Remote Sensing-Based Mapping of Senescent Leaf C:N Ratio in the Sundarbans Reserved Forest Using Machine Learning Techniques
Abstract
1. Introduction
2. Materials and Method
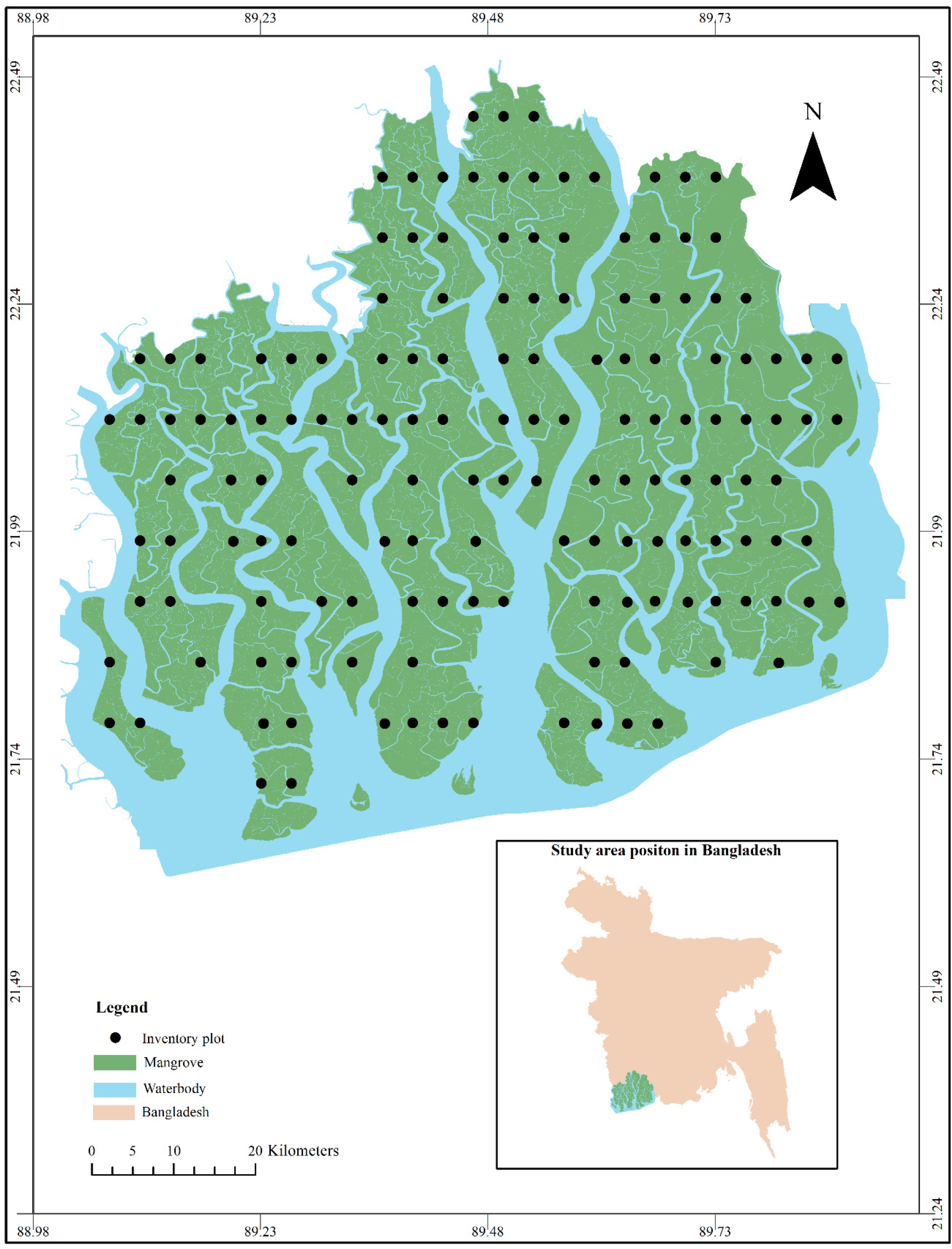
2.1. Description of Study Area
2.2. Field Data
2.3. Remote Sensing Data
3. Modeling Approaches
3.1. Stochastic Gradient Boosting (SGB)
3.2. Random Forest (RF)
3.3. Support Vector Machine (SVM)
3.4. Partial Least Square Regression (PLSR)
3.5. Model Development
3.6. Model Performance
4. Results
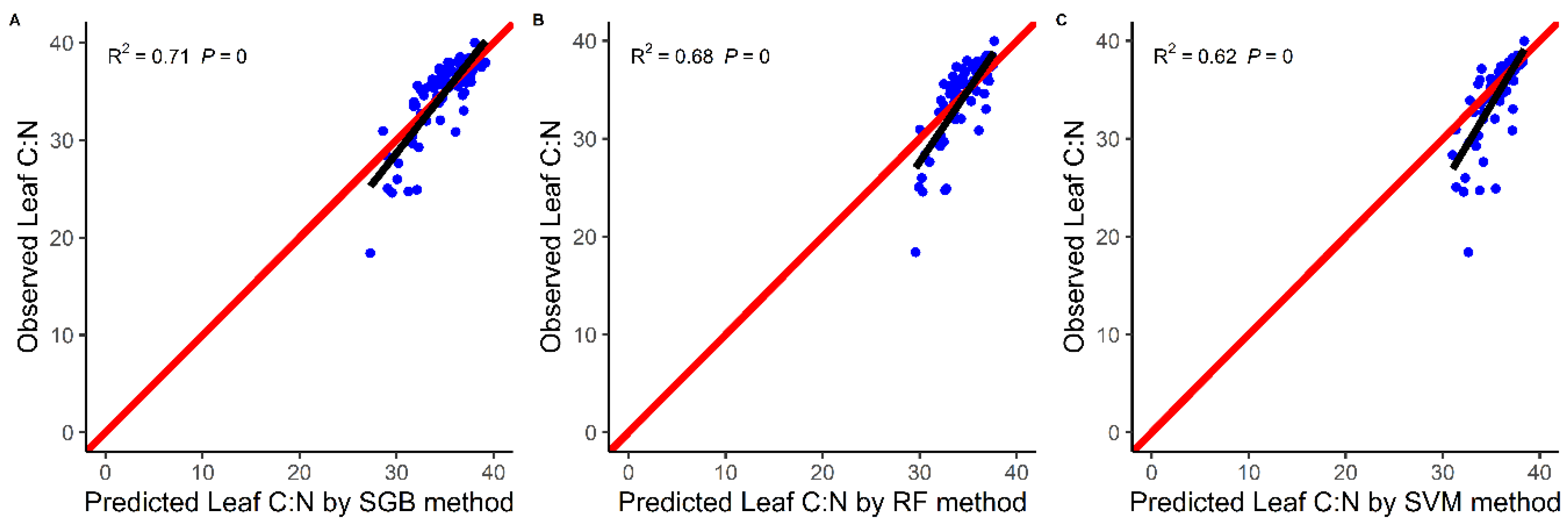
4.1. Models Comparison
4.2. Importance of Predictors
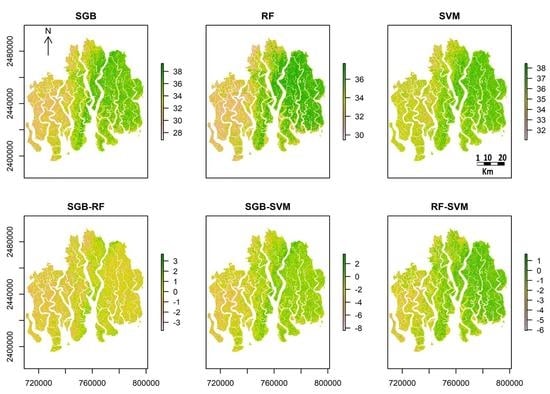
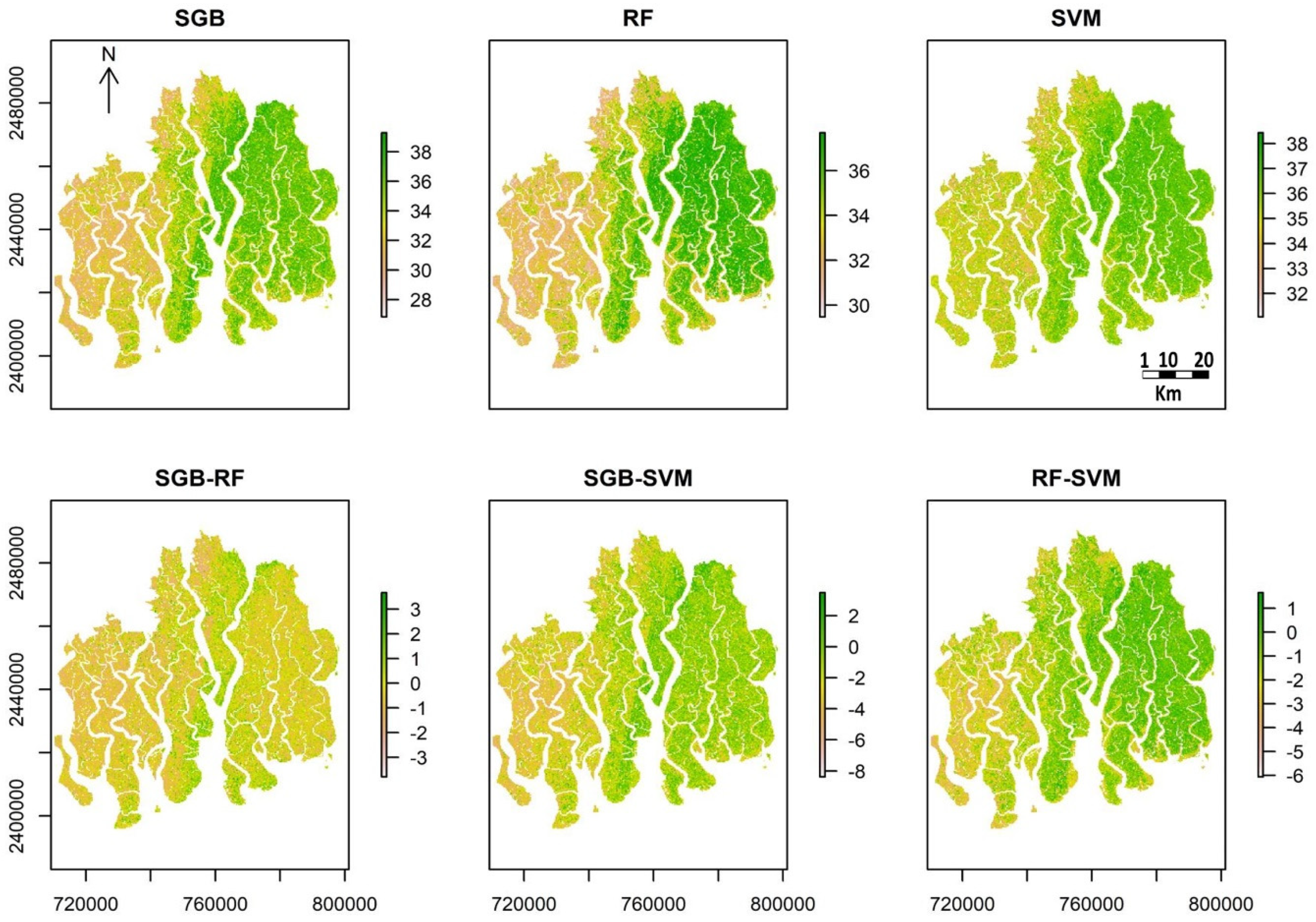
4.3. Spatial and Temporal Distribution of C:N
5. Discussion
5.1. Comparison of Model Performance
5.2. Spatial Distribution and Temporal Change of C:N ratio in SRF
5.3. Implication for Forest Health Monitoring and Policy
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sanderman, J.; Hengl, T.; Fiske, G.; Solvik, K.; Adame, M.F.; Benson, L.; Bukoski, J.J.; Carnell, P.; Cifuentes-Jara, M.; Donato, D.; et al. A global map of mangrove forest soil carbon at 30 m spatial resolution. Environ. Res. Lett. 2018, 13, 55002. [Google Scholar] [CrossRef]
- Donato, D.C.; Kauffman, J.B.; Murdiyarso, D.; Kurnianto, S.; Stidham, M.; Kanninen, M. Mangroves among the most carbon-rich forests in the tropics. Nat. Geosci. 2011, 4, 293–297. [Google Scholar] [CrossRef]
- Alongi, D. Impact of Global Change on Nutrient Dynamics in Mangrove Forests. Forests 2018, 9, 596. [Google Scholar] [CrossRef]
- Alongi, D.M. Carbon sequestration in mangrove forests. Carbon Manag. 2012, 3, 313–322. [Google Scholar] [CrossRef]
- De Deyn, G.B.; Cornelissen, J.H.C.; Bardgett, R.D. Plant functional traits and soil carbon sequestration in contrasting biomes. Ecol. Lett. 2008, 11, 516–531. [Google Scholar] [CrossRef]
- Adame, M.F.; Fry, B. Source and stability of soil carbon in mangrove and freshwater wetlands of the Mexican Pacific coast. Wetl. Ecol. Manag. 2016, 24, 129–137. [Google Scholar] [CrossRef]
- Hossain, M.; Siddique, M.R.H.; Abdullah, S.M.R.; Saha, S.; Ghosh, D.C.; Rahman, M.S.; Limon, S.H. Nutrient Dynamics Associated with Leaching and Microbial Decomposition of Four Abundant Mangrove Species Leaf Litter of the Sundarbans, Bangladesh. Wetlands 2014, 34, 439–448. [Google Scholar] [CrossRef]
- Kristensen, E.; Bouillon, S.; Dittmar, T.; Marchand, C. Organic carbon dynamics in mangrove ecosystems: A review. Aquat. Bot. 2008, 89, 201–219. [Google Scholar] [CrossRef]
- Strickland, M.S.; Osburn, E.; Lauber, C.; Fierer, N.; Bradford, M.A. Litter quality is in the eye of the beholder: Initial decomposition rates as a function of inoculum characteristics. Funct. Ecol. 2009, 23, 627–636. [Google Scholar] [CrossRef]
- Desie, E.; Vancampenhout, K.; Nyssen, B.; van den Berg, L.; Weijters, M.; van Duinen, G.J.; den Ouden, J.; Van Meerbeek, K.; Muys, B. Litter quality and the law of the most limiting: Opportunities for restoring nutrient cycles in acidified forest soils. Sci. Total Environ. 2020, 699, 134383. [Google Scholar] [CrossRef]
- Chen, R.; Twilley, R.R. Patterns of mangrove forest structure and soil nutrient dynamics along the Shark River Estuary, Florida. Estuaries Coasts 1999, 22, 955–970. [Google Scholar] [CrossRef]
- Hättenschwiler, S.; Coq, S.; Barantal, S.; Handa, I.T. Leaf traits and decomposition in tropical rainforests: Revisiting some commonly held views and towards a new hypothesis. New Phytol. 2011, 189, 950–965. [Google Scholar] [CrossRef] [PubMed]
- Chanda, A.; Akhand, A.; Manna, S.; Das, S.; Mukhopadhyay, A.; Das, I.; Hazra, S.; Choudhury, S.B.; Rao, K.H.; Dadhwal, V.K. Mangrove associates versus true mangroves: A comparative analysis of leaf litter decomposition in Sundarban. Wetl. Ecol. Manag. 2016, 24, 293–315. [Google Scholar] [CrossRef]
- Aber, J.D.; Melillo, J.M.; McClaugherty, C.A. Predicting long-term patterns of mass loss, nitrogen dynamics, and soil organic matter formation from initial fine litter chemistry in temperate forest ecosystems. Can. J. Bot. 1990, 68, 2201–2208. [Google Scholar] [CrossRef]
- Jacob, M.; Viedenz, K.; Polle, A.; Thomas, F.M. Leaf litter decomposition in temperate deciduous forest stands with a decreasing fraction of beech (Fagus sylvatica). Oecologia 2010, 164, 1083–1094. [Google Scholar] [CrossRef] [PubMed]
- Mizanur Rahman, M.; Nabiul Islam Khan, M.; Fazlul Hoque, A.K.; Ahmed, I. Carbon stock in the Sundarbans mangrove forest: Spatial variations in vegetation types and salinity zones. Wetl. Ecol. Manag. 2015, 23, 269–283. [Google Scholar] [CrossRef]
- Wahid, S.M.; Babel, M.S.; Bhuiyan, A.R. Hydrologic monitoring and analysis in the Sundarbans mangrove ecosystem, Bangladesh. J. Hydrol. 2007, 332, 381–395. [Google Scholar] [CrossRef]
- Iftekhar, M.S.; Saenger, P. Vegetation dynamics in the Bangladesh Sundarbans mangroves: A review of forest inventories. Wetl. Ecol. Manag. 2008, 16, 291–312. [Google Scholar] [CrossRef]
- Rahman, M.M.; Rahaman, M.M. Impacts of Farakka barrage on hydrological flow of Ganges river and environment in Bangladesh. Sustain. Water Resour. Manag. 2017, 1–14. [Google Scholar] [CrossRef]
- Richards, D.R.; Friess, D.A. Rates and drivers of mangrove deforestation in Southeast Asia, 2000–2012. Proc. Natl. Acad. Sci. USA 2016, 113, 344–349. [Google Scholar] [CrossRef]
- Gilman, E.L.; Ellison, J.; Duke, N.C.; Field, C. Threats to mangroves from climate change and adaptation options: A review. Aquat. Bot. 2008, 89, 237–250. [Google Scholar] [CrossRef]
- Rahman, M.M.; Lagomasino, D.; Lee, S.; Fatoyinbo, T.; Ahmed, I.; Kanzaki, M. Improved assessment of mangrove forests in Sundarbans East Wildlife Sanctuary using WorldView 2 and Tan DEM-X high resolution imagery. Remote Sens. Ecol. Conserv. 2019, 5, 136–149. [Google Scholar] [CrossRef]
- Moreno-Martínez, Á.; Camps-Valls, G.; Kattge, J.; Robinson, N.; Reichstein, M.; van Bodegom, P.; Kramer, K.; Cornelissen, J.H.C.; Reich, P.; Bahn, M.; et al. A methodology to derive global maps of leaf traits using remote sensing and climate data. Remote Sens. Environ. 2018, 218, 69–88. [Google Scholar] [CrossRef]
- Díaz, S.; Lavorel, S.; De Bello, F.; Quétier, F.; Grigulis, K.; Robson, T.M. Incorporating plant functional diversity effects in ecosystem service assessments. Proc. Natl. Acad. Sci. USA 2007, 104, 20684–20689. [Google Scholar] [CrossRef]
- Finegan, B.; Peña-Claros, M.; de Oliveira, A.; Ascarrunz, N.; Bret-Harte, M.S.; Carreño-Rocabado, G.; Casanoves, F.; Díaz, S.; Eguiguren Velepucha, P.; Fernandez, F.; et al. Does functional trait diversity predict above-ground biomass and productivity of tropical forests? Testing three alternative hypotheses. J. Ecol. 2015, 103, 191–201. [Google Scholar] [CrossRef]
- Tilman, D.; Isbell, F.; Cowles, J.M. Biodiversity and Ecosystem Functioning. Annu. Rev. Ecol. Evol. Syst. 2014, 45, 471–493. [Google Scholar] [CrossRef]
- Isbell, F.; Craven, D.; Connolly, J.; Loreau, M.; Schmid, B.; Beierkuhnlein, C.; Bezemer, T.M.; Bonin, C.; Bruelheide, H.; de Luca, E.; et al. Biodiversity increases the resistance of ecosystem productivity to climate extremes. Nature 2015, 526, 574–577. [Google Scholar] [CrossRef]
- Conti, G.; Díaz, S. Plant functional diversity and carbon storage—An empirical test in semi-arid forest ecosystems. J. Ecol. 2013, 101, 18–28. [Google Scholar] [CrossRef]
- Ali, A.; Yan, E.-R.; Chang, S.X.; Cheng, J.-Y.; Liu, X.-Y. Community-weighted mean of leaf traits and divergence of wood traits predict aboveground biomass in secondary subtropical forests. Sci. Total Environ. 2017, 574, 654–662. [Google Scholar] [CrossRef]
- Kröber, W.; Li, Y.; Härdtle, W.; Ma, K.; Schmid, B.; Schmidt, K.; Scholten, T.; Seidler, G.; von Oheimb, G.; Welk, E.; et al. Early subtropical forest growth is driven by community mean trait values and functional diversity rather than the abiotic environment. Ecol. Evol. 2015, 5, 3541–3556. [Google Scholar] [CrossRef]
- Dias, A.T.C.; Berg, M.P.; de Bello, F.; Van Oosten, A.R.; Bílá, K.; Moretti, M. An experimental framework to identify community functional components driving ecosystem processes and services delivery. J. Ecol. 2013, 101, 29–37. [Google Scholar] [CrossRef]
- Cong, W.-F.; van Ruijven, J.; Mommer, L.; De Deyn, G.B.; Berendse, F.; Hoffland, E. Plant species richness promotes soil carbon and nitrogen stocks in grasslands without legumes. J. Ecol. 2014, 102, 1163–1170. [Google Scholar] [CrossRef]
- Abelleira Martínez, O.J.; Fremier, A.K.; Günter, S.; Ramos Bendaña, Z.; Vierling, L.; Galbraith, S.M.; Bosque-Pérez, N.A.; Ordoñez, J.C. Scaling up functional traits for ecosystem services with remote sensing: Concepts and methods. Ecol. Evol. 2016, 6, 4359–4371. [Google Scholar] [CrossRef] [PubMed]
- Wallis, C.I.B.; Homeier, J.; Peña, J.; Brandl, R.; Farwig, N.; Bendix, J. Modeling tropical montane forest biomass, productivity and canopy traits with multispectral remote sensing data. Remote Sens. Environ. 2019, 225, 77–92. [Google Scholar] [CrossRef]
- Homolová, L.; Malenovský, Z.; Clevers, J.G.P.W.; García-Santos, G.; Schaepman, M.E. Review of optical-based remote sensing for plant trait mapping. Ecol. Complex. 2013, 15, 1–16. [Google Scholar] [CrossRef]
- Enquist, B.J.; Norberg, J.; Bonser, S.P.; Violle, C.; Webb, C.T.; Henderson, A.; Sloat, L.L.; Savage, V.M. Scaling from Traits to Ecosystems: Developing a General Trait Driver Theory via Integrating Trait-Based and Metabolic Scaling Theories. In Advances in Ecological Research; Academic Press Inc.: Cambridge, MA, USA, 2015; Volume 52, pp. 249–318. [Google Scholar]
- Chemura, A.; Mutanga, O.; Odindi, J.; Kutywayo, D. Mapping spatial variability of foliar nitrogen in coffee (Coffea arabica L.) plantations with multispectral Sentinel-2 MSI data. ISPRS J. Photogramm. Remote Sens. 2018, 138, 1–11. [Google Scholar] [CrossRef]
- Gara, T.W.; Darvishzadeh, R.; Skidmore, A.K.; Wang, T.; Heurich, M. Accurate modelling of canopy traits from seasonal Sentinel-2 imagery based on the vertical distribution of leaf traits. ISPRS J. Photogramm. Remote Sens. 2019, 157, 108–123. [Google Scholar] [CrossRef]
- Ollinger, S.V.; Richardson, A.D.; Martin, M.E.; Hollinger, D.Y.; Frolking, S.E.; Reich, P.B.; Plourde, L.C.; Katul, G.G.; Munger, J.W.; Oren, R.; et al. Canopy nitrogen, carbon assimilation, and albedo in temperate and boreal forests: Functional relations and potential climate feedbacks. Proc. Natl. Acad. Sci. USA 2008, 105, 19336–19341. [Google Scholar] [CrossRef]
- Woitchik, A.F.; Ohowa, B.; Kazungu, J.M.; Rao, R.G.; Goeyens, L.; Dehairs, F. Nitrogen Enrichment during Decomposition of Mangrove Leaf Litter in an East African Coastal Lagoon (Kenya): Relative Importance of Biological Nitrogen Fixation; Kluwer Academic Publishers: Berlin, Germany, 1997; Volume 39. [Google Scholar]
- Nordhaus, I.; Salewski, T.; Jennerjahn, T.C. Food preferences of mangrove crabs related to leaf nitrogen compounds in the Segara Anakan Lagoon, Java, Indonesia. J. Sea Res. 2011, 65, 414–426. [Google Scholar] [CrossRef]
- Fonte, S.J.; Schowalter, T.D. Decomposition of Greenfall vs. Senescent Foliage in a Tropical Forest Ecosystem in Puerto Rico1. Biotropica 2004, 36, 474. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Ustin, S.L. Modeling canopy water content for carbon estimates from MODIS data at land EOS validation sites. IGARSS 2001 Scanning the Present and Resolving the Future. In Proceedings of the IEEE 2001 International Geoscience and Remote Sensing Symposium 2001, Trento, Italy, 13–14 September 2001; Volume 1, pp. 342–344, (Cat. No.01CH37217). [Google Scholar]
- Clevers, J.G.P.W.; Kooistra, L.; Schaepman, M.E. Using spectral information from the NIR water absorption features for the retrieval of canopy water content. Int. J. Appl. Earth Obs. Geoinf. 2008, 10, 388–397. [Google Scholar] [CrossRef]
- Omer, G.; Mutanga, O.; Abdel-Rahman, E.M.; Peerbhay, K.; Adam, E. Mapping leaf nitrogen and carbon concentrations of intact and fragmented indigenous forest ecosystems using empirical modeling techniques and WorldView-2 data. ISPRS J. Photogramm. Remote Sens. 2017, 131, 26–39. [Google Scholar] [CrossRef]
- Sun, X.; Li, G.; Wang, M.; Fan, Z. Analyzing the uncertainty of estimating forest aboveground biomass using optical imagery and spaceborne LiDAR. Remote Sens. 2019, 11, 722. [Google Scholar] [CrossRef]
- De’Ath, G.; Fabricius, K.E. Classification and regression trees: A powerful yet simple technique for ecological data analysis. Ecology 2000, 81, 3178–3192. [Google Scholar] [CrossRef]
- Lawrence, R.; Bunn, A.; Powell, S.; Zambon, M. Classification of remotely sensed imagery using stochastic gradient boosting as a refinement of classification tree analysis. Remote Sens. Environ. 2004, 90, 331–336. [Google Scholar] [CrossRef]
- Elith, J.; Leathwick, J.R.; Hastie, T. A working guide to boosted regression trees. J. Anim. Ecol. 2008, 77, 802–813. [Google Scholar] [CrossRef]
- Dube, T.; Mutanga, O.; Elhadi, A.; Ismail, R. Intra-and-Inter Species Biomass Prediction in a Plantation Forest: Testing the Utility of High Spatial Resolution Spaceborne Multispectral RapidEye Sensor and Advanced Machine Learning Algorithms. Sensors 2014, 14, 15348–15370. [Google Scholar] [CrossRef]
- Convention on Biological Diversity (CBD). The Strategic Plan for Biodiversity 2011–2020 and the Aichi Biodiversity Targets; CBD: New York, NY, USA; Rio de Janeiro, Brazil, 2011. [Google Scholar]
- Convention on Biological Diversity. National Targets. Available online: https://www.cbd.int/countries/targets/?country=bd (accessed on 20 December 2019).
- MOEF. Integrated Resources Management Plans for the Sundarbans; Ministry of Environment and Forests: Dhaka, Bangladesh, 2010; Volume 1, ISBN 978-3-902762-17-7.
- Rahman, M.S.; Hossain, G.M.; Khan, S.A.; Uddin, S.N. An annotated checklist of the vascular plants of Sundarban Mangrove Forest of Bangladesh. Bangladesh J. Plant. Taxon. 2015, 22, 17–41. [Google Scholar] [CrossRef]
- Islam, S.; Feroz, S.M.; Ahmed, Z.U.; Chowdhury, H.; Khan, R.I.; Al-Mamun, A. Species richness and diversity of the floristic composition of the Sundarbans mangrove reserve forest, Bangladesh in relation to spatial habitats and salinity. Malayian For. 2016, 79, 7–38. [Google Scholar]
- Hamilton, S.E.; Casey, D. Creation of a high spatio-temporal resolution global database of continuous mangrove forest cover for the 21st century (CGMFC-21): CGMFC-21. Glob. Ecol. Biogeogr. 2016, 25, 729–738. [Google Scholar] [CrossRef]
- Forest Department-Government of the People\’s Republic of Bangladesh. Available online: http://www.bforest.gov.bd/site/page/f619019f-14cd-481a-86f4-1d5b4ae40515/- (accessed on 23 April 2020).
- Aziz, A.; Paul, A. Bangladesh Sundarbans: Present Status of the Environment and Biota. Diversity 2015, 7, 242–269. [Google Scholar] [CrossRef]
- Uddin, M.S.; de Ruyter van Steveninck, E.; Stuip, M.; Shah, M.A.R. Economic valuation of provisioning and cultural services of a protected mangrove ecosystem: A case study on Sundarbans Reserve Forest, Bangladesh. Ecosyst. Serv. 2013, 5, 88–93. [Google Scholar] [CrossRef]
- Laliberté, E.; Legendre, P. A distance-based framework for measuring functional diversity from multiple traits. Ecology 2010, 91, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Mensah, S.; Veldtman, R.; Assogbadjo, A.E.; Glèlè Kakaï, R.; Seifert, T. Tree species diversity promotes aboveground carbon storage through functional diversity and functional dominance. Ecol. Evol. 2016, 6, 7546–7557. [Google Scholar] [CrossRef] [PubMed]
- Garnier, E.; Cortez, J.; Billès, G.; Navas, M.L.; Roumet, C.; Debussche, M.; Laurent, G.; Blanchard, A.; Aubry, D.; Bellmann, A.; et al. Plant functional markers capture ecosystem properties during secondary succession. Ecology 2004, 85, 2630–2637. [Google Scholar] [CrossRef]
- Flood, N. Seasonal composite landsat TM/ETM+ Images using the medoid (a multi-dimensional median). Remote Sens. 2013, 5, 6481–6500. [Google Scholar] [CrossRef]
- The Thematic Mapper Landsat Science. Available online: https://landsat.gsfc.nasa.gov/the-thematic-mapper/ (accessed on 5 March 2020).
- Haralick, R.M.; Dinstein, I.; Shanmugam, K. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Conners, R.W.; Trivedi, M.M.; Harlow, C.A. Segmentation of a high-resolution urban scene using texture operators (Sunnyvale, California). Comput. Vis. Graph. Image Process. 1984, 25, 273–310. [Google Scholar] [CrossRef]
- Bello, C.; Galetti, M.; Pizo, M.A.; Magnago, L.F.S.; Rocha, M.F.; Lima, R.A.F.; Peres, C.A.; Ovaskainen, O.; Jordano, P. Defaunation affects carbon storage in tropical forests. Sci. Adv. 2015, 1, 1–11. [Google Scholar] [CrossRef]
- Blackburn, G.A. Spectral indices for estimating photosynthetic pigment concentrations: A test using senescent tree leaves. Int. J. Remote Sens. 1998, 19, 657–675. [Google Scholar] [CrossRef]
- Rama Rao, N.; Garg, P.K.; Ghosh, S.K.; Dadhwal, V.K. Estimation of leaf total chlorophyll and nitrogen concentrations using hyperspectral satellite imagery. J. Agric. Sci. 2008, 146, 65–75. [Google Scholar] [CrossRef]
- Volesky, J.C.; Stern, R.J.; Johnson, P.R. Geological control of massive sulfide mineralization in the Neoproterozoic Wadi Bidah shear zone, southwestern Saudi Arabia, inferences from orbital remote sensing and field studies. Precambrian Res. 2003, 123, 235–247. [Google Scholar] [CrossRef]
- Apan, A.; Held, A.; Scientific, T.C.; Phinn, S. Formulation and Assessment of Narrow-Band Vegetation Indices from EO-1 Hyperion Imagery for Detecting Sugarcane Disease; Spatial Sciences: Adelaide, Australia, 2003. [Google Scholar]
- Los, S.O.; Collatz, G.J.; Bounoua, L.; Sellers, P.J.; Tucker, C.J.; Los, S.O.; Collatz, G.J.; Bounoua, L.; Sellers, P.J.; Tucker, C.J. Global Interannual Variations in Sea Surface Temperature and Land Surface Vegetation, Air Temperature, and Precipitation. J. Clim. 2001, 14, 1535–1549. [Google Scholar] [CrossRef]
- Hewson, R.D.; Cudahy, T.J.; Huntington, J.F. Geologic and alteration mapping at Mt fitton, South Australia, using ASTER satellite-borne data. In Proceedings of the International Geoscience and Remote Sensing Symposium (IGARSS), Trento, Italy, 13–14 September 2001; Volume 2, pp. 724–726. [Google Scholar]
- Fensholt, R.; Sandholt, I. Derivation of a shortwave infrared water stress index from MODIS near- and shortwave infrared data in a semiarid environment. Remote Sens. Environ. 2003, 87, 111–121. [Google Scholar] [CrossRef]
- Xu, H. Modification of normalised difference water index (NDWI) to enhance open water features in remotely sensed imagery. Int. J. Remote Sens. 2006, 27, 3025–3033. [Google Scholar] [CrossRef]
- Al-Khaier, F. Soil Salinity Detection Using Satellite Remotes Sensing; International Institute for Geo-Information Science and Earth Observation: Enschede, The Netherlands, 2003. [Google Scholar]
- Ridgeway, G. Generalized Boosted Models: A guide to the gbm package. Compute 2007, 1, 1–12. [Google Scholar]
- Friedman, J.H. Stochastic gradient boosting. Comput. Stat. Data Anal. 2002, 38, 367–378. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and Regression by Random Forest. R News 2002, 2, 18–22. [Google Scholar]
- Vapnik, V.N. Statistical Learning Theory; Wiley: Hoboken, NJ, USA, 1998; ISBN 9780471030034. [Google Scholar]
- Karatzoglou, A.; Meyer, D.; Hornik, K. Support Vector Algorithm in R. J. Stat. Softw. 2006, 15, 1–28. [Google Scholar] [CrossRef]
- Smola, A.J.; Schölkopf, B. A tutorial on support vector regression. Stat. Comput. 2004, 14, 199–222. [Google Scholar] [CrossRef]
- Peng, J.; Ji, W.; Ma, Z.; Li, S.; Chen, S.; Zhou, L.; Shi, Z. Predicting total dissolved salts and soluble ion concentrations in agricultural soils using portable visible near-infrared and mid-infrared spectrometers. Biosyst. Eng. 2016, 152, 94–103. [Google Scholar] [CrossRef]
- Ali, A.M.; Skidmore, A.K.; Darvishzadeh, R.; van Duren, I.; Holzwarth, S.; Mueller, J. Retrieval of forest leaf functional traits from HySpex imagery using radiative transfer models and continuous wavelet analysis. ISPRS J. Photogramm. Remote Sens. 2016, 122, 68–80. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development for R; RStudio Inc.: Boston, MA, USA, 2019. [Google Scholar]
- Cutler, D.R.; Edwards, T.C.; Beard, K.H.; Cutler, A.; Hess, K.T.; Gibson, J.; Lawler, J.J. Random Forests for Classification in Ecology. Ecology 2007, 88, 2783–2792. [Google Scholar] [CrossRef] [PubMed]
- Breunig, F.M.; Galvão, L.S.; Dalagnol, R.; Dauve, C.E.; Parraga, A.; Santi, A.L.; Della Flora, D.P.; Chen, S. Delineation of management zones in agricultural fields using cover–crop biomass estimates from PlanetScope data. Int. J. Appl. Earth Obs. Geoinf. 2020, 85, 102004. [Google Scholar] [CrossRef]
- Zhang, Y.; Liang, S.; Sun, G. Forest biomass mapping of northeastern china using GLAS and MODIS data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 140–152. [Google Scholar] [CrossRef]
- Shen, W.; Li, M.; Huang, C.; Tao, X.; Wei, A. Annual forest aboveground biomass changes mapped using ICESat/GLAS measurements, historical inventory data, and time-series optical and radar imagery for Guangdong province, China. Agric. For. Meteorol. 2018, 259, 23–28. [Google Scholar] [CrossRef]
- Cao, L.; Pan, J.; Li, R.; Li, J.; Li, Z. Integrating airborne LiDAR and optical data to estimate forest aboveground biomass in arid and semi-arid regions of China. Remote Sens. 2018, 10, 532. [Google Scholar] [CrossRef]
- Houborg, R.; Fisher, J.B.; Skidmore, A.K. Advances in remote sensing of vegetation function and traits. Int. J. Appl. Earth Obs. Geoinf. 2015, 43, 1–6. [Google Scholar] [CrossRef]
- Croft, H.; Arabian, J.; Chen, J.M.; Shang, J.; Liu, J. Mapping within-field leaf chlorophyll content in agricultural crops for nitrogen management using Landsat-8 imagery. Precis. Agric. 2019, 1–25. [Google Scholar] [CrossRef]
- Anderson, M.C.; Neale, C.M.U.; Li, F.; Norman, J.M.; Kustas, W.P.; Jayanthi, H.; Chavez, J. Upscaling ground observations of vegetation water content, canopy height, and leaf area index during SMEX02 using aircraft and Landsat imagery. Remote Sens. Environ. 2004, 92, 447–464. [Google Scholar] [CrossRef]
- Questad, E.J.; Kellner, J.R.; Kinney, K.; Cordell, S.; Asner, G.P.; Thaxton, J.; Diep, J.; Uowolo, A.; Brooks, S.; Inman-Narahari, N.; et al. Mapping habitat suitability for at-risk plant species and its implications for restoration and reintroduction. Ecol. Appl. 2014, 24, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Kabir, M.E.; Ahmed, I. Protected Areas for Climate Change Mitigation and Livelihood Option: A Case Study of the Bangladesh Sundarbans Mangrove Forest; Springer: Tokyo, Japan, 2017; pp. 119–136. [Google Scholar]
- Ball, M.C. Ecophysiology of mangroves. Trees 1988, 2, 129–142. [Google Scholar] [CrossRef]
- Kissling, W.D.; Ahumada, J.A.; Bowser, A.; Fernandez, M.; Fernández, N.; García, E.A.; Guralnick, R.P.; Isaac, N.J.B.; Kelling, S.; Los, W.; et al. Building essential biodiversity variables (EBVs) of species distribution and abundance at a global scale. Biol. Rev. 2018, 93, 600–625. [Google Scholar] [CrossRef] [PubMed]
- Bakker, M.A.; Carreño-Rocabado, G.; Poorter, L. Leaf economics traits predict litter decomposition of tropical plants and differ among land use types. Funct. Ecol. 2011, 25, 473–483. [Google Scholar] [CrossRef]
- Gessner, M.O.; Swan, C.M.; Dang, C.K.; McKie, B.G.; Bardgett, R.D.; Wall, D.H.; Hättenschwiler, S. Diversity meets decomposition. Trends Ecol. Evol. 2010, 25, 372–380. [Google Scholar] [CrossRef]
- McCall, M.K.; Chutz, N.; Skutsch, M. Moving from Measuring, Reporting, Verification (MRV) of Forest Carbon to Community Mapping, Measuring, Monitoring (MMM): Perspectives from Mexico. PLoS ONE 2016, 11, e0146038. [Google Scholar] [CrossRef]
- O’Connor, B.; Secades, C.; Penner, J.; Sonnenschein, R.; Skidmore, A.; Burgess, N.D.; Hutton, J.M. Earth observation as a tool for tracking progress towards the Aichi Biodiversity Targets. Remote Sens. Ecol. Conserv. 2015, 1, 19–28. [Google Scholar] [CrossRef]
- Petrou, Z.I.; Manakos, I.; Stathaki, T. Remote sensing for biodiversity monitoring: A review of methods for biodiversity indicator extraction and assessment of progress towards international targets. Biodivers. Conserv. 2015, 24, 2333–2363. [Google Scholar] [CrossRef]
- Branquinho, C.; Serrano, H.C.; Nunes, A.; Pinho, P.; Matos, P. Essential Biodiversity Change Indicators for Evaluating the Effects of Anthropocene in Ecosystems at a Global Scale; Springer: Cham, Switzerland, 2019; pp. 137–163. [Google Scholar]
- Vihervaara, P.; Auvinen, A.P.; Mononen, L.; Törmä, M.; Ahlroth, P.; Anttila, S.; Böttcher, K.; Forsius, M.; Heino, J.; Heliölä, J.; et al. How Essential Biodiversity Variables and remote sensing can help national biodiversity monitoring. Glob. Ecol. Conserv. 2017, 10, 43–59. [Google Scholar] [CrossRef]
- Latest NBSAPs. Available online: https://www.cbd.int/nbsap/about/latest/#bd (accessed on 20 December 2019).
- Strategic Plan for Biodiversity 2011–2020, Including Aichi Biodiversity Targets. Available online: https://www.cbd.int/sp/ (accessed on 20 December 2019).
- Unit, B. Strategic Plan for Biodiversity 2011–2020, Including Aichi Biodiversity Targets; CBD: New York, NY, USA; Rio de Janeiro, Brazil, 2019. [Google Scholar]


| Species | Frequency (Number of Plots Species Found) | Total Number of Individuals | C:N ± SD |
|---|---|---|---|
| Heritiera fomes | 127 | 9438 | 38.70 ± 1.84 |
| Excoecaria agallocha | 148 | 7241 | 35.73 ± 1.30 |
| Xylocarpus mekongensis | 85 | 434 | 16.07 ± 0.76 |
| Bruguiera sexangula | 23 | 253 | 44.73 ± 2.15 |
| Avicennia officinalis | 35 | 127 | 24.47 ± 0.35 |
| Xylocarpus granatum | 20 | 75 | 19.43 ± 0.68 |
| Amoora cuculata | 31 | 74 | 29.22 ± 0.75 |
| Sonneratia apetala | 7 | 36 | 23.73 ± 0.91 |
| Initia bijuga | 6 | 12 | 29.22 ± 0.75 |
| Sonneratia caseolaris | 2 | 12 | 19.73 ± 0.83 |
| Cynometra ramiflora | 8 | 10 | 29.22 ± 0.75 |
| Aegiceras corniculatum | 6 | 9 | 49.67 ± 2.06 |
| Unknown species | 5 | 9 | 29.22 ± 0.75 |
| Excoecaria. indica | 5 | 8 | 29.22 ± 0.75 |
| Pongamia pinnata | 3 | 7 | 29.22 ± 0.75 |
| Rhizophora mucronata | 3 | 7 | 56.93 ± 5.42 |
| Lumnitzera racemosa | 1 | 4 | 22.43 ± 2.55 |
| Hibiscus tiliaceus | 1 | 2 | 29.22 ± 0.75 |
| Predictors | Source |
|---|---|
| Landsat TM 5 (Band name, wavelength, resolution) | [64] |
| B1 (Visible Blue, 0.45–0.52 µm, 30 m) | |
| B2 (Visible Green, 0.52–0.60 µm, 30 m) | |
| B3 (Visible Red, 0.63–0.69 µm, 30 m) | |
| B4 (NIR, 0.76–0.90 µm, 30 m) | |
| B5 (SWIR 1, 1.55–1.75 µm, 30 m) | |
| B6 (Thermal, 10.40–12.50 µm,120 m) | |
| B7 (SWIR 2, 2.08–2.35 µm, 30 m) | |
| Landsat 8 (Band name, wavelength, resolution) | |
| B2 (Visible Blue, 0.450–0.51 µm, 30 m) | |
| B3 (Visible Green, 0.53–0.59 µm, 30 m) | |
| B4 (Red, 0.64–0.67 µm, 30 m) | |
| B5 (Near-Infrared, 0.85–0.88 µm, 30 m) | |
| B6 (SWIR 1, 1.57–1.65 µm, 30 m) | |
| B7 (SWIR 2, 2.11–2.29 µm, 30 m) | |
| B10 (TIRS 1, 10.6–11.19 µm,100 m) | |
| B11 (TIRS 2, 11.5–12.51 µm,100 m) | |
| GLCM (Grey Level Co-occurrence Metrics) texture | |
| asm (Angular Second Moment) | [65] |
| contrast (Contrast) | [65] |
| corr (Correlation) | [65] |
| variance (Variance) | [65] |
| idm (Inverse Difference Moment) | [65] |
| savg (Sum Average) | [65] |
| svar (Sum Variance) | [65] |
| sent (Sum Entropy) | [65] |
| ent (Entropy) | [65] |
| dvar (Difference variance) | [65] |
| dent (Difference entropy) | [65] |
| imcorr1 (Information Measure of Corr. 1) | [65] |
| imcorr2 (Information Measure of Corr. 2) | [65] |
| maxcorr (Max Corr. Coefficient) | [65] |
| diss (Dissimilarity) | [66] |
| inertia (Inertia) | [66] |
| shade (Cluster Shade) | [66] |
| prom (Cluster prominence) | [66] |
| Vegetation index | |
| MSI (Moisture stress Index) | [67] |
| PSSR (Pigment Specific Simple Ratio) | [68] |
| PBI (Plant biochemical index) | [69] |
| SI (Salinity index) | [70] |
| DWSI (Disease-Water Stress Index) | [71] |
| SRRNI (Simple Ratio Red and NIR) | [72] |
| IO (Simple Ratio Red and Blue) | [73] |
| SRSN1B (Simple index SWIR 1 and blue) | This study |
| SRSN2B (Simple index SWIR 2 and blue) | This study |
| SRSN2G (Simple index SWIR2 and green) | This study |
| SRNB (Simple index NIR and blue) | This study |
| LWC (Leaf water content) | [74] |
| CVI (Chlorophyll Vegetation Index) | [67] |
| SIPI (Structure Intensive Pigment Index 3) | [68] |
| MNDWI (Modified Normalized Difference Water Index) | [75] |
| SCI (Soil Composition Index) | [76] |
| Models | Model Accuracy | Relative Improvement | Error Percentage | |||||
|---|---|---|---|---|---|---|---|---|
| MAE | RMSE | R2 | MAE% | RMSE% | R2% | MAE% | RMSE% | |
| SGB | 2.16 ± 0.58 a | 2.88 ± 0.85 a | 0.51 ± 0.22 a | −11.11 | −11.11 | 37.84 | 6.15 | 8.20 |
| RF | 2.21 ± 0.60 a | 2.99 ± 1.15 a | 0.48 ± 0.24 a | −9.05 | −7.72 | 29.73 | 6.30 | 8.52 |
| SVM | 2.06 ± 0.62 a | 3.12 ± 1.20 a | 0.46 ± 0.24 a | −15.23 | −3.70 | 24.32 | 5.86 | 8.89 |
| PLSR | 2.43 ± 0.68 b | 3.24 ± 1.15 b | 0.37 ± 0.23 b | 0.00 | 0.00 | 0.00 | 6.93 | 9.23 |
| Model Coefficients | Variables | ||
|---|---|---|---|
| y = a + b*x | Obs_C:N vs. Pre_CN_SGB | Obs_C:N vs. Pre_CN_RF | Obs_C:N vs. Pre_CN_SVM |
| a | −9.04 | −16.15 | −24.38 |
| b | 1.26 | 1.46 | 1.65 |
| MAE | 1.58 | 1.55 | 1.51 |
| RMSE | 2.12 | 2.23 | 2.44 |
| MAE% | 4.50 | 4.41 | 4.30 |
| RMSE% | 6.03 | 6.34 | 6.94 |
| Model Coefficients | Variables | ||
|---|---|---|---|
| y = a + b*x | SGB vs. RF | SGB vs. SVM | RF vs. SVM |
| a | −3.65 | −21.10 | −13.86 |
| b | 1.10 | 1.54 | 1.35 |
| R2 | 0.92 | 0.73 | 0.77 |
| N (pixel) | 4499989.00 | 4499989.00 | 4499989.00 |
| F (F-test significance) | 23,384,941.95 | 5,155,875.87 | 6,358,556.93 |
| Mean | Min | Max | SD | |
|---|---|---|---|---|
| Field | 35.15 | 18.4 | 39.99 | 3.96 |
| SGB 2010 | 33.92 | 26.95 | 39.43 | 2.45 |
| RF 2010 | 34.26 | 29.36 | 37.69 | 2.05 |
| SVM2010 | 35.68 | 30.99 | 38.51 | 1.16 |
| SGB 2019 | 32.51 | 27.04 | 37.28 | 1.77 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, M.M.; Zhang, X.; Ahmed, I.; Iqbal, Z.; Zeraatpisheh, M.; Kanzaki, M.; Xu, M. Remote Sensing-Based Mapping of Senescent Leaf C:N Ratio in the Sundarbans Reserved Forest Using Machine Learning Techniques. Remote Sens. 2020, 12, 1375. https://doi.org/10.3390/rs12091375
Rahman MM, Zhang X, Ahmed I, Iqbal Z, Zeraatpisheh M, Kanzaki M, Xu M. Remote Sensing-Based Mapping of Senescent Leaf C:N Ratio in the Sundarbans Reserved Forest Using Machine Learning Techniques. Remote Sensing. 2020; 12(9):1375. https://doi.org/10.3390/rs12091375
Chicago/Turabian StyleRahman, Md Mizanur, Xunhe Zhang, Imran Ahmed, Zaheer Iqbal, Mojtaba Zeraatpisheh, Mamoru Kanzaki, and Ming Xu. 2020. "Remote Sensing-Based Mapping of Senescent Leaf C:N Ratio in the Sundarbans Reserved Forest Using Machine Learning Techniques" Remote Sensing 12, no. 9: 1375. https://doi.org/10.3390/rs12091375
APA StyleRahman, M. M., Zhang, X., Ahmed, I., Iqbal, Z., Zeraatpisheh, M., Kanzaki, M., & Xu, M. (2020). Remote Sensing-Based Mapping of Senescent Leaf C:N Ratio in the Sundarbans Reserved Forest Using Machine Learning Techniques. Remote Sensing, 12(9), 1375. https://doi.org/10.3390/rs12091375