Tree Species Classification of Forest Stands Using Multisource Remote Sensing Data
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Experimental Data
2.2.1. Sentinel-2 Data
2.2.2. Aerial Image and LiDAR Data
2.2.3. Field Data Collection
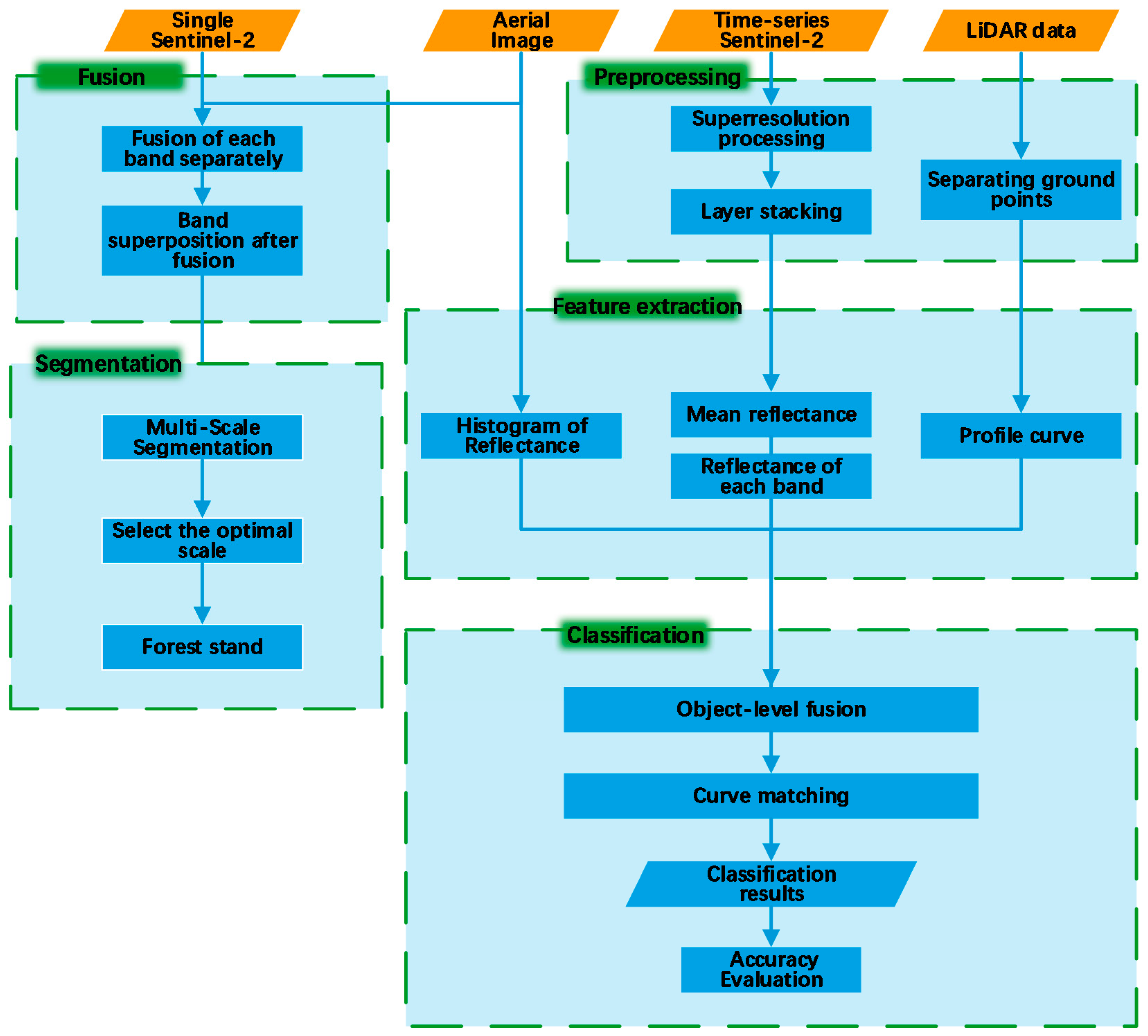
2.3. Methods
2.3.1. Data Preprocessing
2.3.2. Multisource Image Fusion
- (1)
- The spatial details were obtained by the difference between the band and its degraded version:where Mi is the ith band, and Mi,L is an upsampled image using the bicubical method to match the pixel size of the reflective band. The spatial details of the multiple reflective bands can be expressed as follows:where t is the coefficient.
- (2)
- A multivariate regression of a low-resolution image and multiple reflective bands was established.where ci, ai and b are coefficients; e is the residual; and Mlow is the low-spatial-resolution image. Given value t, the coefficients can be estimated using the least squares approach.
- (3)
- The low-spatial-resolution image was fused to the final image with a high spatial resolution by the following equation:
2.3.3. Forest Stand Segmentation
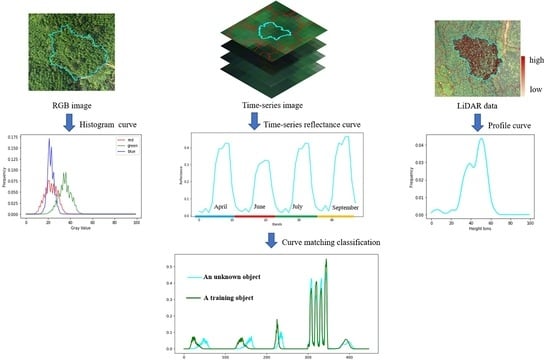
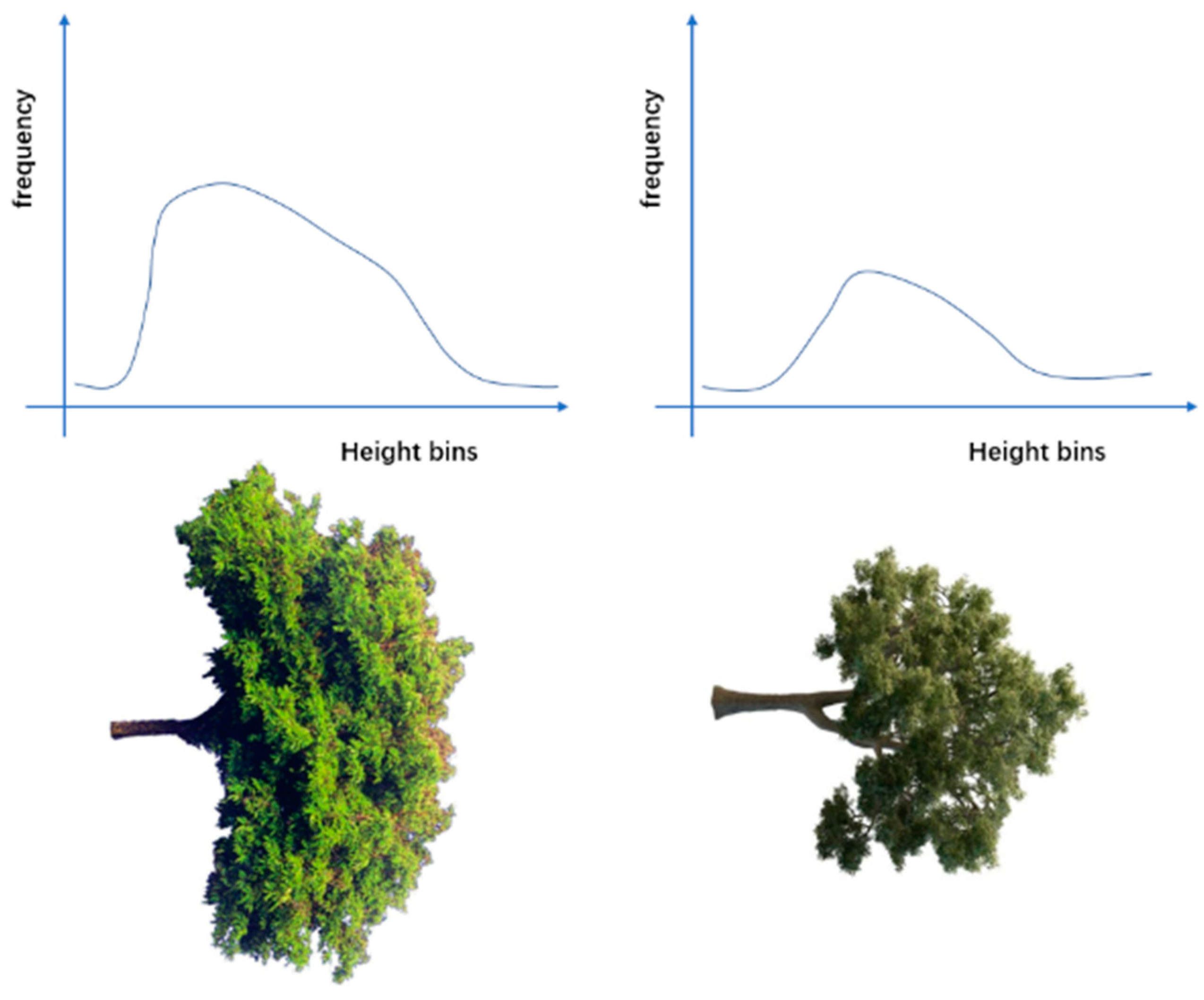
2.3.4. Feature Extraction
2.3.5. Classification
3. Results
3.1. The Results of Fusion and Segmentation
3.2. Feature Extraction Results
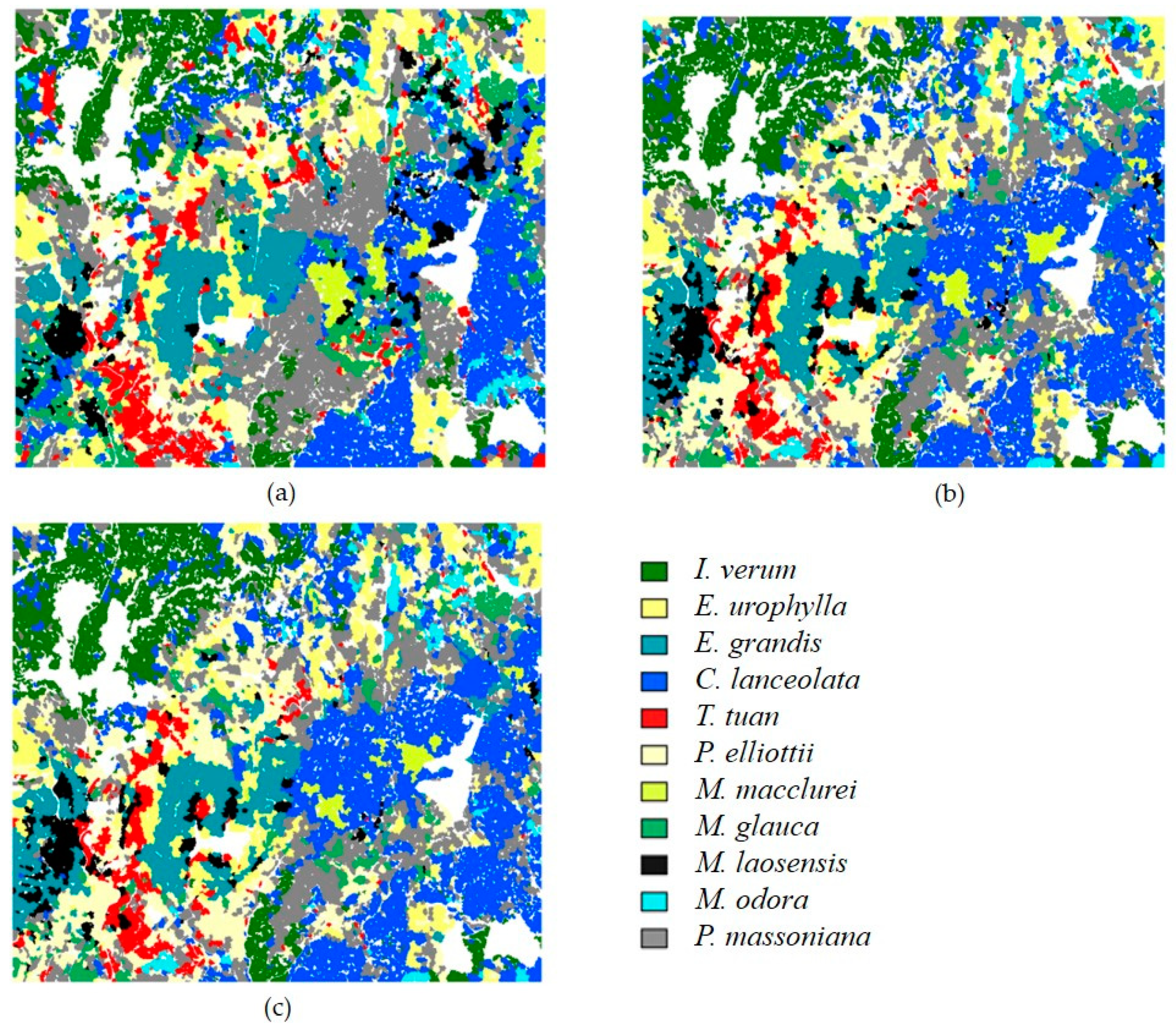
3.3. Classification Results of FSP
3.4. Comparison between Fusion Results of Different Types of Data
3.5. Comparison with Traditional Methods
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schiefer, F.; Kattenborn, T.; Frick, A.; Frey, J.; Schall, P.; Koch, B.; Schmidtlein, S. Mapping forest tree species in high resolution UAV-based RGB-imagery by means of convolutional neural networks. ISPRS J. Photogramm. Remote Sens. 2020, 170, 205–215. [Google Scholar] [CrossRef]
- Puumalainen, J.; Kennedy, P.; Folving, S. Monitoring forest biodiversity: A European perspective with reference to temperate and boreal forest zone. J. Env. Manag. 2003, 67, 5–14. [Google Scholar] [CrossRef]
- Ørka, H.O.; Dalponte, M.; Gobakken, T.; Næsset, E.; Ene, L.T. Characterizing forest species composition using multiple remote sensing data sources and inventory approaches. Scand. J. For. Res. 2013, 28, 677–688. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Tree species classification in the Southern Alps based on the fusion of very high geometrical resolution multispectral/hyperspectral images and LiDAR data. Remote Sens. Environ. 2012, 123, 258–270. [Google Scholar] [CrossRef]
- Li, W.K.; Guo, Q.H.; Jakubowski, M.K.; Kelly, M. A New Method for Segmenting Individual Trees from the Lidar Point Cloud. Photogramm. Eng. Remote Sens. 2012, 78, 75–84. [Google Scholar] [CrossRef]
- Yu, X.W.; Hyyppa, J.; Kaartinen, H.; Maltamo, M. Automatic detection of harvested trees and determination of forest growth using airborne laser scanning. Remote Sens. Environ. 2004, 90, 451–462. [Google Scholar] [CrossRef]
- Torabzadeh, H.; Leiterer, R.; Hueni, A.; Schaepman, M.E.; Morsdorf, F. Tree species classification in a temperate mixed forest using a combination of imaging spectroscopy and airborne laser scanning. Agric. For. Meteorol. 2019, 279, 107744. [Google Scholar] [CrossRef]
- Crabbe, R.A.; Lamb, D.; Edwards, C. Discrimination of species composition types of a grazed pasture landscape using Sentinel-1 and Sentinel-2 data. Int. J. Appl. Earth Obs. Geoinf. 2020, 84, 101978. [Google Scholar] [CrossRef]
- Dechesne, C.; Mallet, C.; Le Bris, A.; Gouet, V.; Hervieu, A. Forest Stand Segmentation Using Airborne Lidar Data and Very High Resolution Multispectral Imagery. ISPRS—Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, XLI-B3, 207–214. [Google Scholar] [CrossRef]
- Liu, J.; Wang, X.; Wang, T. Classification of tree species and stock volume estimation in ground forest images using Deep Learning. Comput. Electron. Agric. 2019, 166, 105012. [Google Scholar] [CrossRef]
- Shi, Y.F.; Skidmore, A.K.; Wang, T.J.; Holzwarth, S.; Heiden, U.; Pinnel, N.; Zhu, X.; Heurich, M. Tree species classification using plant functional traits from LiDAR and hyperspectral data. Int. J. Appl. Earth Obs. Geoinf. 2018, 73, 207–219. [Google Scholar] [CrossRef]
- Lin, Y.; Hyyppä, J. A comprehensive but efficient framework of proposing and validating feature parameters from airborne LiDAR data for tree species classification. Int. J. Appl. Earth Obs. Geoinf. 2016, 46, 45–55. [Google Scholar] [CrossRef]
- Immitzer, M.; Atzberger, C.; Koukal, T. Tree Species Classification with Random Forest Using Very High Spatial Resolution 8-Band WorldView-2 Satellite Data. Remote Sens. 2012, 4, 2661–2693. [Google Scholar] [CrossRef]
- Pu, R.; Landry, S. A comparative analysis of high spatial resolution IKONOS and WorldView-2 imagery for mapping urban tree species. Remote Sens. Environ. 2012, 124, 516–533. [Google Scholar] [CrossRef]
- Wang, K.; Akar, G. Gender gap generators for bike share ridership: Evidence from Citi Bike system in New York City. J. Transp. Geogr. 2019, 76, 1–9. [Google Scholar] [CrossRef]
- Heinzel, J.; Koch, B. Investigating multiple data sources for tree species classification in temperate forest and use for single tree delineation. Int. J. Appl. Earth Obs. Geoinf. 2012, 18, 101–110. [Google Scholar] [CrossRef]
- Clark, M.L.; Roberts, D.A. Species-Level Differences in Hyperspectral Metrics among Tropical Rainforest Trees as Determined by a Tree-Based Classifier. Remote Sens. 2012, 4, 1820–1855. [Google Scholar] [CrossRef]
- Pu, R.; Landry, S. Mapping urban tree species by integrating multi-seasonal high resolution pléiades satellite imagery with airborne LiDAR data. Urban For. Urban Green. 2020, 53, 126675. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E. Airborne spectranomics: Mapping canopy chemical and taxonomic diversity in tropical forests. Front. Ecol. Environ. 2009, 7, 269–276. [Google Scholar] [CrossRef]
- Nagendra, H. Using remote sensing to assess biodiversity. Int. J. Remote Sens. 2010, 22, 2377–2400. [Google Scholar] [CrossRef]
- Wolter, P.T.; Mladenoff, D.J.; Host, G.E.; Crow, T.R. Improved Forest stamped classification in the northern lake states using multi-temporal Landsat imagery. Photogramm. Eng. Remote Sens. 1995, 61, 1129–1143. [Google Scholar]
- Mead, R.A. LANDSAT Digital Data Application to Forest Vegetation and Land Use Classification in Minnesota; Purdue University: West Lafayette, IN, USA, 1977. [Google Scholar]
- Roller, N.E.G.; Visser, L. Accuracy of landsat forest cover type mapping. Cell Biol. Int. 1994, 18, 289–290. [Google Scholar]
- Johansen, K.; Phinn, S. Mapping structural parameters and species composition of riparian vegetation using IKONOS and landsat ETM plus data in Australian tropical savannahs. Photogramm. Eng. Remote Sens. 2006, 72, 71–80. [Google Scholar] [CrossRef]
- Mallinis, G.; Koutsias, N.; Tsakiri-Strati, M.; Karteris, M. Object-based classification using Quickbird imagery for delineating forest vegetation polygons in a Mediterranean test site. ISPRS J. Photogramm. Remote Sens. 2008, 63, 237–250. [Google Scholar] [CrossRef]
- Huang, W.; Li, H.; Lin, G. Classifying Forest Stands Based on Multi-Scale Structure Features Using Quickbird Image. In Proceedings of the 2015 2nd IEEE International Conference on Spatial Data Mining and Geographical Knowledge Services, Fuzhou, China, 8–10 July 2015; pp. 202–208. [Google Scholar]
- Karlson, M.; Ostwald, M.; Reese, H.; Bazié, H.R.; Tankoano, B. Assessing the potential of multi-seasonal WorldView-2 imagery for mapping West African agroforestry tree species. Int. J. Appl. Earth Obs. Geoinf. 2016, 50, 80–88. [Google Scholar] [CrossRef]
- Michez, A.; Piegay, H.; Lisein, J.; Claessens, H.; Lejeune, P. Classification of riparian forest species and health condition using multi-temporal and hyperspatial imagery from unmanned aerial system. Env. Monit. Assess. 2016, 188, 146. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.P.; Wagner, F.H.; Aragão, L.E.O.C.; Shimabukuro, Y.E.; de Souza Filho, C.R. Tree species classification in tropical forests using visible to shortwave infrared WorldView-3 images and texture analysis. ISPRS J. Photogramm. Remote Sens. 2019, 149, 119–131. [Google Scholar] [CrossRef]
- Yu, Q.; Gong, P.; Clinton, N.; Biging, G.; Kelly, M.; Schirokauer, D. Object-based Detailed Vegetation Classification with Airborne High Spatial Resolution Remote Sensing Imagery. Photogramm. Eng. Remote Sens. 2006, 72, 799–811. [Google Scholar] [CrossRef]
- Franklin, S.E.; Ahmed, O.S. Deciduous tree species classification using object-based analysis and machine learning with unmanned aerial vehicle multispectral data. Int. J. Remote Sens. 2017, 39, 5236–5245. [Google Scholar] [CrossRef]
- Zhou, Y.H.; Qiu, F. Fusion of high spatial resolution WorldView-2 imagery and LiDAR pseudo-waveform for object-based image analysis. ISPRS J. Photogramm. Remote Sens. 2015, 101, 221–232. [Google Scholar] [CrossRef]
- Tang, Y.; Qiu, F.; Jing, L.; Shi, F.; Li, X. Integrating spectral variability and spatial distribution for object-based image analysis using curve matching approaches. ISPRS J. Photogramm. Remote Sens. 2020, 169, 320–336. [Google Scholar] [CrossRef]
- Madonsela, S.; Cho, M.A.; Mathieu, R.; Mutanga, O.; Ramoelo, A.; Kaszta, Ż.; Kerchove, R.V.D.; Wolff, E. Multi-phenology WorldView-2 imagery improves remote sensing of savannah tree species. Int. J. Appl. Earth Obs. Geoinf. 2017, 58, 65–73. [Google Scholar] [CrossRef]
- Pu, R.; Landry, S.; Yu, Q. Assessing the potential of multi-seasonal high resolution Pléiades satellite imagery for mapping urban tree species. Int. J. Appl. Earth Obs. Geoinf. 2018, 71, 144–158. [Google Scholar] [CrossRef]
- Cochrane, M.A. Using vegetation reflectance variability for species level classification of hyperspectral data. Int. J. Remote Sens. 2010, 21, 2075–2087. [Google Scholar] [CrossRef]
- Ustin, S.L.; Roberts, D.A.; Gamon, J.A.; Asner, G.P.; Green, R.O. Using imaging spectroscopy to study ecosystem processes and properties. Bioscience 2004, 54, 523–534. [Google Scholar] [CrossRef]
- Fasnacht, L.; Renard, P.; Brunner, P. Robust input layer for neural networks for hyperspectral classification of data with missing bands. Appl. Comput. Geosci. 2020, 8, 100034. [Google Scholar] [CrossRef]
- Zhao, Q.; Jia, S.; Li, Y. Hyperspectral remote sensing image classification based on tighter random projection with minimal intra-class variance algorithm. Pattern Recognit. 2021, 111, 107635. [Google Scholar] [CrossRef]
- Li, W.; Du, Q.; Xiong, M. Kernel Collaborative Representation With Tikhonov Regularization for Hyperspectral Image Classification. IEEE Geosci. Remote Sens. Lett. 2014, 12, 48–52. [Google Scholar]
- Lixin, G.; Weixin, X.; Jihong, P. Segmented minimum noise fraction transformation for efficient feature extraction of hyperspectral images. Pattern Recognit. 2015, 48, 3216–3226. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Fusion of hyperspectral and LIDAR remote sensing data for classification of complex forest areas. IEEE Trans. Geosci. Remote Sens. 2008, 46, 1416–1427. [Google Scholar] [CrossRef]
- Jones, T.G.; Coops, N.C.; Sharma, T. Assessing the utility of airborne hyperspectral and LiDAR data for species distribution mapping in the coastal Pacific Northwest, Canada. Remote Sens. Environ. 2010, 114, 2841–2852. [Google Scholar] [CrossRef]
- Morsdorf, F.; Meier, E.; Kotz, B.; Itten, K.I.; Dobbertin, M.; Allgower, B. LIDAR-based geometric reconstruction of boreal type forest stands at single tree level for forest and wildland fire management. Remote Sens. Environ. 2004, 92, 353–362. [Google Scholar] [CrossRef]
- Vauhkonen, J.; Ørka, H.O.; Holmgren, J.; Dalponte, M.; Heinzel, J.; Koch, B. Tree Species Recognition Based on Airborne Laser Scanning and Complementary Data Sources. In Forestry Applications of Airborne Laser Scanning; Springer: Berlin/Heidelberg, Germany, 2014; pp. 135–156. [Google Scholar]
- Ke, Y.H.; Quackenbush, L.J.; Im, J. Synergistic use of QuickBird multispectral imagery and LIDAR data for object-based forest species classification. Remote Sens. Environ. 2010, 114, 1141–1154. [Google Scholar] [CrossRef]
- Nilsson, M.; Nordkvist, K.; Jonzén, J.; Lindgren, N.; Axensten, P.; Wallerman, J.; Egberth, M.; Larsson, S.; Nilsson, L.; Eriksson, J.; et al. A nationwide forest attribute map of Sweden predicted using airborne laser scanning data and field data from the National Forest Inventory. Remote Sens. Environ. 2017, 194, 447–454. [Google Scholar] [CrossRef]
- Popescu, S.C.; Wynne, R.H.; Nelson, R.F. Measuring individual tree crown diameter with lidar and assessing its influence on estimating forest volume and biomass. Can. J. Remote Sens. 2003, 29, 564–577. [Google Scholar] [CrossRef]
- Morsdorf, F.; Kotz, B.; Meier, E.; Itten, K.I.; Allgower, B. Estimation of LAI and fractional cover from small footprint airborne laser scanning data based on gap fraction. Remote Sens. Environ. 2006, 104, 50–61. [Google Scholar] [CrossRef]
- Piiroinen, R.; Fassnacht, F.E.; Heiskanen, J.; Maeda, E.; Mack, B.; Pellikka, P. Invasive tree species detection in the Eastern Arc Mountains biodiversity hotspot using one class classification. Remote Sens. Environ. 2018, 218, 119–131. [Google Scholar] [CrossRef]
- Karasiak, N.; Sheeren, D.; Fauvel, M.; Willm, J.; Monteil, C. Mapping tree species of forests in southwest France using Sentinel-2 image time series. In Proceedings of the 2017 9th International Workshop on the Analysis of Multitemporal Remote Sensing Images, Brugge, Belgium, 27–29 June 2017. [Google Scholar]
- Shi, Y.; Wang, T.; Skidmore, A.K.; Heurich, M. Important LiDAR metrics for discriminating forest tree species in Central Europe. ISPRS J. Photogramm. Remote Sens. 2018, 137, 163–174. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Sterenczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Plourde, L.C.; Ollinger, S.V.; Smith, M.L.; Martin, M.E. Martin. Estimating Species Abundance in a Northern Temperate Forest Using Spectral Mixture Analysis. Photogramm. Eng. Remote Sens. 2007, 73, 829–840. [Google Scholar] [CrossRef]
- Andrew, M.E.; Ustin, S.L. Habitat suitability modelling of an invasive plant with advanced remote sensing data. Divers. Distrib. 2009, 15, 627–640. [Google Scholar] [CrossRef]
- Lucas, R.M.; Lee, A.C.; Bunting, P.J. Retrieving forest biomass through integration of CASI and LiDAR data. Int. J. Remote Sens. 2008, 29, 1553–1577. [Google Scholar] [CrossRef]
- Asner, G.P.; Knapp, D.E.; Kennedy-Bowdoin, T.; Jones, M.O.; Martin, R.E.; Boardman, J.; Hughes, R.F. Invasive species detection in Hawaiian rainforests using airborne imaging spectroscopy and LiDAR. Remote Sens. Environ. 2008, 112, 1942–1955. [Google Scholar] [CrossRef]
- Hill, R.A.; Thomson, A.G. Mapping woodland species composition and structure using airborne spectral and LiDAR data. Int. J. Remote Sens. 2011, 26, 3763–3779. [Google Scholar] [CrossRef]
- Naidoo, L.; Cho, M.A.; Mathieu, R.; Asner, G. Classification of savanna tree species, in the Greater Kruger National Park region, by integrating hyperspectral and LiDAR data in a Random Forest data mining environment. ISPRS J. Photogramm. Remote Sens. 2012, 69, 167–179. [Google Scholar] [CrossRef]
- Hudak, A.T.; Evans, J.S.; Smith, A.M.S. LiDAR Utility for Natural Resource Managers. Remote Sens. 2009, 1, 934–951. [Google Scholar] [CrossRef]
- Machala, M.; Zejdová, L. Forest Mapping Through Object-based Image Analysis of Multispectral and LiDAR Aerial Data. Eur. J. Remote Sens. 2017, 47, 117–131. [Google Scholar] [CrossRef]
- Sridharan, H.; Qiu, F. Developing an Object-based Hyperspatial Image Classifier with a Case Study Using WorldView-2 Data. Photogramm. Eng. Remote Sens. 2013, 79, 1027–1036. [Google Scholar] [CrossRef]
- Blair, J.B.; Hofton, M.A. Modeling laser altimeter return waveforms over complex vegetation using high-resolution elevation data. Geophys. Res. Lett. 1999, 26, 2509–2512. [Google Scholar] [CrossRef]
- Lovell, J.L.; Jupp, D.L.B.; Culvenor, D.S.; Coops, N.C. Using airborne and ground-based ranging lidar to measure canopy structure in Australian forests. Can. J. Remote Sens. 2003, 29, 607–622. [Google Scholar] [CrossRef]
- Farid, A.; Goodrich, D.C.; Bryant, R.; Sorooshian, S. Using airborne lidar to predict Leaf Area Index in cottonwood trees and refine riparian water-use estimates. J. Arid Environ. 2008, 72, 1–15. [Google Scholar] [CrossRef]
- Muss, J.D.; Mladenoff, D.J.; Townsend, P.A. A pseudo-waveform technique to assess forest structure using discrete lidar data. Remote Sens. Environ. 2011, 115, 824–835. [Google Scholar] [CrossRef]
- Popescu, S.C.; Zhao, K.G.; Neuenschwander, A.; Lin, C.S. Satellite lidar vs. small footprint airborne lidar: Comparing the accuracy of aboveground biomass estimates and forest structure metrics at footprint level. Remote Sens. Environ. 2011, 115, 2786–2797. [Google Scholar] [CrossRef]
- Pang, Y.; Li, Z.Y.; Ju, H.B.; Lu, H.; Jia, W.; Si, L.; Guo, Y.; Liu, Q.W.; Li, S.M.; Liu, L.X.; et al. LiCHy: The CAF’s LiDAR, CCD and Hyperspectral Integrated Airborne Observation System. Remote Sens. 2016, 8, 398. [Google Scholar] [CrossRef]
- Brodu, N. Super-Resolving Multiresolution Images With Band-Independent Geometry of Multispectral Pixels. IEEE Trans. Geosci. Remote Sens. 2017, 55, 4610–4617. [Google Scholar] [CrossRef]
- Zhao, X.Q.; Guo, Q.H.; Su, Y.J.; Xue, B.L. Improved progressive TIN densification filtering algorithm for airborne LiDAR data in forested areas. ISPRS J. Photogramm. Remote Sens. 2016, 117, 79–91. [Google Scholar] [CrossRef]
- Jing, L.H.; Cheng, Q.M. A technique based on non-linear transform and multivariate analysis to merge thermal infrared data and higher-resolution multispectral data. Int. J. Remote Sens. 2010, 31, 6459–6471. [Google Scholar] [CrossRef]
- Schäpe, M.B.A. Multiresolution Segmentation: An optimization approach for high quality multi-scale image segmentation. In Beutrage zum AGIT-Symposium. Salzburg, Heidelberg; Wichmann: Lotte, Germany, 2000; pp. 12–23. [Google Scholar]
- Hamada, Y.; Stow, D.A.; Coulter, L.L.; Jafolla, J.C.; Hendricks, L.W. Detecting Tamarisk species (Tamarix spp.) in riparian habitats of Southern California using high spatial resolution hyperspectral imagery. Remote Sens. Environ. 2007, 109, 237–248. [Google Scholar] [CrossRef]
- Stow, D.A.; Toure, S.I.; Lippitt, C.D.; Lippitt, C.L.; Lee, C.R. Frequency distribution signatures and classification of within-object pixels. Int. J. Appl. Earth Obs. Geoinf. 2012, 15, 49–56. [Google Scholar] [CrossRef]
- Wessel, M.; Brandmeier, M.; Tiede, D. Evaluation of Different Machine Learning Algorithms for Scalable Classification of Tree Types and Tree Species Based on Sentinel-2 Data. Remote Sens. 2018, 10, 1419. [Google Scholar] [CrossRef]
- Diamand, M. The Solution to Overpopulation, The Depletion of Resources and Global Warming. J. Neurol. Neurosci. 2016, 7, 140. [Google Scholar] [CrossRef]













| CCD: DigiCAM-60 | LiDAR: Riegl LMS-Q680i | ||
|---|---|---|---|
| Frame size | 8956 × 6708 | Wavelength | 1550 nm |
| Pixel size | 6 µm | Laser beam divergence | 0.5 mrad |
| Imaging sensor size | 40.30 mm × 53.78 mm | Laser pulse length | 3 ns |
| Feld of view (FOV) | 56.2° | Cross-track FOV | ±30° |
| Ground resolution @1000 m altitude | 0.12 m | Vertical resolution | 0.15 m |
| Focal length | 50 mm | Point density @1000 m altitude | 3.6 pts/m2 |
| —— | —— | Waveform Sampling interval | 1 ns |
| —— | —— | Maximum scanning speed | 200 lines/s |
| —— | —— | Maximum laser pulse repetition rate | 400 kHz |
| Species | Illicium verum | Tilia tuan | Eucalyptus urophylla | Michelia odora |
| Abbreviation | I. verum | T. tuan | E. urophylla | M. odora |
| Species | Eucalyptus grandis | Pinus massoniana | Mytilaria laosensis | Cunninghamia lanceolata |
| Abbreviation | E. grandis | P. massoniana | M. laosensis | C. lanceolata |
| Species | Manglietia glauca | Michelia macclurei | Pinus elliottii | —— |
| Abbreviation | M. glauca | M. macclurei | P. elliottii | —— |
| FSP Method | KL-Based | CAM-Based | RSSDA-Based | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Class | UA | PA | F1-Score | UA | PA | F1-Score | UA | PA | F1-Score |
| I. verum | 1 | 0.933 | 0.966 | 1 | 0.867 | 0.929 | 0.938 | 1 | 0.968 |
| T. tuan | 1 | 1 | 1 | 1 | 0.941 | 0.970 | 1 | 0.824 | 0.903 |
| E. urophylla | 0.818 | 0.692 | 0.750 | 0.800 | 0.615 | 0.696 | 0.909 | 0.769 | 0.833 |
| M. odora | 1 | 1 | 1 | 1 | 0.800 | 0.889 | 1.000 | 1 | 1 |
| M. glauca | 1 | 0.933 | 0.966 | 1 | 0.933 | 0.966 | 0.875 | 0.933 | 0.903 |
| M. macclurei | 1 | 1 | 1 | 0.889 | 0.889 | 0.889 | 1 | 1 | 1 |
| E. grandis | 0.885 | 0.920 | 0.902 | 0.800 | 0.960 | 0.873 | 0.958 | 0.920 | 0.939 |
| P. massoniana | 0.980 | 0.877 | 0.926 | 0.942 | 0.860 | 0.899 | 0.943 | 0.877 | 0.909 |
| M. laosensis | 0.905 | 1 | 0.950 | 0.900 | 0.947 | 0.923 | 1 | 0.947 | 0.973 |
| C. lanceolata | 0.86 | 1 | 0.993 | 0.932 | 0.971 | 0.951 | 0.920 | 0.986 | 0.952 |
| P. elliottii | 0.562 | 0.900 | 0.692 | 0.571 | 0.800 | 0.667 | 0.643 | 0.900 | 0.750 |
| Overall accuracy: 0.937 Kappa coefficient: 0.926 | Overall accuracy: 0.902 Kappa coefficient: 0.884 | Overall accuracy: 0.925 Kappa coefficient: 0.911 | |||||||
| UA: user accuracy; PA: product accuracy | |||||||||
| Aerial Alone | Fusion of Aerial Image and Time-Series Images | Fusion of Aerial Image, Time-Series Images, and LiDAR Data | |||||||
|---|---|---|---|---|---|---|---|---|---|
| AVG | SD | MAX | AVG | SD | MAX | AVG | SD | MAX | |
| KL | 0.795 | 0.023 | 0.835 | 0.805 | 0.021 | 0.839 | 0.911 | 0.017 | 0.937 |
| CAM | 0.788 | 0.016 | 0.808 | 0.788 | 0.017 | 0.812 | 0.900 | 0.014 | 0.925 |
| RSSDA | 0.794 | 0.019 | 0.820 | 0.797 | 0.017 | 0.824 | 0.913 | 0.017 | 0.945 |
| Algorithm | AVG | SD | MAX | |
|---|---|---|---|---|
| Traditional methods | SVM | 0.814 | 0.025 | 0.855 |
| RF | 0.824 | 0.034 | 0.875 | |
| XGBoost | 0.817 | 0.025 | 0.855 | |
| FSP | KL | 0.911 | 0.017 | 0.937 |
| CAM | 0.900 | 0.014 | 0.925 | |
| RSSDA | 0.913 | 0.017 | 0.945 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wan, H.; Tang, Y.; Jing, L.; Li, H.; Qiu, F.; Wu, W. Tree Species Classification of Forest Stands Using Multisource Remote Sensing Data. Remote Sens. 2021, 13, 144. https://doi.org/10.3390/rs13010144
Wan H, Tang Y, Jing L, Li H, Qiu F, Wu W. Tree Species Classification of Forest Stands Using Multisource Remote Sensing Data. Remote Sensing. 2021; 13(1):144. https://doi.org/10.3390/rs13010144
Chicago/Turabian StyleWan, Haoming, Yunwei Tang, Linhai Jing, Hui Li, Fang Qiu, and Wenjin Wu. 2021. "Tree Species Classification of Forest Stands Using Multisource Remote Sensing Data" Remote Sensing 13, no. 1: 144. https://doi.org/10.3390/rs13010144
APA StyleWan, H., Tang, Y., Jing, L., Li, H., Qiu, F., & Wu, W. (2021). Tree Species Classification of Forest Stands Using Multisource Remote Sensing Data. Remote Sensing, 13(1), 144. https://doi.org/10.3390/rs13010144