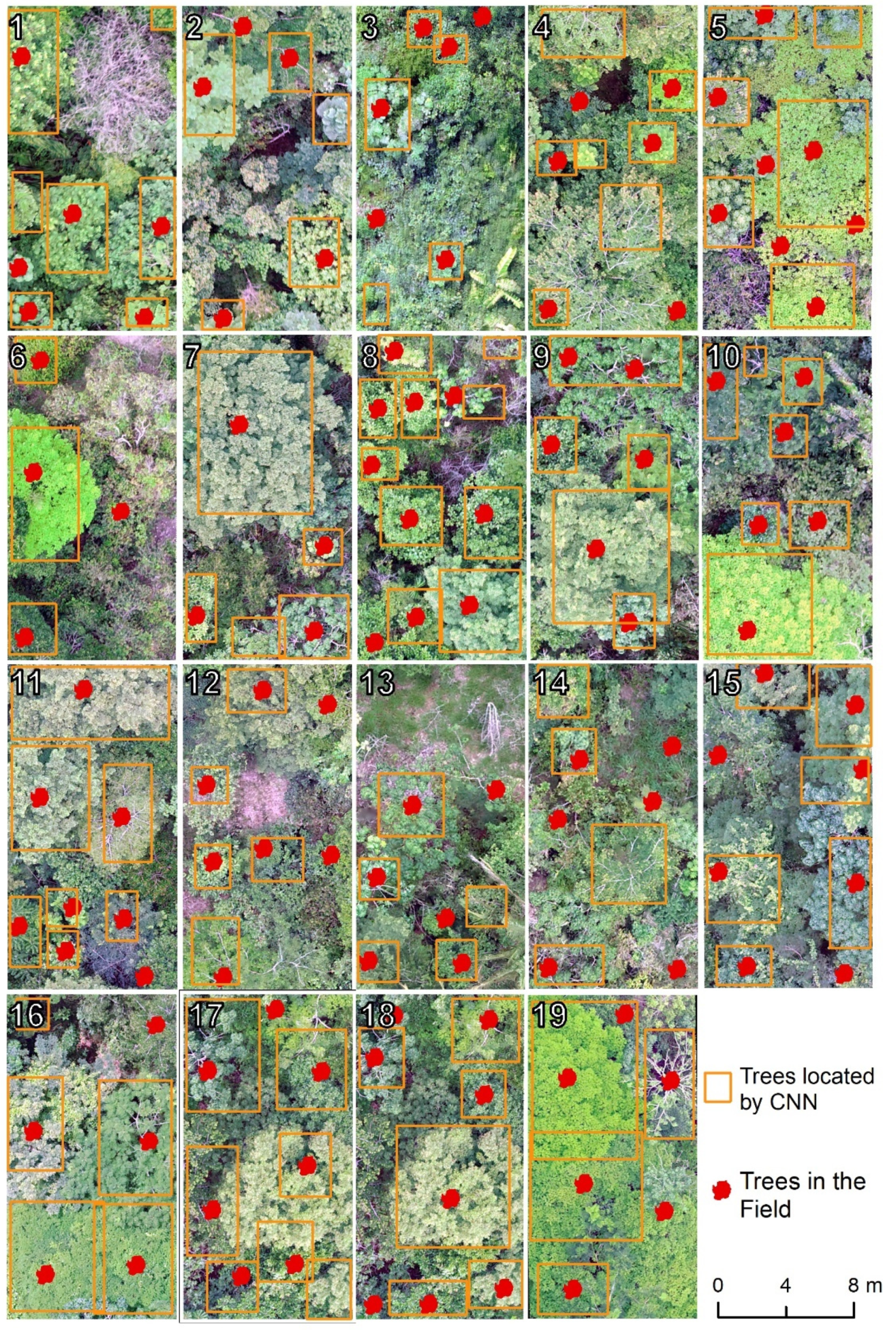
2.4. Classification Validation
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Natesan, S.; Armenakis, C.; Vepakomma, U. Individual tree species identification using Dense Convolutional Network (DenseNet) on multitemporal RGB images from UAV. J. Unmanned Veh. Syst. 2020, 8, 310–333. [Google Scholar] [CrossRef]
- Casapia, X.T.; Falen, L.; Bartholomeus, H.; Cárdenas, R.; Flores, G.; Herold, M.; Coronado, E.N.H.; Baker, T.R. Identifying and Quantifying the Abundance of Economically Important Palms in Tropical Moist Forest Using UAV Imagery. Remote Sens. 2019, 12, 9. [Google Scholar] [CrossRef] [Green Version]
- Cao, D.; Chen, Z.; Gao, L. An improved object detection algorithm based on multi-scaled and deformable convolutional neural networks. Hum. Cent. Comput. Inf. Sci. 2020, 10, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Wang, J.; Qian, K.; Liu, Y.; Guo, R.; Hu, S.; Yuancong, W.; Chen, T.; Rong, L. An energy-efficient deep convolutional neural networks coprocessor for multi-object detection. Microelectron. J. 2020, 98, 104737. [Google Scholar] [CrossRef]
- Zhang, M.; Chen, Y.; Liu, X.; Lv, B.; Wang, J. Adaptive Anchor Networks for Multi-Scale Object Detection in Remote Sensing Images. IEEE Access 2020, 8, 57552–57565. [Google Scholar] [CrossRef]
- Yao, Q.; Hu, X.; Lei, H. Multiscale Convolutional Neural Networks for Geospatial Object Detection in VHR Satellite Images. IEEE Geosci. Remote Sens. Lett. 2021, 18, 23–27. [Google Scholar] [CrossRef]
- Pi, Y.; Nath, N.D.; Behzadan, A.H. Convolutional neural networks for object detection in aerial imagery for disaster response and recovery. Adv. Eng. Inform. 2020, 43, 101009. [Google Scholar] [CrossRef]
- Qiao, R.; Ghodsi, A.; Wu, H.; Chang, Y.; Wang, C. Simple weakly supervised deep learning pipeline for detecting individual red-attacked trees in VHR remote sensing images. Remote Sens. Lett. 2020, 11, 650–658. [Google Scholar] [CrossRef]
- Xiao, C.; Qin, R.; Huang, X. Treetop detection using convolutional neural networks trained through automatically generated pseudo labels. Int. J. Remote Sens. 2019, 41, 3010–3030. [Google Scholar] [CrossRef]
- Roslan, Z.; Awang, Z.; Husen, M.N.; Ismail, R.; Hamzah, R. Deep Learning for Tree Crown Detection in Tropical Forest. In Proceedings of the 14th International Conference on Ubiquitous Information Management and Communication (IMCOM), Taichung, Taiwan, 3–5 January 2020; pp. 1–7. [Google Scholar]
- Fromm, M.; Schubert, M.; Castilla, G.; Linke, J.; McDermid, G. Automated Detection of Conifer Seedlings in Drone Imagery Using Convolutional Neural Networks. Remote Sens. 2019, 11, 2585. [Google Scholar] [CrossRef] [Green Version]
- Syrris, V.; Hasenohr, P.; Delipetrev, B.; Kotsev, A.; Kempeneers, P.; Soille, P. Evaluation of the Potential of Convolutional Neural Networks and Random Forests for Multi-Class Segmentation of Sentinel-2 Imagery. Remote Sens. 2019, 11, 907. [Google Scholar] [CrossRef] [Green Version]
- Sanquetta, C.R.; Behling, A.; Corte, A.P.D.; Cadori, G.C.; Junior, S.C.; Macedo, J.H.P. Eficiência de conversão da radiação fotossintética interceptada em Fitomassa de mudas de Eucalyptus dunii Maiden em função da densidade de plantas e do ambiente de cultivo. Sci. For. Sci. 2014, 42, 573–580. [Google Scholar]
- Sanquetta, C.R.; Behling, A.; Dalla Corte, A.P.; Péllico Netto, S.; Rodrigues, A.L.; Simon, A.A. A model based on environmental factors for diameter distribution in black wattle in Brazil. PLoS ONE 2014, 9, 100093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albawi, S.; Mohammed, T.A.; Al-Zawi, S. Understanding of a convolutional neural network. In Proceedings of the 2017 International Conference on Engineering and Technology (ICET), Antalya, Turkey, 21–23 August 2017; pp. 1–6. [Google Scholar]
- Shakya, A.; Biswas, M.; Pal, M. Parametric study of convolutional neural network based remote sensing image classification. Int. J. Remote Sens. 2021, 42, 2663–2685. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, J.; Tao, D. Recursive Context Routing for Object Detection. Int. J. Comput. Vis. 2021, 129, 142–160. [Google Scholar] [CrossRef]
- Jia, F.; Liu, J.; Tai, X.-C. A regularized convolutional neural network for semantic image segmentation. Anal. Appl. 2021, 19, 147–165. [Google Scholar] [CrossRef]
- Shivappriya, S.; Priyadarsini, M.; Stateczny, A.; Puttamadappa, C.; Parameshachari, B. Cascade Object Detection and Remote Sensing Object Detection Method Based on Trainable Activation Function. Remote Sens. 2021, 13, 200. [Google Scholar] [CrossRef]
- Safonova, A.; Guirado, E.; Maglinets, Y.; Alcaraz-Segura, D.; Tabik, S. Olive Tree Biovolume from UAV Multi-Resolution Image Segmentation with Mask R-CNN. Sensors 2021, 21, 1617. [Google Scholar] [CrossRef]
- Kattenborn, T.; Leitloff, J.; Schiefer, F.; Hinz, S. Review on Convolutional Neural Networks (CNN) in vegetation remote sensing. ISPRS J. Photogramm. Remote Sens. 2021, 173, 24–49. [Google Scholar] [CrossRef]
- Flores, D.; González-Hernández, I.; Lozano, R.; Vazquez-Nicolas, J.M.; Toral, J.L.H. Automated Agave Detection and Counting Using a Convolutional Neural Network and Unmanned Aerial Systems. Drones 2021, 5, 4. [Google Scholar] [CrossRef]
- Yan, S.; Jing, L.; Wang, H. A New Individual Tree Species Recognition Method Based on a Convolutional Neural Network and High-Spatial Resolution Remote Sensing Imagery. Remote Sens. 2021, 13, 479. [Google Scholar] [CrossRef]
- Korznikov, K.A.; Kislov, D.E.; Altman, J.; Doležal, J.; Vozmishcheva, A.S.; Krestov, P.V. Using U-Net-Like Deep Convolutional Neural Networks for Precise Tree Recognition in Very High Resolution RGB (Red, Green, Blue) Satellite Images. Forests 2021, 12, 66. [Google Scholar] [CrossRef]
- Jassmann, T.J.; Tashakkori, R.; Parry, R.M. Leaf classification utilizing a convolutional neural network. In Proceedings of the SoutheastCon 2015, Fort Lauderdale, FL, USA, 9–12 April 2015. [Google Scholar]
- Gu, J.; Wang, Z.; Kuen, J.; Ma, L.; Shahroudy, A.; Shuai, B.; Liu, T.; Wang, X.; Wang, G.; Cai, J.; et al. Recent advances in convolutional neural networks. Pattern Recognit. 2018, 77, 354–377. [Google Scholar] [CrossRef] [Green Version]
- Woźniak, M. Advanced Computational Intelligence for Object Detection, Feature Extraction and Recognition in Smart Sensor Environments. Sensors 2020, 21, 45. [Google Scholar] [CrossRef] [PubMed]
- Pullanagari, R.; Dehghan-Shoar, M.; Yule, I.J.; Bhatia, N. Field spectroscopy of canopy nitrogen concentration in temperate grasslands using a convolutional neural network. Remote Sens. Environ. 2021, 257, 112353. [Google Scholar] [CrossRef]
- Jaihuni, M.; Khan, F.; Lee, D.; Basak, J.K.; Bhujel, A.; Moon, B.E.; Park, J.; Kim, H.T. Determining Spatiotemporal Distribution of Macronutrients in a Cornfield Using Remote Sensing and a Deep Learning Model. IEEE Access 2021, 9, 30256–30266. [Google Scholar] [CrossRef]
- Wulder, M. Optical remote-sensing techniques for the assessment of forest inventory and biophysical parameters. Prog. Phys. Geogr. Earth Environ. 1998, 22, 449–476. [Google Scholar] [CrossRef]
- Hyyppä, J.; Hyyppä, H.; Inkinen, M.; Engdahl, M.; Linko, S.; Zhu, Y.-H. Accuracy comparison of various remote sensing data sources in the retrieval of forest stand attributes. For. Ecol. Manag. 2000, 128, 109–120. [Google Scholar] [CrossRef]
- Silva, C.A.; Klauberg, C.; Hentz, Â.M.K.; Corte, A.P.D.; Ribeiro, U.; Liesenberg, V. Comparing the performance of ground filtering algorithms for terrain modeling in a forest environment using airborne LiDAR data. Floresta Ambiente 2018, 25, e20160150. [Google Scholar] [CrossRef] [Green Version]
- Da Cunha Neto, E.M.; Rex, F.E.; Veras, H.F.P.; Moura, M.M.; Sanquetta, C.R.; Käfer, P.S.; Sanquetta, M.N.I.; Zambrano, A.M.A.; Broadbent, E.N.; Corte, A.P.D. Using high-density UAV-Lidar for deriving tree height of Araucaria Angustifolia in an Urban Atlantic Rain Forest. Urban For. Urban Green 2021, 63, 127197. [Google Scholar] [CrossRef]
- Miraki, M.; Sohrabi, H.; Fatehi, P.; Kneubuehler, M. Individual tree crown delineation from high-resolution UAV images in broadleaf forest. Ecol. Inform. 2021, 61, 101207. [Google Scholar] [CrossRef]
- Zheng, J.; Fu, H.; Li, W.; Wu, W.; Yu, L.; Yuan, S.; Tao, W.Y.W.; Pang, T.K.; Kanniah, K.D. Growing status observation for oil palm trees using Unmanned Aerial Vehicle (UAV) images. ISPRS J. Photogramm. Remote Sens. 2021, 173, 95–121. [Google Scholar] [CrossRef]
- Kopačková-Strnadová, V.; Koucká, L.; Jelének, J.; Lhotáková, Z.; Oulehle, F. Canopy Top, Height and Photosynthetic Pigment Estimation Using Parrot Sequoia Multispectral Imagery and the Unmanned Aerial Vehicle (UAV). Remote Sens. 2021, 13, 705. [Google Scholar] [CrossRef]
- Neuville, R.; Bates, J.; Jonard, F. Estimating Forest Structure from UAV-Mounted LiDAR Point Cloud Using Machine Learning. Remote Sens. 2021, 13, 352. [Google Scholar] [CrossRef]
- Fassnacht, F.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Berg, C.C. Espécies de Cecropia da Amazônia Brasileira. Acta Amaz. 1978, 8, 149–182. [Google Scholar] [CrossRef] [Green Version]
- Pinto, R.B.; Tozzi, A.M.G.A.; Mansano, V. Hymenaea in Flora do Brasil. Available online: http://floradobrasil.jbrj.gov.br/reflora/floradobrasil/FB22976 (accessed on 18 June 2021).
- CNCFlora. Hymenaea courbaril in Lista Vermelha da Flora Brasileira Versão. Available online: http://cncflora.jbrj.gov.br/portal/pt-br/profile/Hymenaea%20courbaril (accessed on 18 June 2021).
- Vaz, A.M.S.F. Bauhinia in Flora do Brasil. 2020. Available online: http://floradobrasil.jbrj.gov.br/reflora/PrincipalUC/PrincipalUC.do (accessed on 18 June 2021).
- Silva-Luz, C.L.; Pirani, J.R.; Pell, S.K.; Mitchell, J.D. Anacardiaceae in Flora do Brasil. Available online: http://floradobrasil.jbrj.gov.br/reflora/PrincipalUC/PrincipalUC.do (accessed on 18 June 2021).
- Lorenzi, H. Árvores Brasileiras: Manual de Identificação e Cultivo de Plantas Arbóreas Nativas do Brasil; Instituto Plantarum: Nova Odessa, Brazil, 2000. [Google Scholar]
- Lohmann, L.G. Handroanthus in Flora do Brasil. Available online: http://floradobrasil.jbrj.gov.br/reflora/PrincipalUC/PrincipalUC.do (accessed on 18 June 2021).
- Morim, M.P. Anadenanthera in Flora do Brasil. Available online: http://floradobrasil.jbrj.gov.br/reflora/PrincipalUC/PrincipalUC.do (accessed on 18 June 2021).
- Tzutalin Git Code. Available online: https://github.com/tzutalin/labelImg (accessed on 30 March 2021).
- Frankle, J.; Schwab, D.J.; Morcos, A.S. The Early Phase of Neural Network Training. arXiv 2020, arXiv:2002.10365. [Google Scholar]
- Taqi, A.M.; Awad, A.; Al-Azzo, F.; Milanova, M. The Impact of Multi-Optimizers and Data Augmentation on TensorFlow Convolutional Neural Network Performance. In Proceedings of the 2018 IEEE Conference on Multimedia Information Processing and Retrieval (MIPR), Miami, FL, USA, 10–12 April 2018; pp. 140–145. [Google Scholar]
- Cohen, J. A Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Sammut, C.; Webb, G.I. Encyclopedia of Machine Learning; Springer: New York, NY, USA, 2010. [Google Scholar]
- Maratea, A.; Petrosino, A.; Manzo, M. Adjusted F-measure and kernel scaling for imbalanced data learning. Inf. Sci. 2014, 257, 331–341. [Google Scholar] [CrossRef]
- Matthews, B. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta (BBA) Protein Struct. 1975, 405, 442–451. [Google Scholar] [CrossRef]
- Ballabio, D.; Grisoni, F.; Todeschini, R. Multivariate comparison of classification performance measures. Chemom. Intell. Lab. Syst. 2018, 174, 33–44. [Google Scholar] [CrossRef]
- Ferreira, M.P.; Wagner, F.H.; Aragão, L.E.; Shimabukuro, Y.E.; Filho, C.R.D.S. Tree species classification in tropical forests using visible to shortwave infrared WorldView-3 images and texture analysis. ISPRS J. Photogramm. Remote Sens. 2019, 149, 119–131. [Google Scholar] [CrossRef]
- Mubin, N.A.; Nadarajoo, E.; Shafri, H.Z.M.; Hamedianfar, A. Young and mature oil palm tree detection and counting using convolutional neural network deep learning method. Int. J. Remote Sens. 2019, 40, 7500–7515. [Google Scholar] [CrossRef]
- Freudenberg, M.; Nölke, N.; Agostini, A.; Urban, K.; Wörgötter, F.; Kleinn, C. Large Scale Palm Tree Detection in High Resolution Satellite Images Using U-Net. Remote Sens. 2019, 11, 312. [Google Scholar] [CrossRef] [Green Version]
- Gollob, C.; Ritter, T.; Wassermann, C.; Nothdurft, A. Influence of Scanner Position and Plot Size on the Accuracy of Tree Detection and Diameter Estimation Using Terrestrial Laser Scanning on Forest Inventory Plots. Remote Sens. 2019, 11, 1602. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Feng, Y.; Attwood, K.; Tian, L. Optimal threshold selection methods under tree or umbrella ordering. J. Biopharm. Stat. 2018, 29, 98–114. [Google Scholar] [CrossRef]
- Sarabia, R.; Aquino, A.; Ponce, J.M.; López, G.; Andújar, J.M. Automated Identification of Crop Tree Crowns from UAV Multispectral Imagery by Means of Morphological Image Analysis. Remote Sens. 2020, 12, 748. [Google Scholar] [CrossRef] [Green Version]
- Xiong, J.; Liu, Z.; Chen, S.; Liu, B.; Zheng, Z.; Zhong, Z.; Yang, Z.; Peng, H. Visual detection of green mangoes by an unmanned aerial vehicle in orchards based on a deep learning method. Biosyst. Eng. 2020, 194, 261–272. [Google Scholar] [CrossRef]
- Wagner, F.H.; Sanchez, A.; Tarabalka, Y.; Lotte, R.G.; Ferreira, M.P.; Aidar, M.P.M.; Gloor, E.; Phillips, O.; Aragão, L.E.O.C. Using the U-net convolutional network to map forest types and disturbance in the Atlantic rainforest with very high resolution images. Remote Sens. Ecol. Conserv. 2019, 5, 360–375. [Google Scholar] [CrossRef] [Green Version]
- Mohan, M.; Richardson, G.; Gopan, G.; Aghai, M.M.; Bajaj, S.; Galgamuwa, G.A.P.; Vastaranta, M.; Arachchige, P.S.P.; Amorós, L.; Corte, A.P.D.; et al. UAV-Supported Forest Regeneration: Current Trends, Challenges and Implications. Remote Sens. 2021, 13, 2596. [Google Scholar] [CrossRef]
| Characteristics | Type |
|---|---|
| Width | 5.472 pixels |
| Height | 3.648 pixels |
| Camera | FC6310 |
| Exposure time | 1/400s |
| Aperture value | 4.7EV (f/5,6) |
| Speed rate (ISO) | 100 |
| Image type | .jpeg |
| Focal distance | 8.8 mm |
| Pixel resolution | 2.2 cm |
| Species | Samples | Subsamples | |
|---|---|---|---|
| Trees | Test (30%) | Training (70%) | |
| Cecropia juranyiana | 157 | 158 | 369 |
| Hymenaea courbaril | 102 | 126 | 294 |
| Bauhinia acreana | 96 | 108 | 252 |
| Anacardium occidentale | 90 | 80 | 186 |
| Handroanthus serratifolius | 106 | 116 | 270 |
| Anadenanthera sp. | 132 | 144 | 336 |
| Total | 683 | 731 | 1706 |
| Statistical Index | Equation | Description | Reference |
|---|---|---|---|
| Kappa | Ranges from 1 to −1, where 1 indicates a perfect rating and −1 represents an unreliable rating | [50] | |
| Accuracy | Number of correct classifications of all predictions | [51] | |
| Adjusted F-score | Use all confusion matrix elements and provide more weights to samples that are correctly classified in the lowest class. | [52] | |
| Mathews Correlation Coefficient | Correlation coefficient of observed and predicted binary classifications, where: Negligible—<0.3 Poor—0.3–0.5 Moderate—0.5–0.7 Strong—0.7–0.9 Very strong—>0.9 | [53] | |
| Similarity | It ranges from 0 to 1, where: 0—no correct ratings 1—perfect classification | [54] |
| Cecropia | Hymenaea | Bauhinia | Anacardium | Handroanthus | Anadenanthera | Total | User | |
|---|---|---|---|---|---|---|---|---|
| Cecropia | 356 | 0 | 26 | 4 | 0 | 0 | 386 | 92.23% |
| Hymenaea | 0 | 281 | 0 | 0 | 5 | 12 | 298 | 94.30% |
| Bauhinia | 4 | 12 | 201 | 4 | 5 | 5 | 235 | 87.01% |
| Anacardium | 4 | 0 | 8 | 171 | 0 | 1 | 184 | 92.93% |
| Handroanthus | 0 | 1 | 12 | 7 | 240 | 9 | 269 | 89.22% |
| Anadenanthera | 5 | 0 | 5 | 0 | 11 | 318 | 335 | 93.81% |
| Total | 369 | 294 | 252 | 186 | 261 | 345 | 1707 | - |
| Producer accuracy | 96.48% | 95.58% | 79.76% | 91.94% | 91.95% | 92.17% | - | 1567 |
| Global accuracy | 91.80% | k | g | |||||
| Index | Cecropia | Hymenaea | Bauhinia | Anacardium | Handroanthus | Anadenanthera |
|---|---|---|---|---|---|---|
| Kappa | 0.9006 | |||||
| Accuracy | 0.9748 | 0.9824 | 0.9525 | 0.9836 | 0.9707 | 0.9718 |
| Adjusted F-score | 0.9706 | 0.9667 | 0.9135 | 0.9587 | 0.9594 | 0.9589 |
| Matthews correlation | 0.92721 | 0.9387 1 | 0.8057 2 | 0.9151 1 | 0.8884 2 | 0.9123 1 |
| Sensitivity | 0.9222 | 0.9429 | 0.8701 | 0.9293 | 0.8921 | 0.938 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moura, M.M.; de Oliveira, L.E.S.; Sanquetta, C.R.; Bastos, A.; Mohan, M.; Corte, A.P.D. Towards Amazon Forest Restoration: Automatic Detection of Species from UAV Imagery. Remote Sens. 2021, 13, 2627. https://doi.org/10.3390/rs13132627
Moura MM, de Oliveira LES, Sanquetta CR, Bastos A, Mohan M, Corte APD. Towards Amazon Forest Restoration: Automatic Detection of Species from UAV Imagery. Remote Sensing. 2021; 13(13):2627. https://doi.org/10.3390/rs13132627
Chicago/Turabian StyleMoura, Marks Melo, Luiz Eduardo Soares de Oliveira, Carlos Roberto Sanquetta, Alexis Bastos, Midhun Mohan, and Ana Paula Dalla Corte. 2021. "Towards Amazon Forest Restoration: Automatic Detection of Species from UAV Imagery" Remote Sensing 13, no. 13: 2627. https://doi.org/10.3390/rs13132627
APA StyleMoura, M. M., de Oliveira, L. E. S., Sanquetta, C. R., Bastos, A., Mohan, M., & Corte, A. P. D. (2021). Towards Amazon Forest Restoration: Automatic Detection of Species from UAV Imagery. Remote Sensing, 13(13), 2627. https://doi.org/10.3390/rs13132627








