Early Identification of Root Rot Disease by Using Hyperspectral Reflectance: The Case of Pathosystem Grapevine/Armillaria
Abstract
:1. Introduction
1.1. Root Symptoms
1.2. Foliar Symptoms
1.3. Disease Diagnosis
1.4. The Potential of Hyperspectral Sensors
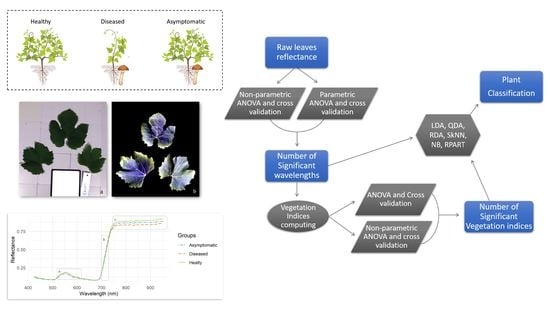
2. Materials and Methods
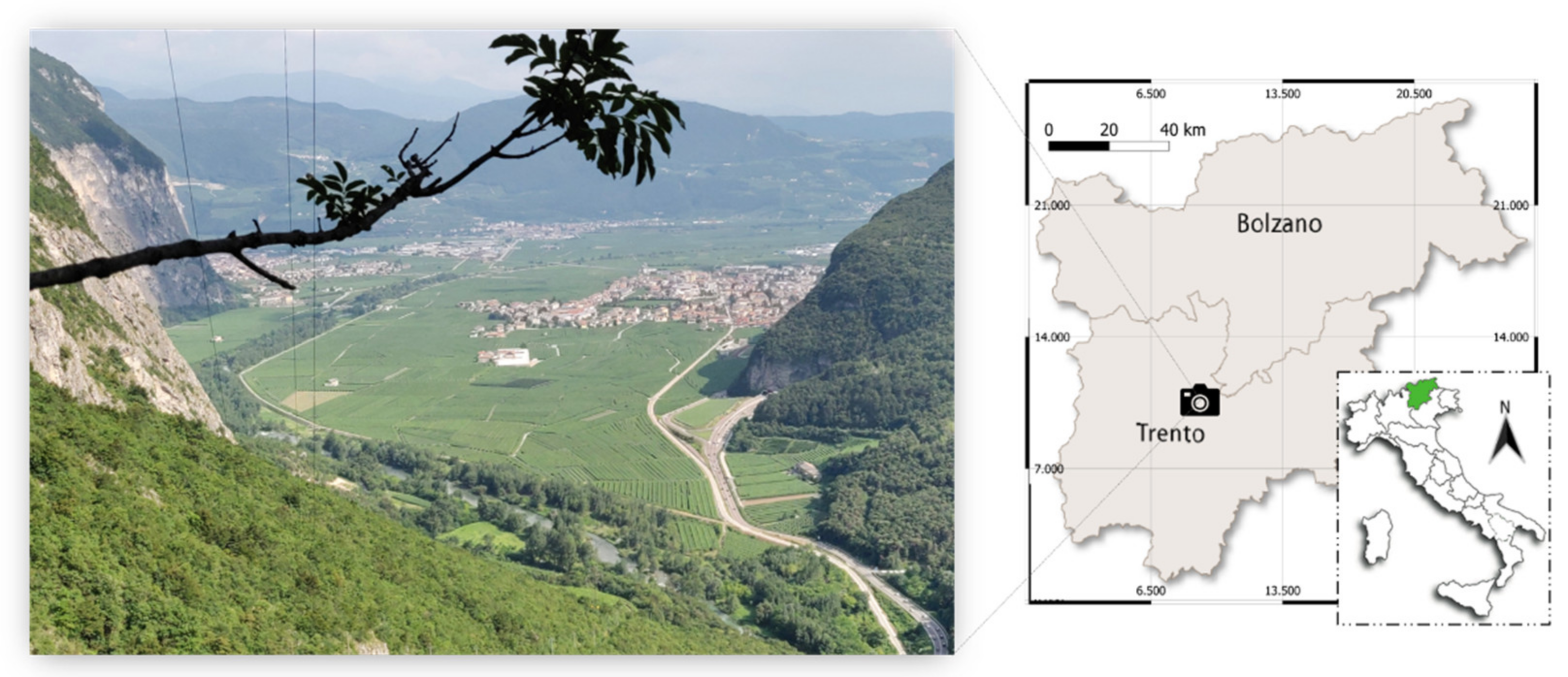
2.1. Study Site
2.2. Foliar Sampling
2.3. Root Sampling and Inspection
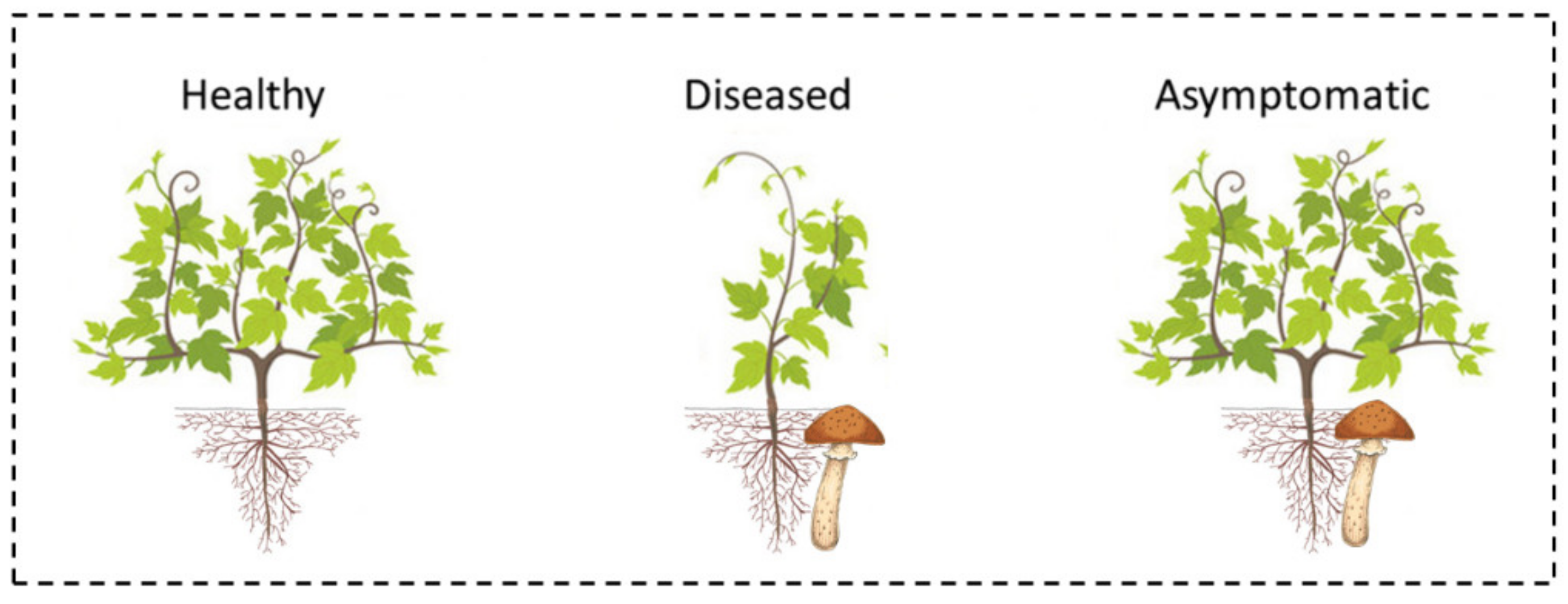
2.4. Plant Groups
2.5. Hyperspectral Data Acquisition
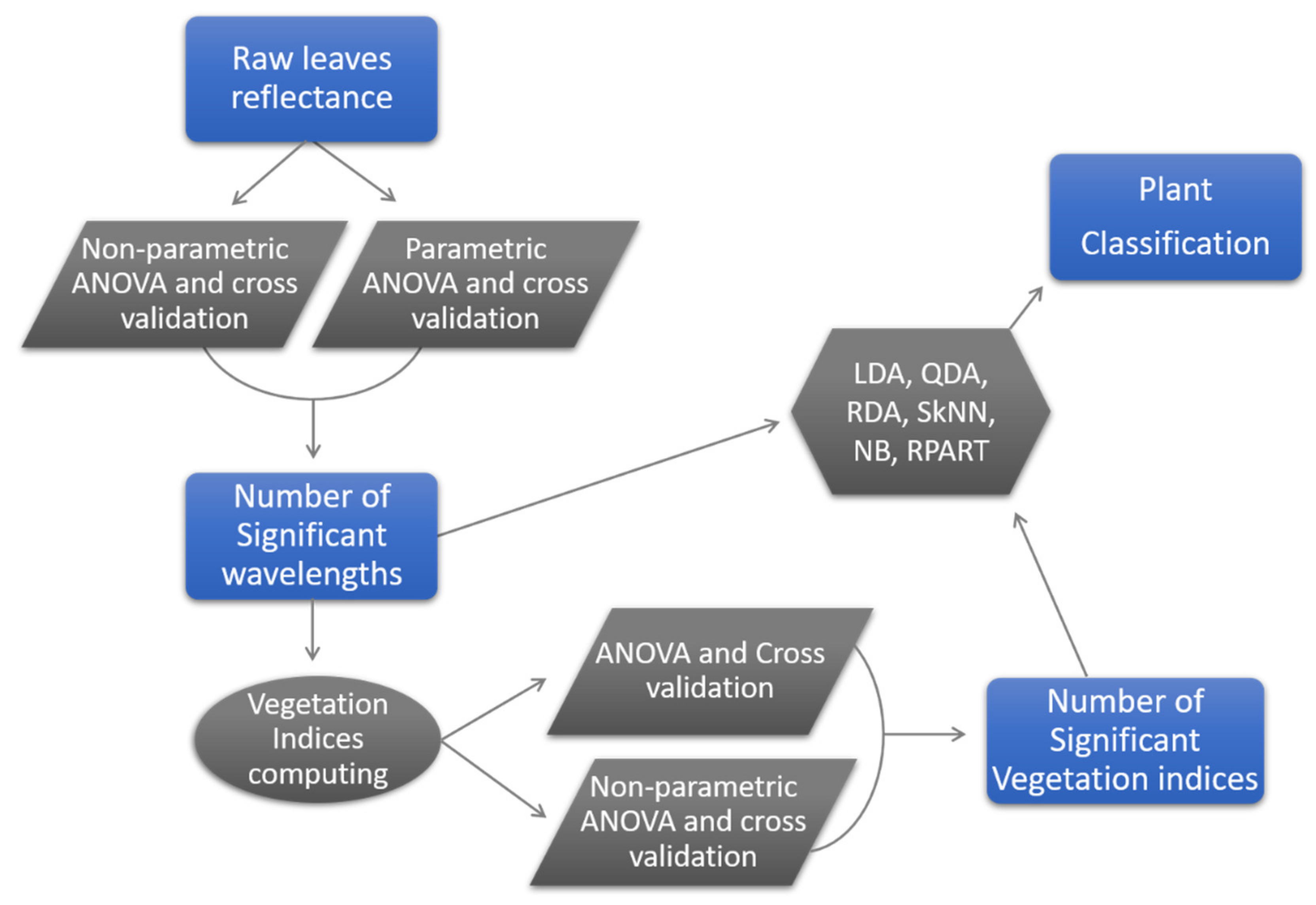
2.6. Statistical Analyses
2.7. Accuracy Assessment
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| No. | Index | p-Value | Diseased vs. Asymptomatic | Healthy vs. Asymptomatic | Healthy vs. Diseased |
|---|---|---|---|---|---|
| Cross-Validation P-adj | |||||
| 1 | GDVI | 0.000004 | 0.00005 | 0.52294 | 0.00002 |
| 2 | MGVI | 0.00004 | 0.00130 | 0.27417 | 0.00009 |
| 3 | OSAVI | 0.00009 | 0.00016 | 1 | 0.00259 |
| 4 | NDchl | 0.00011 | 0.00009 | 0.05128 | 0.35577 |
| 5 | mARI | 0.00012 | 0.00004 | 0.87102 | 0.05249 |
| 6 | Ctr4 | 0.00016 | 0.00019 | 0.02559 | 0.60489 |
| 7 | Chlred-edge | 0.00016 | 0.00018 | 0.03003 | 0.58745 |
| 8 | REIP3 | 0.00019 | 0.00045 | 0.00916 | 0.95927 |
| 9 | SR750/710 | 0.00020 | 0.00026 | 0.02886 | 0.58745 |
| 10 | GNDVI | 0.00021 | 0.00014 | 0.19992 | 0.15410 |
| 11 | NDVI | 0.00021 | 0.00008 | 0.49839 | 0.13671 |
| 12 | LCI | 0.00023 | 0.00023 | 0.05320 | 0.43261 |
| 13 | AVI | 0.00025 | 0.03044 | 0.09668 | 0.00014 |
| 14 | DVI | 0.00039 | 0.00014 | 0.88900 | 0.11608 |
| 15 | Vog2 | 0.00122 | 0.00155 | 0.04942 | 1.00000 |
| 16 | mCRIRE | 0.00219 | 0.05001 | 0.25925 | 0.00276 |
| 17 | ARI | 0.00908 | 0.00451 | 0.98258 | 0.62269 |
| 18 | WBI | 0.15533 | 0.22688 | 0.70442 | 1 |
| No. | Index | p-Value | F-Value | Diseased vs. Asymptomatic | Healthy vs. Asymptomatic | Healthy vs. Diseased |
|---|---|---|---|---|---|---|
| Cross-Validation P-adj | ||||||
| 1 | GDVI | 6.6 × 10−7 | 16.42 | 1.4 × 10−5 | 5.0 × 10−1 | 8.7 × 10−6 |
| 2 | NDchl | 1.2 × 10−5 | 12.69 | 6.7 × 10−6 | 6.3 × 10−2 | 1.6 × 10−1 |
| 3 | MGVI | 1.3 × 10−5 | 12.63 | 4.3 × 10−4 | 3.0 × 10−1 | 3.9 × 10−5 |
| 4 | OSAVI | 1.4 × 10−5 | 12.50 | 9.6 × 10−5 | 7.4 × 10−1 | 1.7 × 10−4 |
| 5 | GNDVI | 1.5 × 10−5 | 12.42 | 7.6 × 10−6 | 1.7 × 10−1 | 6.6 × 10−2 |
| 6 | NDVI | 1.7 × 10−5 | 12.24 | 9.00 × 10−6 | 2.50 × 10−1 | 4.26 × 10−2 |
| 7 | LCI | 1.8 × 10−5 | 12.17 | 1.0 × 10−5 | 7.2 × 10−2 | 1.7 × 10−1 |
| 8 | Ctr4 | 2.0 × 10−5 | 12.07 | 1.3 × 10−5 | 3.8 × 10−2 | 2.8 × 10−1 |
| 9 | REIP3 | 2.2 × 10−5 | 11.94 | 1.5 × 10−5 | 3.5 × 10−2 | 3.1 × 10−1 |
| 10 | Chlred.edge | 2.6 × 10−5 | 11.71 | 2.0 × 10−5 | 3.1 × 10−2 | 3.6 × 10−1 |
| 11 | mARI | 2.8 × 10−5 | 11.62 | 1.7 × 10−5 | 4.3 × 10−1 | 2.4 × 10−2 |
| 12 | SR750.710 | 3.7 × 10−5 | 11.30 | 2.8 × 10−5 | 3.2 × 10−2 | 4.0 × 10−1 |
| 13 | AVI | 7.3 × 10−5 | 10.47 | 7.6 × 10−3 | 1.0 × 10−1 | 7.2 × 10−5 |
| 14 | DVI | 9.1 × 10−5 | 10.21 | 5.4 × 10−5 | 4.4 × 10−1 | 4.3 × 10−2 |
| 15 | Vog2 | 2.7 × 10−4 | 8.90 | 2.6 × 10−4 | 3.9 × 10−2 | 6.3 × 10−1 |
| 16 | mCRIRE | 2.5 × 10−3 | 6.35 | 6.5 × 10−2 | 1.9 × 10−1 | 2.1 × 10−3 |
| 17 | ARI | 5.5 × 10−3 | 5.47 | 3.7 × 10−3 | 4.7 × 10−1 | 2.8 × 10−1 |
| 18 | WBI | 2.8 × 10−1 | 1.28 | 3.0 × 10−1 | 5.0 × 10−1 | 9.9 × 10−1 |
References
- Watling, R.; Kile, G.A.; Burdsall, H.H., Jr. Nomenclature, Taxonomy and Identification. In Armillaria Root Desease; Agriculture Handbook 691; USDA For. Serv.: Washington, DC, USA, 1991; pp. 1–9. [Google Scholar]
- Coetzee, M.P.A.; Wingfield, B.D.; Wingfield, M.J. Armillaria Root-Rot Pathogens: Species Boundaries and Global Distribution. Pathogens 2018, 7, 83. [Google Scholar] [CrossRef] [Green Version]
- Yafetto, L.; Davis, D.J.; Money, N.P. Biomechanics of Invasive Growth by Armillaria Rhizomorphs. Fungal Genet. Biol. 2009, 46, 688–694. [Google Scholar] [CrossRef]
- Yafetto, L. The Structure of Mycelial Cords and Rhizomorphs of Fungi: A Mini-Review. Mycosphere 2018, 9, 984–998. [Google Scholar] [CrossRef]
- Heinzelmann, R.; Dutech, C.; Tsykun, T.; Labbé, F.; Soularue, J.P.; Prospero, S. Latest Advances and Future Perspectives in Armillaria Research. Can. J. Plant Pathol. 2019, 41, 1–23. [Google Scholar] [CrossRef]
- La Porta, N.; Capretti, P.; Thomsen, I.M.; Kasanen, R.; Hietala, A.M.; von Weissenberg, K. Forest Pathogens with Higher Damage Potential Due to Climate Change in Europe. Can. J. Plant Pathol. 2008, 30, 177–195. [Google Scholar] [CrossRef]
- Baumgartner, K.; Coetzee, M.P.A.; Hoffmeister, D. Secrets of the Subterranean Pathosystem of Armillaria. Mol. Plant Pathol. 2011, 12, 515–534. [Google Scholar] [CrossRef]
- Cromey, M.G.; Drakulic, J.; Beal, E.J.; Waghorn, I.A.G.; Perry, J.N.; Clover, G.R.G. Susceptibility of Garden Trees and Shrubs to Armillaria Root Rot. Plant Dis. 2020, 104, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Marsh, R.W. Field Observations on the Spread of Armillaria Mellea in Apple Orchards and in a Blackcurrant Plantation. Trans. Br. Mycol. Soc. 1952, 35, 201–207. [Google Scholar] [CrossRef]
- Rizzo, D.M.; Whiting, E.C.; Elkins, R.B. Spatial Distribution of Armillaria mellea in Pear Orchards. Plant Dis. 1998, 82, 1226–1231. [Google Scholar] [CrossRef] [PubMed]
- Beckman, T.G.; Okie, W.R.; Nyczepir, A.P.; Pusey, P.L.; Reilly, C.C. Relative Susceptibility of Peach and Plum Germplasm to Armillaria Root Rot. HortScience 1998, 33, 1062–1065. [Google Scholar] [CrossRef] [Green Version]
- Miller, S.B.; Gasic, K.; Reighard, G.L.; Henderson, W.G.; Rollins, P.A.; Vassalos, M.; Schnabel, G. Preventative Root-Collar Excavation Reduces Peach Tree Mortality Caused by Armillaria Root Rot on Replant Sites. Plant Dis. 2020, 104, 1274–1279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donati, I.; Cellini, A.; Sangiorgio, D.; Caldera, E.; Sorrenti, G.; Spinelli, F. Pathogens Associated to Kiwifruit Vine Decline in Italy. Agriculture 2020, 10, 119. [Google Scholar] [CrossRef] [Green Version]
- Baumgartner, K.; Rizzo, D.M. Spread of Armillaria Root Disease in a California Vineyard. Am. J. Enol. Vitic. 2002, 53, 197–203. [Google Scholar]
- Baumgartner, K. Root Collar Excavation for Postinfection Control of Armillaria Root Disease of Grapevine. Plant Dis. 2004, 88, 1235–1240. [Google Scholar] [CrossRef] [Green Version]
- Prodorutti, D.; de Luca, F.; Michelon, L.; Pertot, I. Susceptibility to Armillaria mellea Root Rot in Grapevine Rootstocks Commonly Grafted onto Teroldego Rotaliano. Phytopathol. Mediterr. 2009, 48, 285–290. [Google Scholar] [CrossRef]
- Aguín, O.; Abuín, M.; Lozano, F.; Ferreiroa, V.; Corral, M.; Mansilla, J.P. Incidencia y Distribución Del Género Armillaria En Viñedos de Las Cinco Denominaciones de Origen de Vino de Galicia (Noroeste de España). Rev. Iberoam. Micol. 2015, 32, 13–19. [Google Scholar] [CrossRef]
- Ricciolini, M.; Rizzo, D. Avversità Della Vite e Strategie Di Difesa Integrata in Toscana; Press Service srl: Sesto Fiorentino, Italy, 2007. [Google Scholar]
- Nieuwenhuis, B.P.S.; Billiard, S.; Vuilleumier, S.; Petit, E.; Hood, M.E.; Giraud, T. Evolution of Uni- and Bifactorial Sexual Compatibility Systems in Fungi. Heredity 2013, 111, 445–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prodorutti, D.; de Luca, F.; Pellegrini, A.; Pertot, I. I Marciumi Radicali Della Vite; Safe Crop: San Michele all’Adige, Italy, 2007. [Google Scholar]
- Kwaśna, H.; Szynkiewicz-Wronek, A. Culturable Microfungi Inhibitory to Armillaria Rhizomorph Formation from Fagus Sylvatica Stump Roots and Soil. J. Phytopathol. 2018, 166, 314–323. [Google Scholar] [CrossRef]
- Bendel, N.; Kicherer, A.; Backhaus, A.; Köckerling, J.; Maixner, M.; Bleser, E.; Klück, H.C.; Seiffert, U.; Voegele, R.T.; Töpfer, R. Detection of Grapevine Leafroll-Associated Virus 1 and 3 in White and Red Grapevine Cultivars Using Hyperspectral Imaging. Remote Sens. 2020, 12, 1693. [Google Scholar] [CrossRef]
- Toler, R.W.; Smith, B.D.; Harlan, J.C. Use of Aerial Color Infrared Photography to Evaluate Crop Disease. Plant Dis. 1981, 65, 24–31. [Google Scholar] [CrossRef]
- Behmann, J.; Steinrücken, J.; Plümer, L. Detection of Early Plant Stress Responses in Hyperspectral Images. ISPRS J. Photogramm. Remote Sens. 2014, 93, 98–111. [Google Scholar] [CrossRef]
- Wang, T.; Thomasson, J.A.; Yang, C.; Isakeit, T.; Nichols, R.L. Automatic Classification of Cotton Root Rot Disease Based on UAV Remote Sensing. Remote Sens. 2020, 12, 1310. [Google Scholar] [CrossRef] [Green Version]
- Behmann, J.; Acebron, K.; Emin, D.; Bennertz, S.; Matsubara, S.; Thomas, S.; Bohnenkamp, D.; Kuska, M.T.; Jussila, J.; Salo, H.; et al. Specim IQ: Evaluation of a New, Miniaturized Handheld Hyperspectral Camera and Its Application for Plant Phenotyping and Disease Detection. Sensors 2018, 18, 441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahlein, A.K.; Steiner, U.; Hillnhütter, C.; Dehne, H.W.; Oerke, E.C. Hyperspectral Imaging for Small-Scale Analysis of Symptoms Caused by Different Sugar Beet Diseases. Plant Methods 2012, 8, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naidu, R.A.; Perry, E.M.; Pierce, F.J.; Mekuria, T. The Potential of Spectral Reflectance Technique for the Detection of Grapevine Leafroll-Associated Virus-3 in Two Red-Berried Wine Grape Cultivars. Comput. Electron. Agric. 2009, 66, 38–45. [Google Scholar] [CrossRef]
- Gao, Z.; Khot, L.R.; Naidu, R.A.; Zhang, Q. Early Detection of Grapevine Leafroll Disease in a Red-Berried Wine Grape Cultivar Using Hyperspectral Imaging. Comput. Electron. Agric. 2020, 179, 105807. [Google Scholar] [CrossRef]
- Junges, A.H.; Ducati, J.R.; Cristian, S.L.; Almança, M.A.K. Detection of Grapevine Leaf Stripe Disease Symptoms by Hyperspectral Sensor. Phytopathol. Mediterr. 2015, 54, 241–252. [Google Scholar] [CrossRef]
- Bendel, N.; Kicherer, A.; Backhaus, A.; Klück, H.C.; Seiffert, U.; Fischer, M.; Voegele, R.T.; Töpfer, R. Evaluating the Suitability of Hyper- and Multispectral Imaging to Detect Foliar Symptoms of the Grapevine Trunk Disease Esca in Vineyards. Plant Methods 2020, 16, 1–19. [Google Scholar] [CrossRef]
- Junges, A.H.; Almança, M.A.K.; Fajardo, T.V.M.; Ducati, J.R. Leaf Hyperspectral Reflectance as a Potential Tool to Detect Diseases Associated with Vineyard Decline. Trop. Plant Pathol. 2020, 45, 522–533. [Google Scholar] [CrossRef]
- Boulent, J.; St-Charles, P.-L.; Foucher, S.; Théau, J. Automatic Detection of Flavescence Dorée Symptoms Across White Grapevine Varieties Using Deep Learning. Front. Artif. Intell. 2020, 3. [Google Scholar] [CrossRef]
- Albetis, J.; Duthoit, S.; Guttler, F.; Jacquin, A.; Goulard, M.; Poilvé, H.; Féret, J.-B.; Dedieu, G. Detection of Flavescence Dorée Grapevine Disease Using Unmanned Aerial Vehicle (UAV) Multispectral Imagery. Remote Sens. 2017, 9, 308. [Google Scholar] [CrossRef] [Green Version]
- Bendel, N.; Backhaus, A.; Kicherer, A.; Köckerling, J.; Maixner, M.; Jarausch, B.; Biancu, S.; Klück, H.-C.; Seiffert, U.; Voegele, R.T.; et al. Detection of Two Different Grapevine Yellows in Vitis Vinifera Using Hyperspectral Imaging. Remote Sens. 2020, 12, 4151. [Google Scholar] [CrossRef]
- Oberti, R.; Marchi, M.; Tirelli, P.; Calcante, A.; Iriti, M.; Borghese, A.N. Automatic Detection of Powdery Mildew on Grapevine Leaves by Image Analysis: Optimal View-Angle Range to Increase the Sensitivity. Comput. Electron. Agric. 2014, 104, 1–8. [Google Scholar] [CrossRef]
- Alt, V.V.; Gurova, T.A.; Elkin, O.V.; Klimenko, D.N.; Maximov, L.V.; Pestunov, I.A.; Dubrovskaya, O.A.; Genaev, M.A.; Erst, T.V.; Genaev, K.A.; et al. The Use of Specim IQ, a Hyperspectral Camera, for Plant Analysis. Vavilovskii Zhurnal Genet. Selektsii 2020, 24, 259–266. [Google Scholar] [CrossRef]
- Barreto, A.; Paulus, S.; Varrelmann, M.; Mahlein, A.K. Hyperspectral Imaging of Symptoms Induced by Rhizoctonia Solani in Sugar Beet: Comparison of Input Data and Different Machine Learning Algorithms. J. Plant Dis. Prot. 2020, 127, 441–451. [Google Scholar] [CrossRef]
- Trento Province. Provincia Autonoma Di Trento; Provincia Autonoma di Trento: Trento, Italy, 2020. [Google Scholar]
- Pertot, I.; Gobbin, D.; de Luca, F.; Prodorutti, D. Methods of Assessing the Incidence of Armillaria Root Rot across Viticultural Areas and the Pathogen’s Genetic Diversity and Spatial-Temporal Pattern in Northern Italy. Crop Prot. 2008, 27, 1061–1070. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Vegetation Stress: An Introduction to the Stress Concept in Plants. J. Plant Physiol. 1996, 148, 4–14. [Google Scholar] [CrossRef]
- Peñuelas, J.; Gamon, J.A.; Fredeen, A.L.; Merino, J.; Field, C.B. Reflectance Indices Associated with Physiological Changes in Nitrogen- and Water-Limited Sunflower Leaves. Remote Sens. Environ. 1994, 48, 135–146. [Google Scholar] [CrossRef]
- Steele, M.R.; Gitelson, A.A.; Rundquist, D.C.; Merzlyak, M.N. Nondestructive Estimation of Anthocyanin Content in Grapevine Leaves. Am. J. Enol. Vitic. 2009, 60, 87–92. [Google Scholar]
- Gitelson, A.A.; Keydan, G.P.; Merzlyak, M.N. Three-Band Model for Noninvasive Estimation of Chlorophyll, Carotenoids, and Anthocyanin Contents in Higher Plant Leaves. Geophys. Res. Lett. 2006, 33, 2–6. [Google Scholar] [CrossRef] [Green Version]
- Singh, D.; Singh, S. Geospatial Modeling of Canopy Chlorophyll Content Using High Spectral Resolution Satellite Data in Himalayan Forests. Clim. Chang. Environ. Sustain. 2018, 6, 20. [Google Scholar] [CrossRef]
- Vogelmann, J.E.; Rock, B.N.; Moss, D.M. Red Edge Spectral Measurements from Sugar Maple Leaves. Int. J. Remote Sens. 1993, 14, 1563–1575. [Google Scholar] [CrossRef]
- Pu, R.; Gong, P.; Yu, Q. Comparative Analysis of EO-1 ALI and Hyperion, and Landsat ETM+ Data for Mapping Forest Crown Closure and Leaf Area Index. Sensors 2008, 8, 3744–3766. [Google Scholar] [CrossRef] [Green Version]
- Main, R.; Cho, M.A.; Mathieu, R.; O’Kennedy, M.M.; Ramoelo, A.; Koch, S. An Investigation into Robust Spectral Indices for Leaf Chlorophyll Estimation. ISPRS J. Photogramm. Remote Sens. 2011, 66, 751–761. [Google Scholar] [CrossRef]
- Bannari, A.; Morin, D.; Bonn, F.; Huete, A.R. A Review of Vegetation Indices. Remote Sens. Rev. 1995, 13, 95–120. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a Green Channel in Remote Sensing of Global Vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Wu, W. The Generalized Difference Vegetation Index (GDVI) for Dryland Characterization. Remote Sens. 2014, 6, 1211–1233. [Google Scholar] [CrossRef] [Green Version]
- Carter, G.A. Ratios of Leaf Reflectances in Narrow Wavebands as Indicators of Plant Stress. Int. J. Remote Sens. 1994, 15, 517–520. [Google Scholar] [CrossRef]
- Manevski, K.; Jabloun, M.; Gupta, M.; Kalaitzidis, C. Field-Scale Sensitivity of Vegetation Discrimination to Hyperspectral Reflectance and Coupled Statistics. In Sensitivity Analysis in Earth Observation Modelling; Elsevier Inc.: Amsterdam, The Netherlands, 2017. [Google Scholar] [CrossRef]
- Avola, G.; Di Gennaro, S.F.; Cantini, C.; Riggi, E.; Muratore, F.; Tornambè, C.; Matese, A. Remotely Sensed Vegetation Indices to Discriminate Field-Grown Olive Cultivars. Remote Sens. 2019, 11, 1242. [Google Scholar] [CrossRef] [Green Version]
- Kozak, M.; Piepho, H.P. What’s Normal Anyway? Residual Plots Are More Telling than Significance Tests When Checking ANOVA Assumptions. J. Agron. Crop Sci. 2018, 204, 86–98. [Google Scholar] [CrossRef]
- Layard, M.W.J. Robust Large-Sample Tests for Homogeneity of Variances. J. Am. Stat. Assoc. 1973, 68, 195–198. [Google Scholar] [CrossRef]
- Zapolska, A.; Kalaitzidis, C.; Markakis, E.; Ligoxigakis, E.; Koubouris, G. Linear Discriminant Analysis of Spectral Measurements for Discrimination between Healthy and Diseased Trees of Olea Europaea L. Artificially Infected by Fomitiporia Mediterranea. Int. J. Remote Sens. 2020, 41, 5388–5398. [Google Scholar] [CrossRef]
- Sankaran, S.; Ehsani, R.; Inch, S.A.; Ploetz, R.C. Evaluation of Visible-near Infrared Reflectance Spectra of Avocado Leaves as a Non-Destructive Sensing Tool for Detection of Laurel Wilt. Plant Dis. 2012, 96, 1683–1689. [Google Scholar] [CrossRef]
- Furlanetto, R.H.; Moriwaki, T.; Falcioni, R.; Pattaro, M.; Vollmann, A.; Sturion Junior, A.C.; Antunes, W.C.; Nanni, M.R. Hyperspectral Reflectance Imaging to Classify Lettuce Varieties by Optimum Selected Wavelengths and Linear Discriminant Analysis. Remote Sens. Appl. Soc. Environ. 2020, 20, 100400. [Google Scholar] [CrossRef]
- Moshou, D.; Bravo, C.; West, J.; Wahlen, S.; McCartney, A.; Ramon, H. Automatic Detection of “yellow Rust” in Wheat Using Reflectance Measurements and Neural Networks. Comput. Electron. Agric. 2004, 44, 173–188. [Google Scholar] [CrossRef]
- Sankaran, S.; Mishra, A.; Maja, J.M.; Ehsani, R. Visible-Near Infrared Spectroscopy for Detection of Huanglongbing in Citrus Orchards. Comput. Electron. Agric. 2011, 77, 127–134. [Google Scholar] [CrossRef]
- Chan, A.H.Y.; Barnes, C.; Swinfield, T.; Coomes, D.A. Monitoring Ash Dieback (Hymenoscyphus fraxineus) in British Forests Using Hyperspectral Remote Sensing. Remote Sens. Ecol. Conserv. 2020, 1–15. [Google Scholar] [CrossRef]
- Feret, J.B.; Asner, G.P. Tree Species Discrimination in Tropical Forests Using Airborne Imaging Spectroscopy. IEEE Trans. Geosci. Remote Sens. 2013, 51, 73–84. [Google Scholar] [CrossRef]
- Berge, A.; Solberg, A.S. A Comparison of Methods for Improving Classification of Hyperspectral Data. Int. Geosci. Remote Sens. Symp. 2004, 2, 945–948. [Google Scholar] [CrossRef]
- Chen, Y.; Crawford, M.M.; Ghosh, J. Applying Nonlinear Manifold Learning to Hyperspectral Data for Land Cover Classification. In Proceedings of the 2005 IEEE International Geoscience and Remote Sensing Symposium, IGARSS ’05, Seoul, Korea, 29 July 2005; IEEE: Piscataway, NJ, USA, 2005; Volume 6, pp. 4311–4314. [Google Scholar] [CrossRef] [Green Version]
- Abdulridha, J.; Batuman, O.; Ampatzidis, Y. UAV-Based Remote Sensing Technique to Detect Citrus Canker Disease Utilizing Hyperspectral Imaging and Machine Learning. Remote Sens. 2019, 11, 1373. [Google Scholar] [CrossRef] [Green Version]
- Ravikanth, L.; Singh, C.B.; Jayas, D.S.; White, N.D.G. Classification of Contaminants from Wheat Using Near-Infrared Hyperspectral Imaging. Biosyst. Eng. 2015, 135, 73–86. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.; Stone, C.J.; Olshen, R.A. Classification and Regression Trees; CRC Press: Boca Raton, FL, USA, 1984. [Google Scholar]
- Moghadam, P.; Ward, D.; Goan, E.; Jayawardena, S.; Sikka, P.; Hernandez, E. Plant Disease Detection Using Hyperspectral Imaging. In Proceedings of the DICTA 2017—2017 International Conference on Digital Image Computing: Techniques and Applications, 29 November–1 December 2017; pp. 1–8. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, Y.; Pu, R.; Gonzalez-Moreno, P.; Yuan, L.; Wu, K.; Huang, W. Monitoring Plant Diseases and Pests through Remote Sensing Technology: A Review. Comput. Electron. Agric. 2019, 165, 104943. [Google Scholar] [CrossRef]
- Martinelli, F.; Scalenghe, R.; Davino, S.; Panno, S.; Scuderi, G.; Ruisi, P.; Villa, P.; Stroppiana, D.; Boschetti, M.; Goulart, L.R.; et al. Advanced Methods of Plant Disease Detection. A Review. Agron. Sustain. Dev. 2015, 35, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Kong, W.; Zhang, C.; Huang, W.; Liu, F.; He, Y. Application of Hyperspectral Imaging to Detect Sclerotinia Sclerotiorum on Oilseed Rape Stems. Sensors 2018, 18, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morel, J.; Jay, S.; Féret, J.B.; Bakache, A.; Bendoula, R.; Carreel, F.; Gorretta, N. Exploring the Potential of PROCOSINE and Close-Range Hyperspectral Imaging to Study the Effects of Fungal Diseases on Leaf Physiology. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Garhwal, A.S.; Pullanagari, R.R.; Li, M.; Reis, M.M.; Archer, R. Hyperspectral Imaging for Identification of Zebra Chip Disease in Potatoes. Biosyst. Eng. 2020, 197, 306–317. [Google Scholar] [CrossRef]
- Zhang, M.; Qin, Z.; Liu, X. Remote Sensed Spectral Imagery to Detect Late Blight in Field Tomatoes. Precis. Agric. 2005, 6, 489–508. [Google Scholar] [CrossRef]
- Vescovo, L.; Wohlfahrt, G.; Balzarolo, M.; Pilloni, S.; Sottocornola, M.; Rodeghiero, M.; Gianelle, D. New Spectral Vegetation Indices Based on the Near-Infrared Shoulder Wavelengths for Remote Detection of Grassland Phytomass. Int. J. Remote Sens. 2012, 33, 2178–2195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Huang, W.; Pu, R.; Wang, J. Detection of Internal Leaf Structure Deterioration Using a New Spectral Ratio Index in the Near-Infrared Shoulder Region. J. Integr. Agric. 2014, 13, 760–769. [Google Scholar] [CrossRef]
- Gausman, H.W. Leaf Reflectance of Near-Infrared. Photogramm. Eng. 1974, 40, 183–191. [Google Scholar]
- Reynolds, G.J.; Windels, C.E.; MacRae, I.V.; Laguette, S. Remote Sensing for Assessing Rhizoctonia Crown and Root Rot Severity in Sugar Beet. Plant. Dis. 2012, 96, 497–505. [Google Scholar] [CrossRef]
- Imran, H.A.; Gianelle, D.; Rocchini, D.; Dalponte, M.; Martín, M.P.; Sakowska, K.; Wohlfahrt, G.; Vescovo, L. VIS-NIR, Red-Edge and NIR-Shoulder Based Normalized Vegetation Indices Response to Co-Varying Leaf and Canopy Structural Traits in Heterogeneous Grasslands. Remote Sens. 2020, 12, 2254. [Google Scholar] [CrossRef]
- Nogales, A.; Aguirreolea, J.; Santa María, E.; Camprubí, A.; Calvet, C. Response of Mycorrhizal Grapevine to Armillaria Mellea Inoculation: Disease Development and Polyamines. Plant Soil 2009, 317, 177–187. [Google Scholar] [CrossRef] [Green Version]
- Camprubi, A.; Solari, J.; Bonini, P.; Garcia-Figueres, F.; Colosimo, F.; Cirino, V.; Lucini, L.; Calvet, C. Plant Performance and Metabolomic Profile of Loquat in Response to Mycorrhizal Inoculation, Armillaria Mellea and Their Interaction. Agronomy 2020, 10, 899. [Google Scholar] [CrossRef]
- Perazzolli, M.; Bampi, F.; Faccin, S.; Moser, M.; de Luca, F.; Ciccotti, A.M.; Velasco, R.; Gessler, C.; Pertot, I.; Moser, C. Armillaria mellea Induces a Set of Defense Genes in Grapevine Roots and One of Them Codifies a Protein with Antifungal Activity. Mol. Plant. Microbe Interact. 2010, 23, 485–496. [Google Scholar] [CrossRef]
- Mahlein, A.-K.; Kuska, M.T.; Behmann, J.; Polder, G.; Walter, A. Hyperspectral Sensors and Imaging Technologies in Phytopathology: State of the Art. Annu. Rev. Phytopathol. 2018, 56, 535–558. [Google Scholar] [CrossRef]
- Vergara-Diaz, O.; Vatter, T.; Kefauver, S.C.; Obata, T.; Fernie, A.R.; Araus, J.L. Assessing Durum Wheat Ear and Leaf Metabolomes in the Field through Hyperspectral Data. Plant. J. 2020, 102, 615–630. [Google Scholar] [CrossRef] [PubMed]
- Heritage, C. Spectral Remote Sensing of Tomato Plants (Lycopersicon esculentum L.) Infected with Tomato Mosaic Virus (ToMV). In Proceedings of the 30th EARSeL Symposium: Remote Sensing for Science, Education and Culture, Paris, France, 31 May–4 June 2010. [Google Scholar]
- Mutanga, O.; Skidmore, A.K. Red Edge Shift and Biochemical Content in Grass Canopies. ISPRS J. Photogramm. Remote Sens. 2007, 62, 34–42. [Google Scholar] [CrossRef]
- Steele, M.; Gitelson, A.A.; Rundquist, D. Nondestructive Estimation of Leaf Chlorophyll Content in Grapes. Am. J. Enol. Vitic. 2008, 59, 299–305. [Google Scholar]
- Pinar, A.; Curran, P.J. Technical Note: Grass Chlorophyll and the Reflectance Red Edge. Int. J. Remote Sens. 1996, 17, 351–357. [Google Scholar] [CrossRef]
- Rizzuti, A.; Aguilera-Sáez, L.M.; Santoro, F.; Valentini, F.; Gualano, S.; D’onghia, A.M.; Gallo, V.; Mastrorilli, P.; Latronico, M. Detection of Erwinia Amylovora in Pear Leaves Using a Combined Approach by Hyperspectral Reflectance and Nuclear Magnetic Resonance Spectroscopy. Phytopathol. Mediterr. 2018, 57, 296–306. [Google Scholar]
- Huang, R.; Hong, D.; Xu, Y.; Yao, W.; Stilla, U. Multi-Scale Local Context Embedding for LiDAR Point Cloud Classification. IEEE Geosci. Remote Sens. Lett. 2020, 17, 721–725. [Google Scholar] [CrossRef]
- Hang, R.; Hang, R.; Li, Z.; Ghamisi, P.; Hong, D.; Hong, D.; Xia, G.; Liu, Q. Classification of Hyperspectral and LiDAR Data Using Coupled CNNs. IEEE Trans. Geosci. Remote Sens. 2020, 58, 4939–4950. [Google Scholar] [CrossRef] [Green Version]
- Hong, D.; Gao, L.; Hang, R.; Zhang, B.; Chanussot, J. Deep Encoder-Decoder Networks for Classification of Hyperspectral and LiDAR Data. IEEE Geosci. Remote Sens. Lett. 2020, 1–5. [Google Scholar] [CrossRef]
- Hong, D.; Yokoya, N.; Chanussot, J.; Zhu, X.X. An Augmented Linear Mixing Model to Address Spectral Variability for Hyperspectral Unmixing. IEEE Trans. Image Process. 2019, 28, 1923–1938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Candiago, S.; Remondino, F.; de Giglio, M.; Dubbini, M.; Gattelli, M. Evaluating Multispectral Images and Vegetation Indices for Precision Farming Applications from UAV Images. Remote Sens. 2015, 7, 4026–4047. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Bueno, M.L.; Pineda, M.; Vida, C.; Fernández-Ortuño, D.; Torés, J.A.; de Vicente, A.; Cazorla, F.M.; Barón, M. Detection of White Root Rot in Avocado Trees by Remote Sensing. Plant. Dis. 2019, 103, 1119–1125. [Google Scholar] [CrossRef]



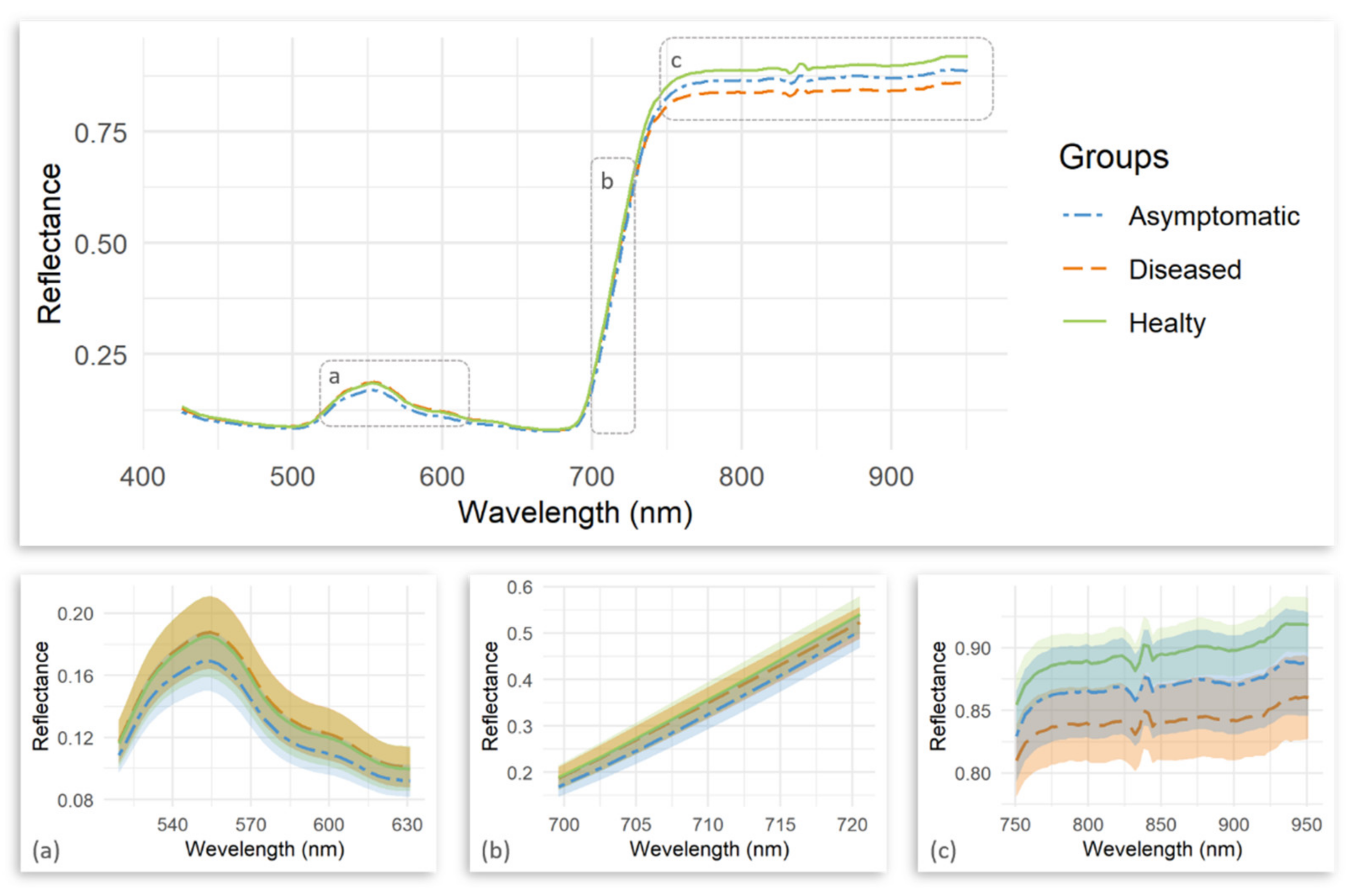
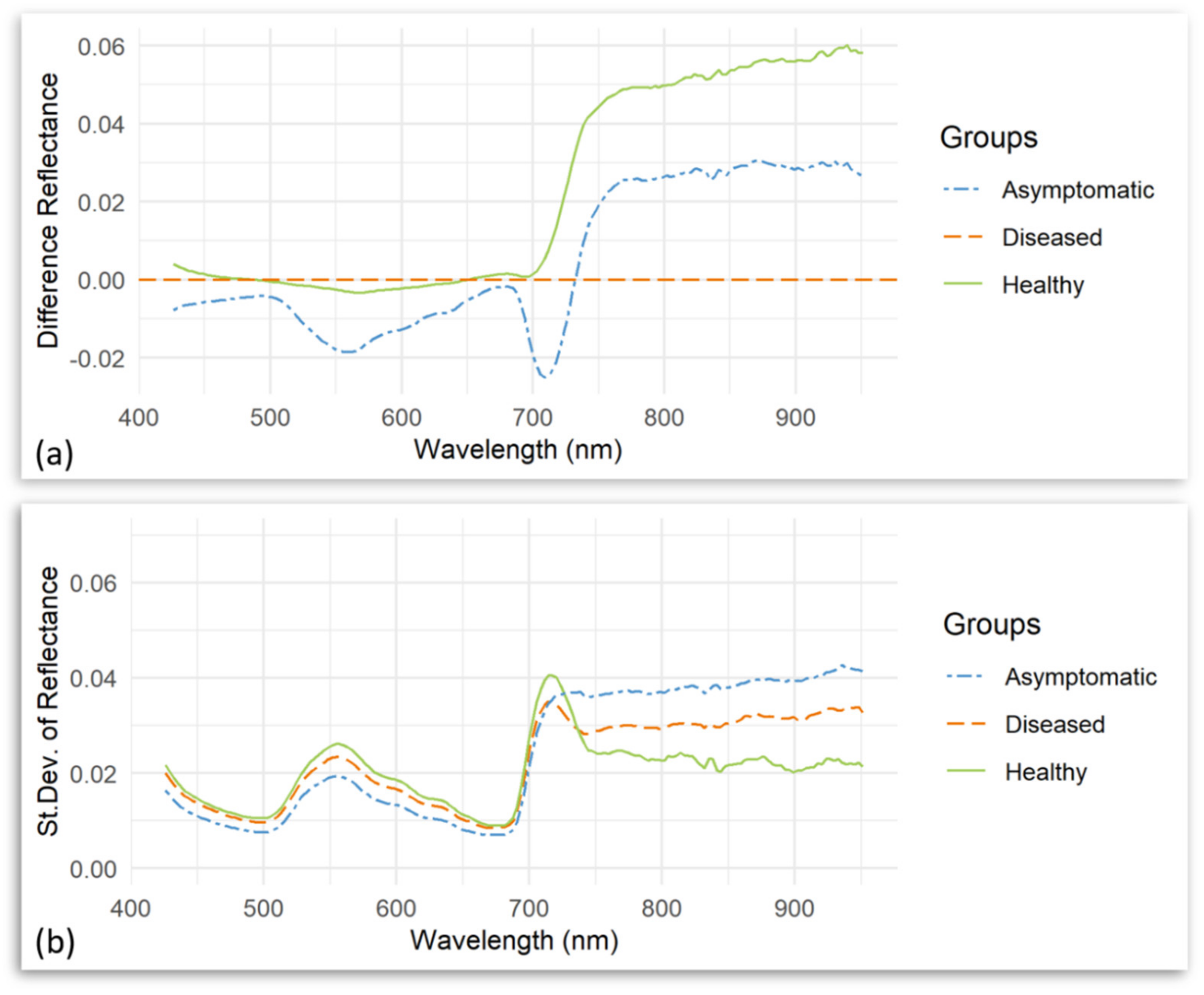
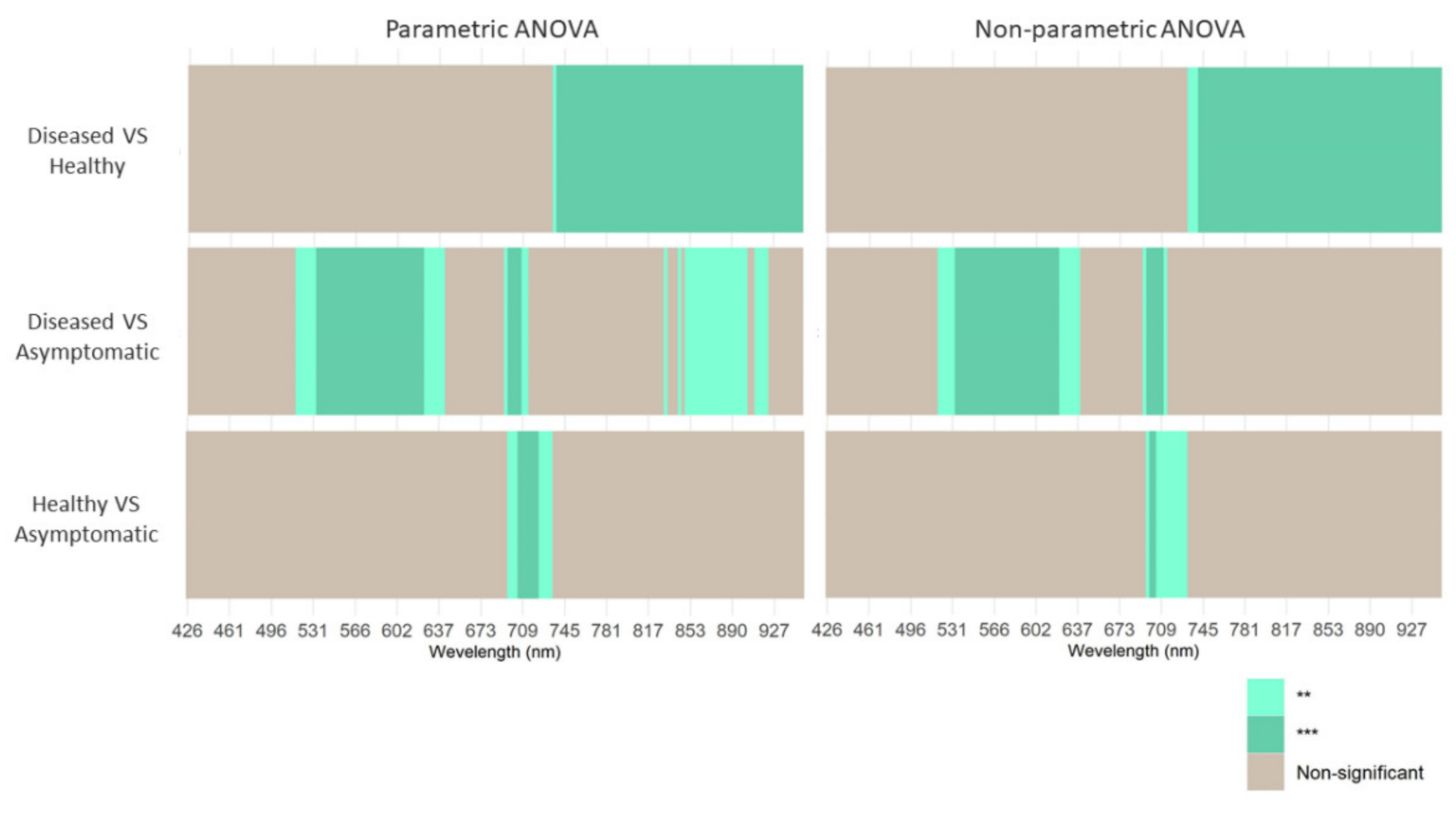
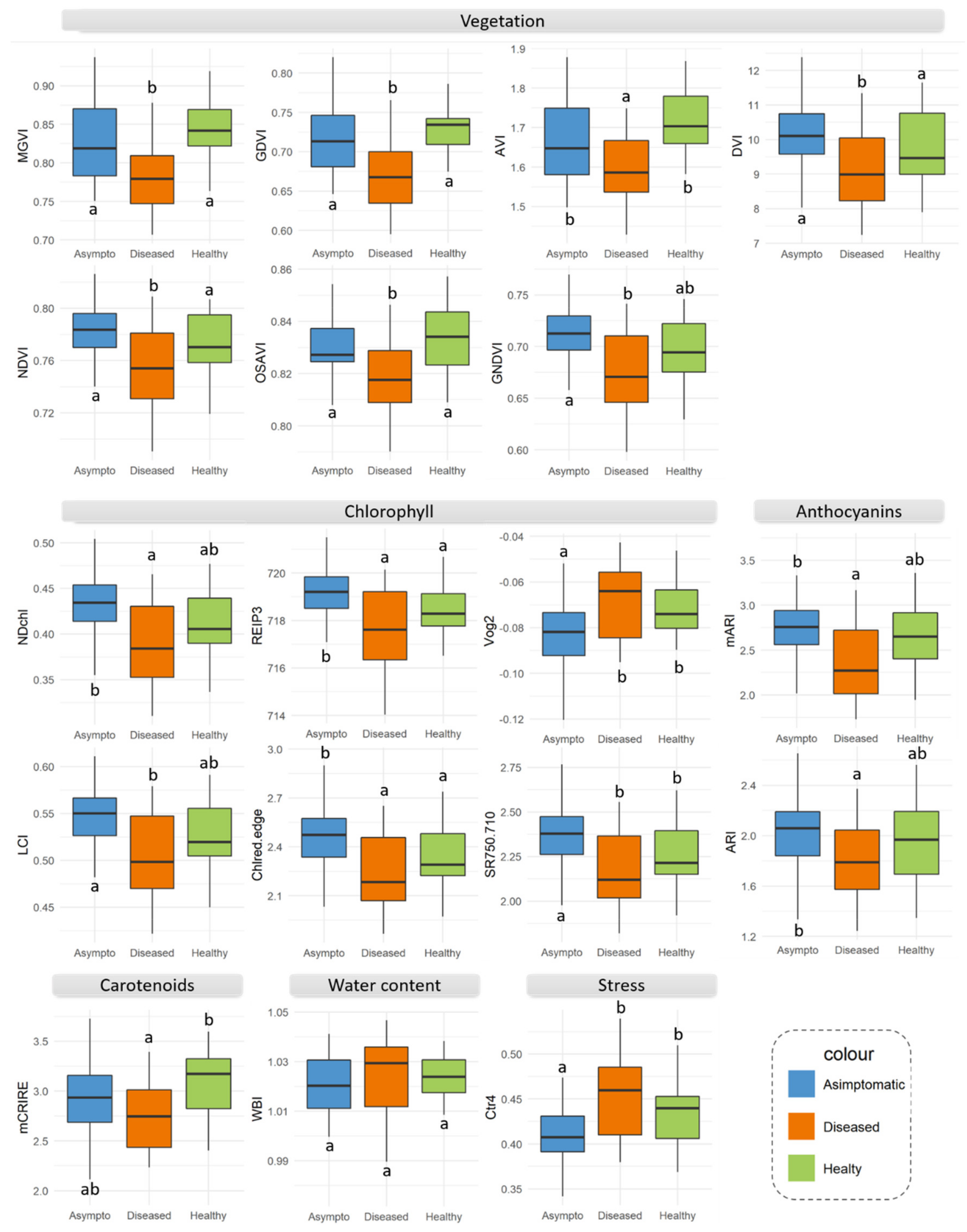
| Number | Vegetation Index | Abbreviation | Equation | Related to | Reference |
|---|---|---|---|---|---|
| 1 | Anthocyanin Reflectance Index | ARI | anthocyanins | [43] | |
| 2 | Modified Anthocyanin Reflectance Index | mARI | anthocyanins | [44] | |
| 3 | Carotenoid Reflectance Index Red Edge | mCRIRE | carotenoid | [44] | |
| 4 | Normalized Difference Chlorophyll | NDchl | chlorophyll | [45] | |
| 5 | Red Edge Inflection Point 3 | REIP3 | chlorophyll | [46] | |
| 6 | Leaf Chlorophyll Index | LCI | chlorophyll | [47] | |
| 7 | Vogelmann Indices 2 | Vog2 | chlorophyll | [24] | |
| 8 | Zarco-Tejada and Miller | SR750/710 | chlorophyll | [48] | |
| 9 | Chlorophyll Red Edge | Chlred-edge | chlorophyll | [44] | |
| 10 | Difference Vegetation Index | DVI | vegetation | [49] | |
| 11 | Normalized Difference Vegetation Index | NDVI | vegetation | [49] | |
| 12 | Misra Green Vegetation Index | MGVI | vegetation | [49] | |
| 13 | Green Normalized Difference Vegetation Index | GNDVI | vegetation | [50] | |
| 14 | Ashburn Vegetation Index | AVI | vegetation | [49] | |
| 15 | Green Difference Vegetation Index | GDVI | vegetation | [51] | |
| 16 | Optimized Soil-Adjusted Vegetation Index | OSAVI | vegetation | [48] | |
| 17 | Simple Ratio Carter4 | Ctr4 | stress | [52] | |
| 18 | Water Band Index | WBI | water content | [42] |
| Model | Groups | Variables Used |
|---|---|---|
| 1 | Healthy vs. Diseased | GDVI, MGVI, AVI, OSAVI, R920 |
| 2 | Healthy vs. Asymptomatic | R705, R711, R708, R714, R717 |
| 3 | Healthy vs. Asymptomatic vs. Diseased | GDVI, NDchl, MGVI, OSAVI, GNDVI |
| Overall Accuracy | Kappa Coefficient | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | SkNN | LDA | QDA | RDA | NB | RPART | Mean | Std.Dev. | Model | SkNN | LDA | QDA | RDA | NB | RPART | Mean | Std.Dev. |
| 1 | 80% | 90% | 80% | 90% | 90% | 95% | 88% | 0.061 | 1 | 0.6 | 0.78 | 0.57 | 0.78 | 0.76 | 0.89 | 0.73 | 0.122 |
| 2 | 72% | 72% | 80% | 76% | 76% | 72% | 75% | 0.033 | 2 | 0.29 | 0.23 | 0.49 | 0.31 | 0.31 | 0.34 | 0.33 | 0.087 |
| 3 | 60% | 57% | 54% | 54% | 76% | 68% | 62% | 0.088 | 3 | 0.32 | 0.3 | 0.27 | 0.27 | 0.6 | 0.42 | 0.36 | 0.128 |
| Mean | 71% | 73% | 71% | 73% | 81% | 78% | Mean | 0.40 | 0.44 | 0.44 | 0.45 | 0.56 | 0.55 | ||||
| Std.Dev. | 0.101 | 0.165 | 0.150 | 0.181 | 0.081 | 0.146 | Std.Dev. | 0.171 | 0.299 | 0.155 | 0.284 | 0.228 | 0.297 | ||||
| Error of Omission | Error of Commission | ||||||||||||||||
| Model | SkNN | LDA | QDA | RDA | NB | RPART | Mean | Std.Dev. | Model | SkNN | LDA | QDA | RDA | NB | RPART | Mean | Std.Dev. |
| 1 d | 29% | 14% | 21% | 14% | 7% | 7% | 15% | 0.085 | 1 d | 0% | 0% | 17% | 0% | 6% | 0% | 4% | 0.069 |
| 2 a | 12% | 6% | 6% | 0% | 0% | 17% | 7% | 0.067 | 2 a | 63% | 75% | 50% | 75% | 75% | 50% | 65% | 0.123 |
| 3 a | 32% | 42% | 42% | 47% | 21% | 5% | 32% | 0.160 | 3 a | 50% | 39% | 33% | 39% | 22% | 50% | 39% | 0.106 |
| 3 d | 30% | 20% | 30% | 20% | 20% | 30% | 25% | 0.055 | 3 d | 18% | 26% | 26% | 30% | 15% | 11% | 21% | 0.074 |
| 3 h | 75% | 75% | 75% | 75% | 37% | 100% | 73% | 0.202 | 3 h | 3% | 7% | 14% | 7% | 37% | 0% | 11% | 0.134 |
| Mean | 36% | 31% | 35% | 31% | 17% | 32% | Mean | 27% | 29% | 28% | 30% | 31% | 22% | ||||
| Std.Dev. | 0.234 | 0.278 | 0.260 | 0.298 | 0.143 | 0.394 | Std.Dev. | 0.283 | 0.298 | 0.144 | 0.297 | 0.271 | 0.258 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calamita, F.; Imran, H.A.; Vescovo, L.; Mekhalfi, M.L.; La Porta, N. Early Identification of Root Rot Disease by Using Hyperspectral Reflectance: The Case of Pathosystem Grapevine/Armillaria. Remote Sens. 2021, 13, 2436. https://doi.org/10.3390/rs13132436
Calamita F, Imran HA, Vescovo L, Mekhalfi ML, La Porta N. Early Identification of Root Rot Disease by Using Hyperspectral Reflectance: The Case of Pathosystem Grapevine/Armillaria. Remote Sensing. 2021; 13(13):2436. https://doi.org/10.3390/rs13132436
Chicago/Turabian StyleCalamita, Federico, Hafiz Ali Imran, Loris Vescovo, Mohamed Lamine Mekhalfi, and Nicola La Porta. 2021. "Early Identification of Root Rot Disease by Using Hyperspectral Reflectance: The Case of Pathosystem Grapevine/Armillaria" Remote Sensing 13, no. 13: 2436. https://doi.org/10.3390/rs13132436