1. Introduction
With the rising potential of remote-sensing applications in real life, research in remote-sensing analysis is increasingly necessary [
1,
2]. Hyperspectral imaging is commonly used in remote sensing. A hyperspectral image (HSI) is obtained by collecting tens or hundreds of spectrum bands in an identical region of the Earth’s surface by an imaging spectrometer [
3,
4]. In an HSI, each pixel in the scene includes a sequential spectrum, which can be analyzed by its reflectance or emissivity to identify the type of material in each pixel [
5,
6]. Owing to the subtle differences among HSI spectra, HSIs have been applied in many fields. For instance, hydrological science [
7], ecological science [
8,
9], geological science [
10,
11], precision agriculture [
12,
13], and military applications [
14].
In recent decades, the classification of HSIs has become a popular field of research for the hyperspectral community. While the abundant spectral information is useful for improving classification accuracy compared to natural images, the high dimensionality presents new difficulties [
15,
16]. The HSI classification task has the following challenges: (1) HSI has high intra-class variability and inter-class diversity. These are influenced by many factors, such as changes in lighting, environment, atmosphere, and temporal conditions. (2) The available training samples are limited in relation to the high dimensionality of HSIs. As the dimension of HSIs increases, the required training samples also keep increasing, while the available samples of HSIs are limited. Therefore, these factors can result in an unsuitable methodology, reducing the classifier’s ability for generalization.
In early HSI classification studies, most approaches focused on the influence of HSI spectral features on classification results. Therefore, several existing methods are based on pixel-level HSI classification, for instance, multinomial logistic regression [
17], support vector machines (SVM) [
18,
19,
20], K-nearest neighbor (KNN) [
21], neural networks [
22], linear discriminative analysis [
23,
24,
25], and maximum likelihood methods [
26]. SVM is mainly dedicated to the transformation of linearly inseparable problems into linearly separable problems by finding the optimal hyperplane (such as the radial basis kernel and composite kernel [
19]), which finally completes the classification task. Since these methods utilize the spatial context information insufficiently, the classification results obtained by these pixel classifiers using only spectral features are unsatisfactory. Recently, researchers have found that spatial feature-based classification methods have significantly improved the representation of hyperspectral data and classification accuracy [
27,
28]. Thus, more researchers are combining spectral-spatial features into pixel classifiers to exploit the information of HSIs completely and improve the classification results. For example, multiple kernel learning uses various kernel functions to extract different features separately, which are fed into the classifier to generate a map of classification results. In addition, researchers in [
29,
30] segmented HSIs into multiple superpixels to obtain similar spatial pixels based on intensity or texture similarity. Although these methods have achieved sufficient performance, hand-crafted filters extract limited features, and most can only extract shallow features. The hand-crafted features depend on the expert’s experience in setting parameters, which limits the development and applicability of these methods. Therefore, for HSI classification, the extraction of deeper and more easily discernible features is the key.
In recent decades, deep learning [
31,
32,
33] has been extensively adopted in computer vision, for instance, in image classification [
34,
35,
36], object detection [
37,
38,
39,
40], natural language processing [
41], and has obtained remarkable performance in HSI classification. In contrast to traditional algorithms, deep learning extracts deep information from input data through a range of hierarchical structures. In detail, some simple line and shape features can be extracted at shallow layers, while deeper layers can extract abstract and complex features. The deep learning process is fully automatic without human intervention and can extract different feature types depending on the network; therefore, deep learning methods are suitable for handling various situations.
At present, there are various deep-learning-based approaches for HSI classification, including deep belief networks (DBNs) [
42], stacked auto-encoders (SAEs) [
43], recurrent neural networks (RNNs) [
44,
45], convolutional neural networks (CNNs) [
46,
47], residual networks [
48], and generative adversarial networks (GANs) [
49]. The SAEs consist of multiple auto-encoder (AE) units that use the output of one layer as input to subsequent layers. Li et al. [
50] used active learning techniques to enhance the parameter training of SAEs. Guo et al. [
51] reduce the dimensionality by fusing principal component analysis (PCA) and kernel PCA to optimize the standard training process of DBNs. Although these methods have adequate classification performance, the number of model parameters is large. In addition, the HSI cube data are vectorized, and the spatial structure can be corrupted, which leads to inaccurate classification.
The CNN can extract local two-dimensional (2-D) spatial features of images, and the weight-sharing mechanism of a CNN can effectively decrease the number of network parameters. Therefore, CNNs are widely used in HSI classification. Hu et al. [
52] proposed a deep CNN with five one-dimensional (1-D) layers, which receives pixel vectors as input data and classifies HSI data in the spectral domain only. However, this method loses spatial information, and the network depth is shallow, limiting the extraction of complex features. Zhao et al. [
53] proposed a CNN2D architecture, in which multi-scale, convolutional AEs based on Laplace pyramids obtain a series of deep spatial features, while the PCA extracts three principal components. Then, logistic regression is used as a classifier that connects the extracted spatial features and spectral information. However, the method does not consider spectral features and the classification effect on improvement. To extract the spatial–spectral information, Chen et al. [
54] proposed three convolutional models for creating input blocks of their CNN3D model using full-pixel vectors from the original HSI. This method extracts spectral, spatial, and spatial–spectral features, which generate data redundancy. In addition, Liu et al. [
55] proposed a bidirectional-convolutional long short-term memory (Bi-CLSTM) network with which the convolutional operators across spatial domains are combined into a bidirectional long short-term memory (Bi-LSTM) network to obtain spatial features while fully incorporating spectral contextual information.
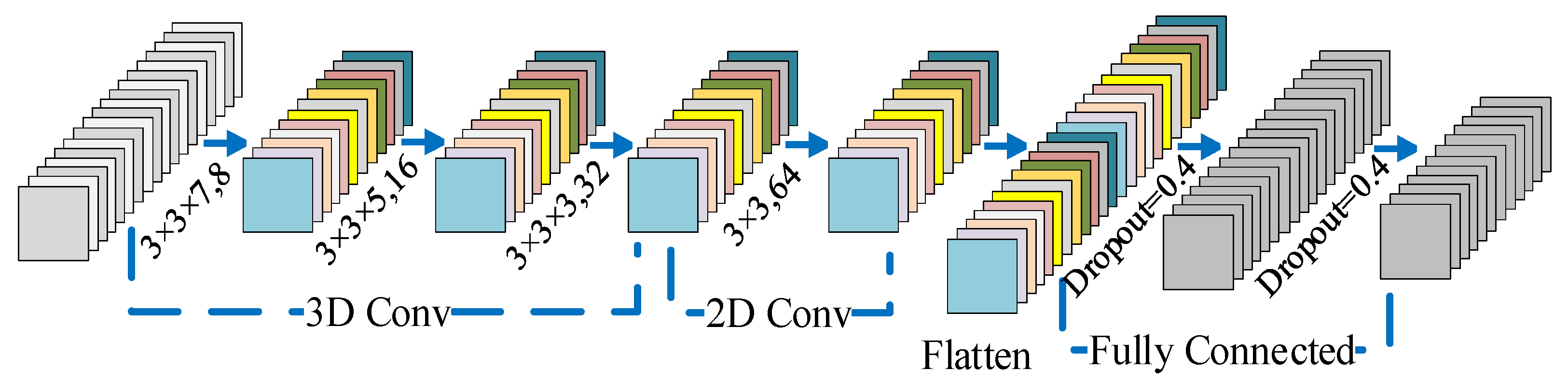
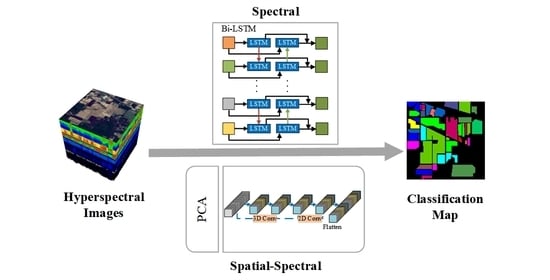
In summary, sufficiently exploiting features of HSI data and minimizing computational burden are the keys to HSI classification. This paper proposes a joint unified network operating in the spatial–spectral domain for the HSI classification. The network uses three layers of 3-D convolution for extracting the spatial–spectral feature of HSI, and subsequently adds a layer of 2-D convolution to further extract spatial features. For spectral feature extraction, this network treats all spectral bands as a sequence of images and enhances the interactions between spectral bands using Bi-LSTM. Finally, two fully connected (FC) layers are combined and use the softmax function for classification, which forms a unified neural network. We list the major contributions of our proposed method.
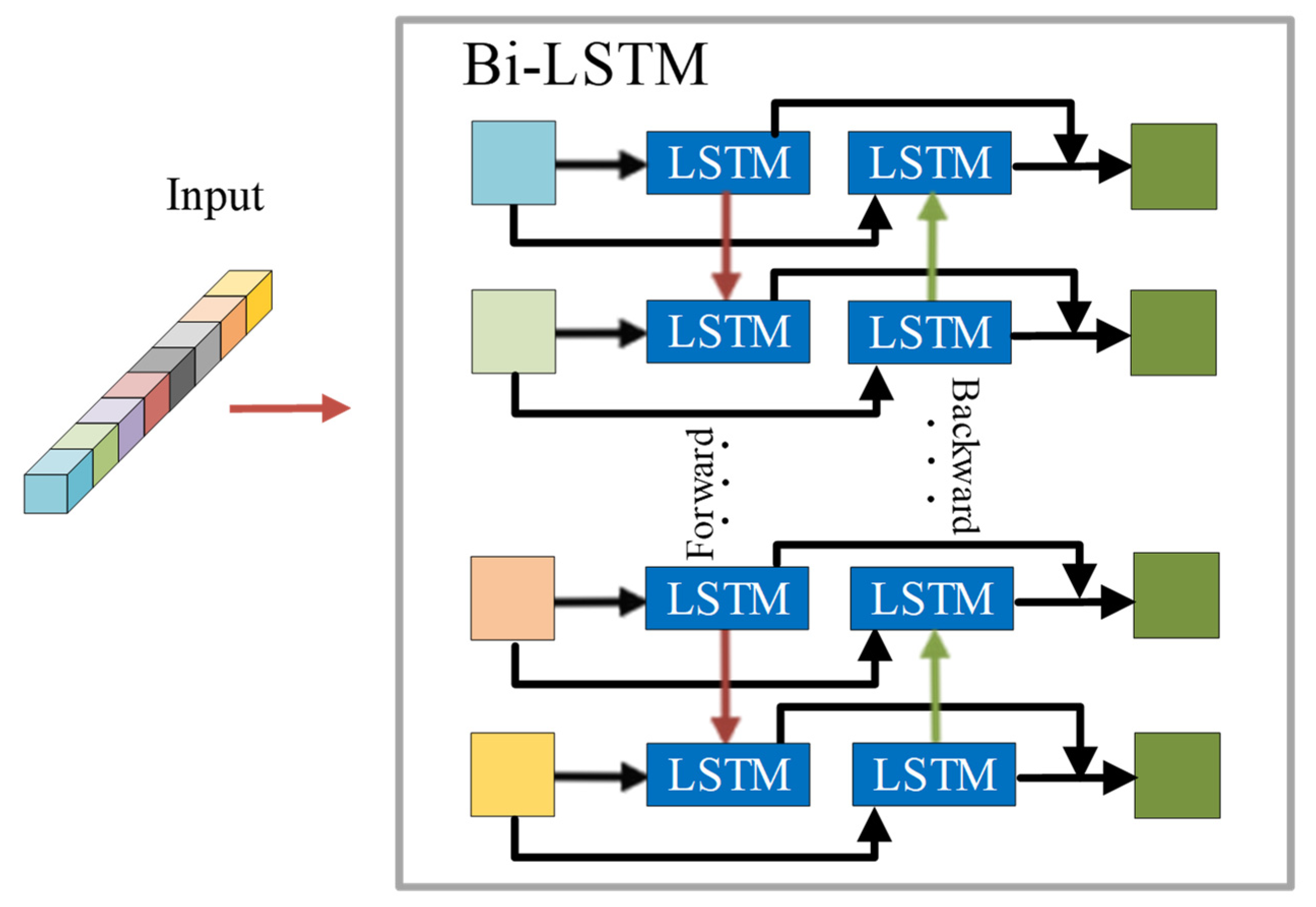
A Bi-LSTM framework based on band grouping is proposed for extracting spectral features. Bi-LSTM can obtain better performance in learning contextual features between adjacent spectral bands. In contrast to the general recurrent neural network, this framework can better adapt to a deeper network for HSI classification.
The proposed method adopts 3-D CNN for extracting the spatial–spectral features. To reduce the computational complexity of the whole framework, PCA is used before the convolutional layer of the 3-D CNN to reduce the data dimensionality.
A unified framework named the Bi-LSTM-CNN is proposed which integrates two subnetworks into a unified network by sharing the loss function. In addition, the framework adds the auxiliary loss function, which balances the effects of spectral and spatial-spectral features for the classification results to increase the classification accuracy.
The structure of the remaining part is as follows.
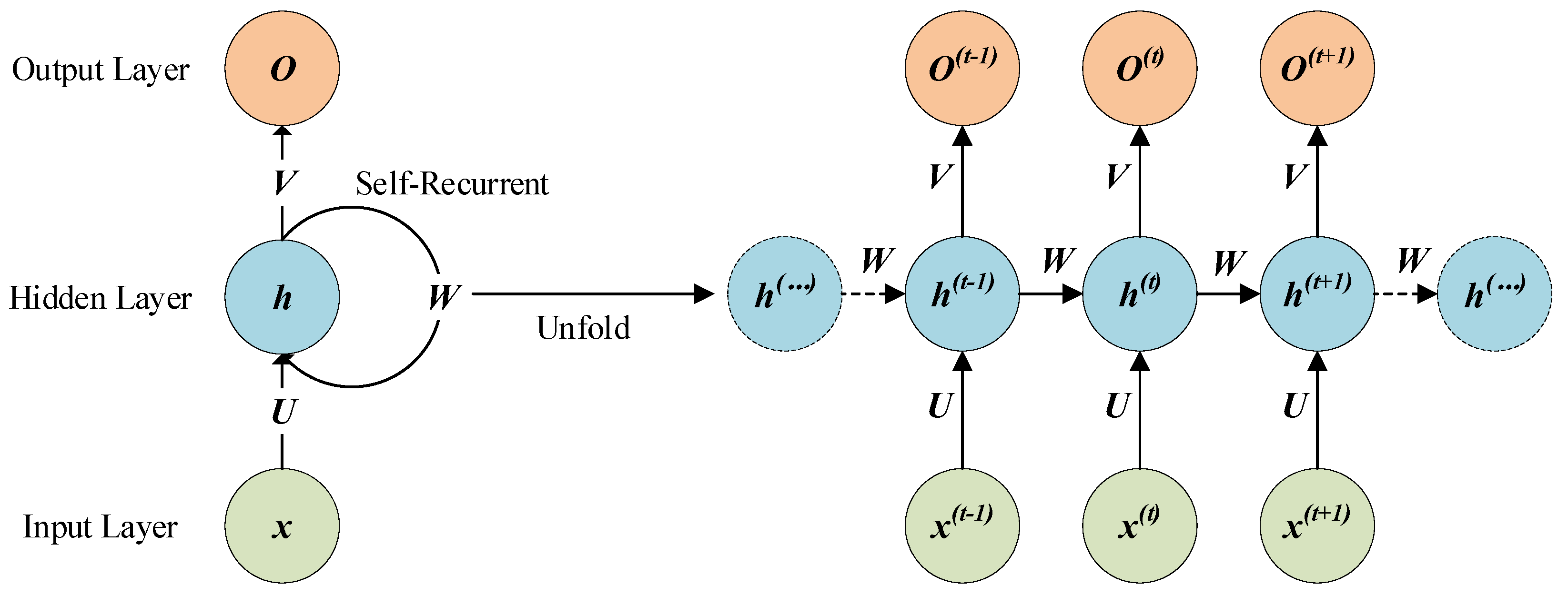
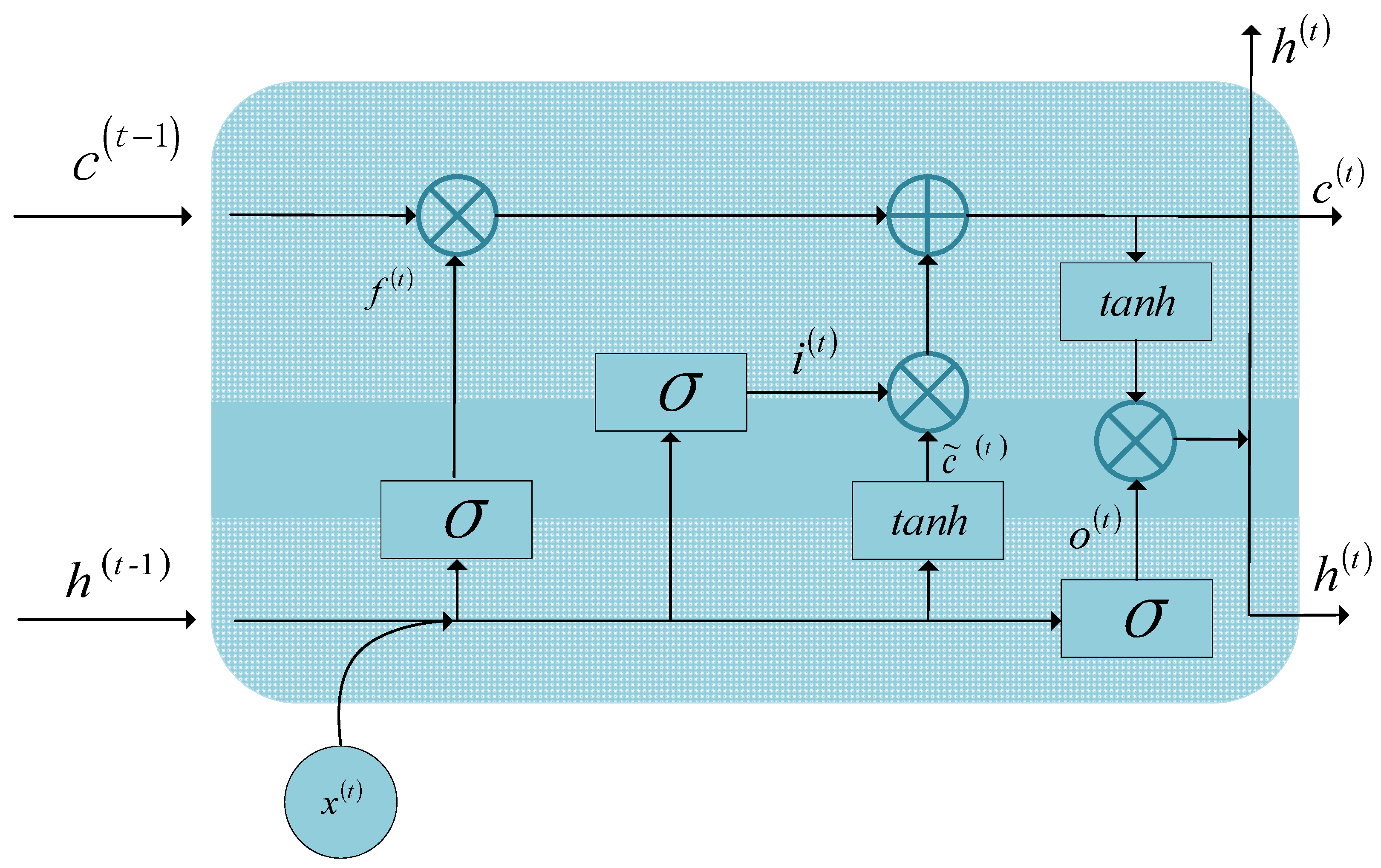
Section 2 describes long short-term memory (LSTM), a 3-D CNN, and the framework of the Bi-LSTM-CNN.
Section 3 introduces the HSI datasets, experimental configuration, and experimental results.
Section 4 provides a detailed analysis and interpretation of the experimental results. Finally, conclusions are summarized in
Section 5.
4. Discussion
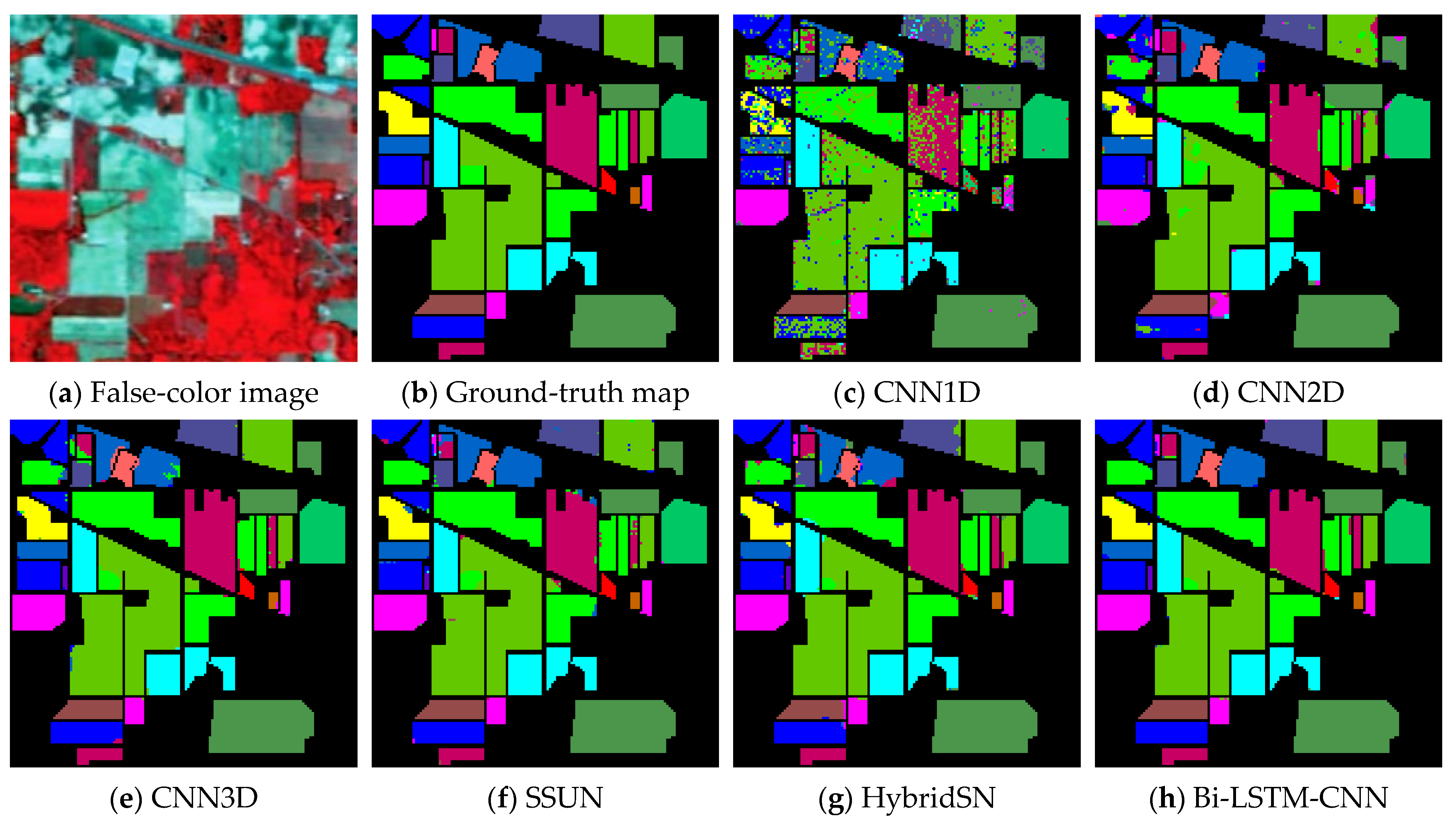
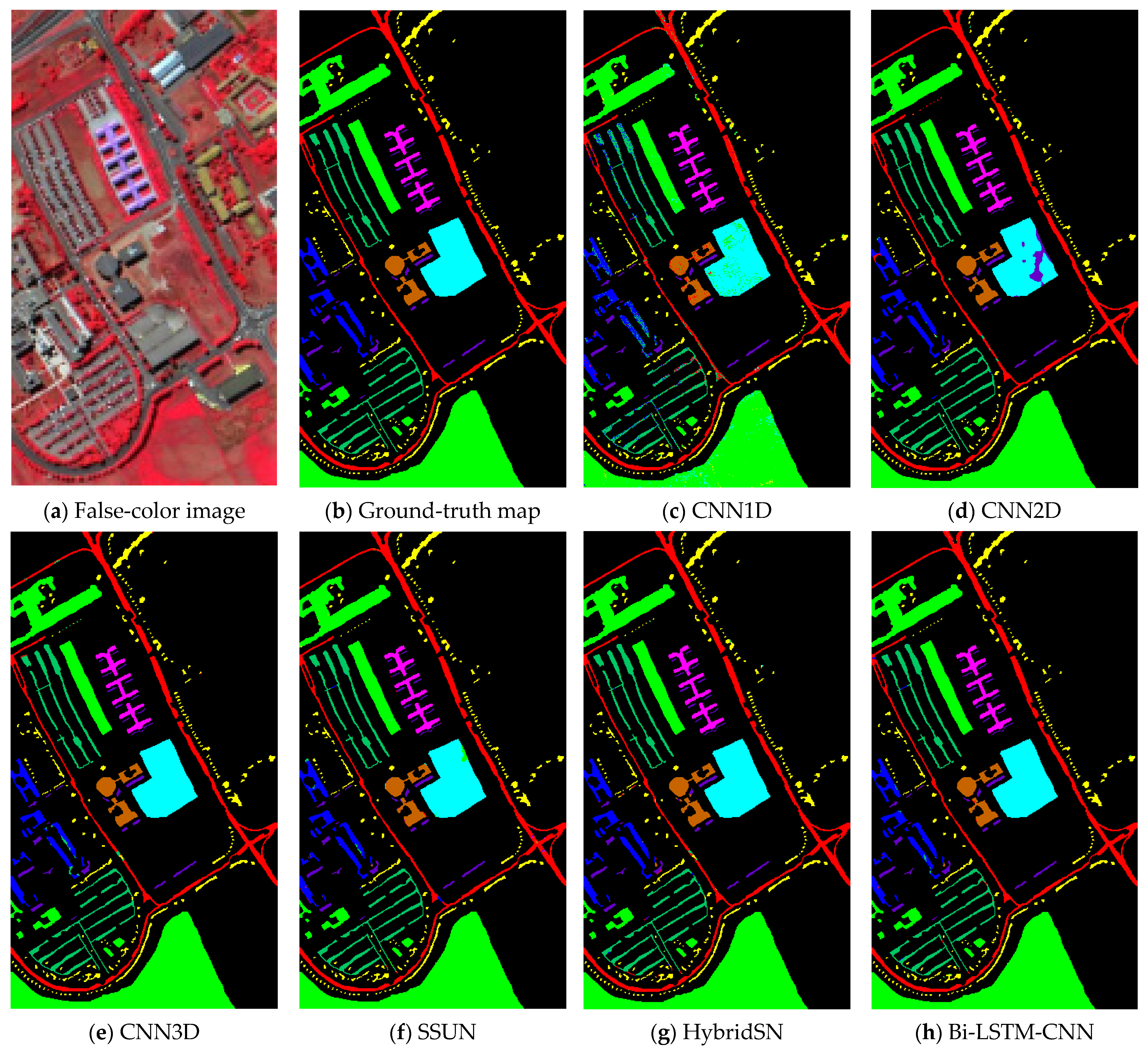
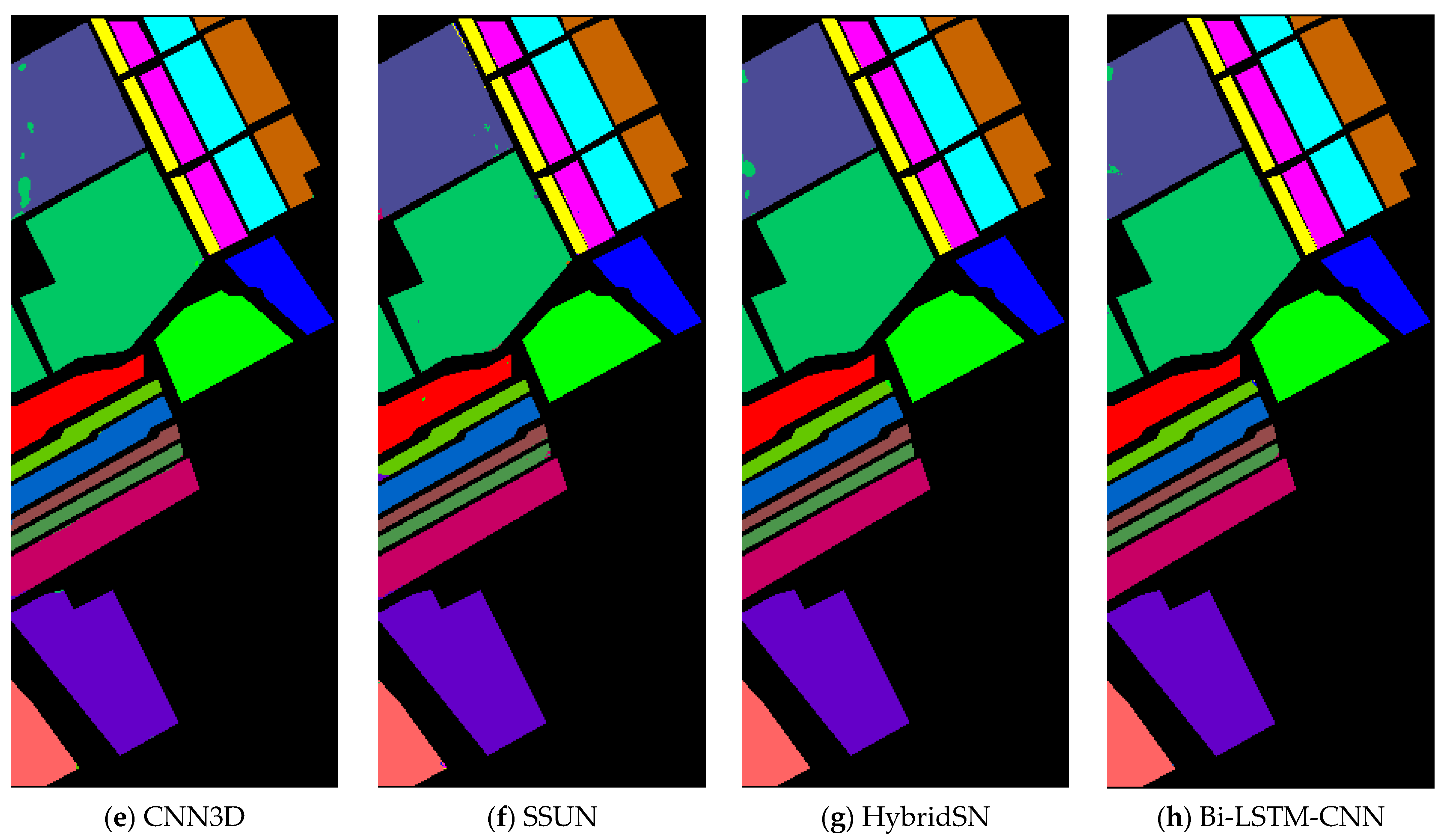
In the experiment result, it is obvious that the Bi-LSTM-CNN significantly outperforms the other methods. The OA of the CNN1D method did not exceed 94% in all the considered datasets. Since the input data of CNN1D is a 1-D vector, spatial information of the input data is lost, resulting in the worst classification results of CNN1D among all methods. The CNN2D model considers the spatial information, which makes the classification results an improvement compared to CNN1D. Thus, it shows that spatial information is critical for HSI classification.
However, the CNN2D model has problems, which usually result in degraded shapes of some objects and materials. The union of spatial and spectral information can address this issue, and the other methods (CNN3D, SSUN, HybridSN, and Bi-LSTM-CNN) all achieve more similar classification results to the corresponding ground-truth maps. The SSUN model extracts spatial and spectral features separately, which are integrated and then sent to the classifier for classification. As spatial features dominate the classification results, SSUN is unable to effectively balance the two features, thus resulting in a little contribution of spectral features to the classification results. The CNN3D model directly extracts the spatial-spectral features of the HSI, but to decline the computational complexity of the convolutional layers, the PCA dimensionality reduction is performed on the input data. Hence, a small amount of spectral information is lost. Despite this, CNN3D still spends a lot of time in the testing phase on the PU and SV dataset compared to HybridSN and Bi-LSTM-CNN.
The HybridSN model replaces the final 3-D convolutional layer with 2-D convolution, decreasing the number of parameters in the network while maintaining accuracy. However, in the PU dataset experiments, the OA of the HybridSN model is lower than the CNN3D model, and the generalizability of the HybridSN method is slightly worse. In the Bi-LSTM-CNN, the lack of 3-D CNN processing spectral information is compensated, and the experimental results after adding Bi-LSTM are significantly better than the other methods.
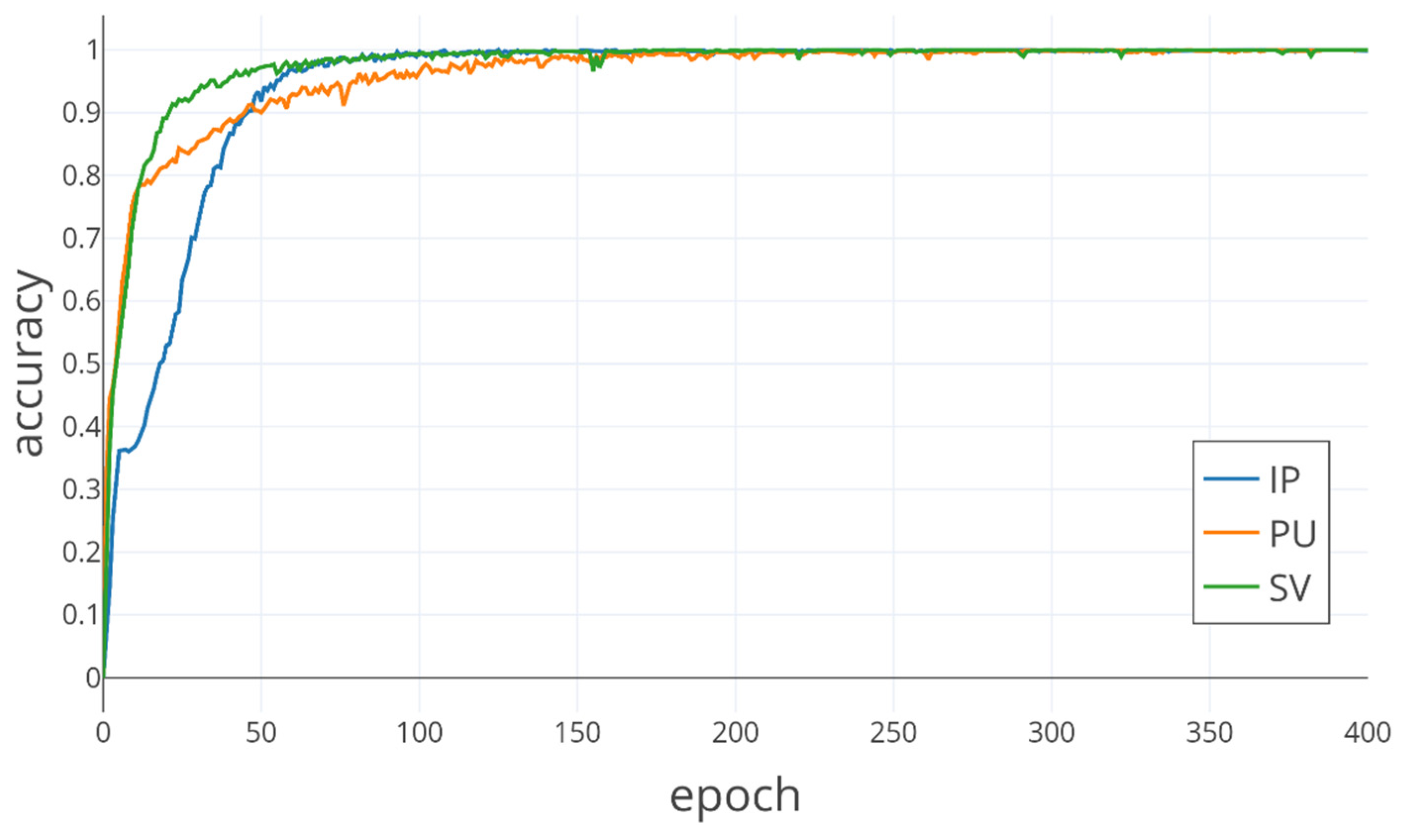
In the classes with a small number of samples, the Bi-LSTM-CNN method also obtains better classification results. In the IP dataset, due to the very small number of labeled samples in some classes, the number of available training samples is extremely small. For example, the number of samples for C1, C7, C9, C16 is not more than ten, which greatly increases the learning difficulty for these classes. Except for C1, the best classification accuracy is obtained for several other categories. Except for C1, the Bi-LSTM-CNN method obtains a higher OA in the other classes than other methods. In the PU and SV datasets, the number of training samples for each class is sufficient for the Bi-LSTM-CNN method, although there is a large difference in the number of samples for different classes.