Predicting Soil Organic Carbon and Soil Nitrogen Stocks in Topsoil of Forest Ecosystems in Northeastern China Using Remote Sensing Data
Abstract
1. Introduction
2. Materials and Methods
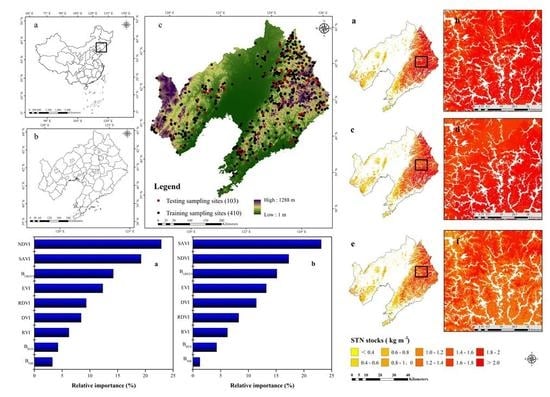
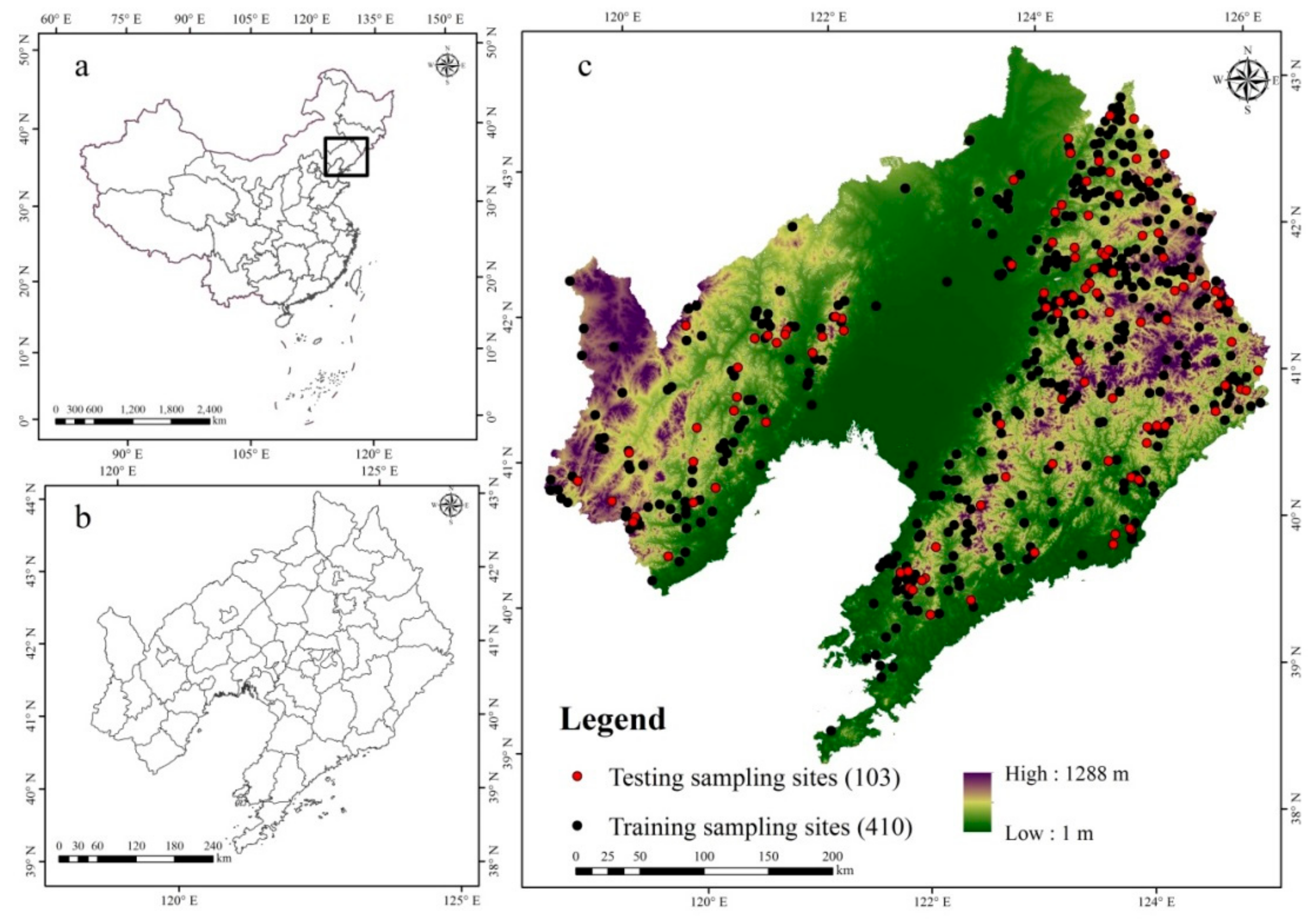
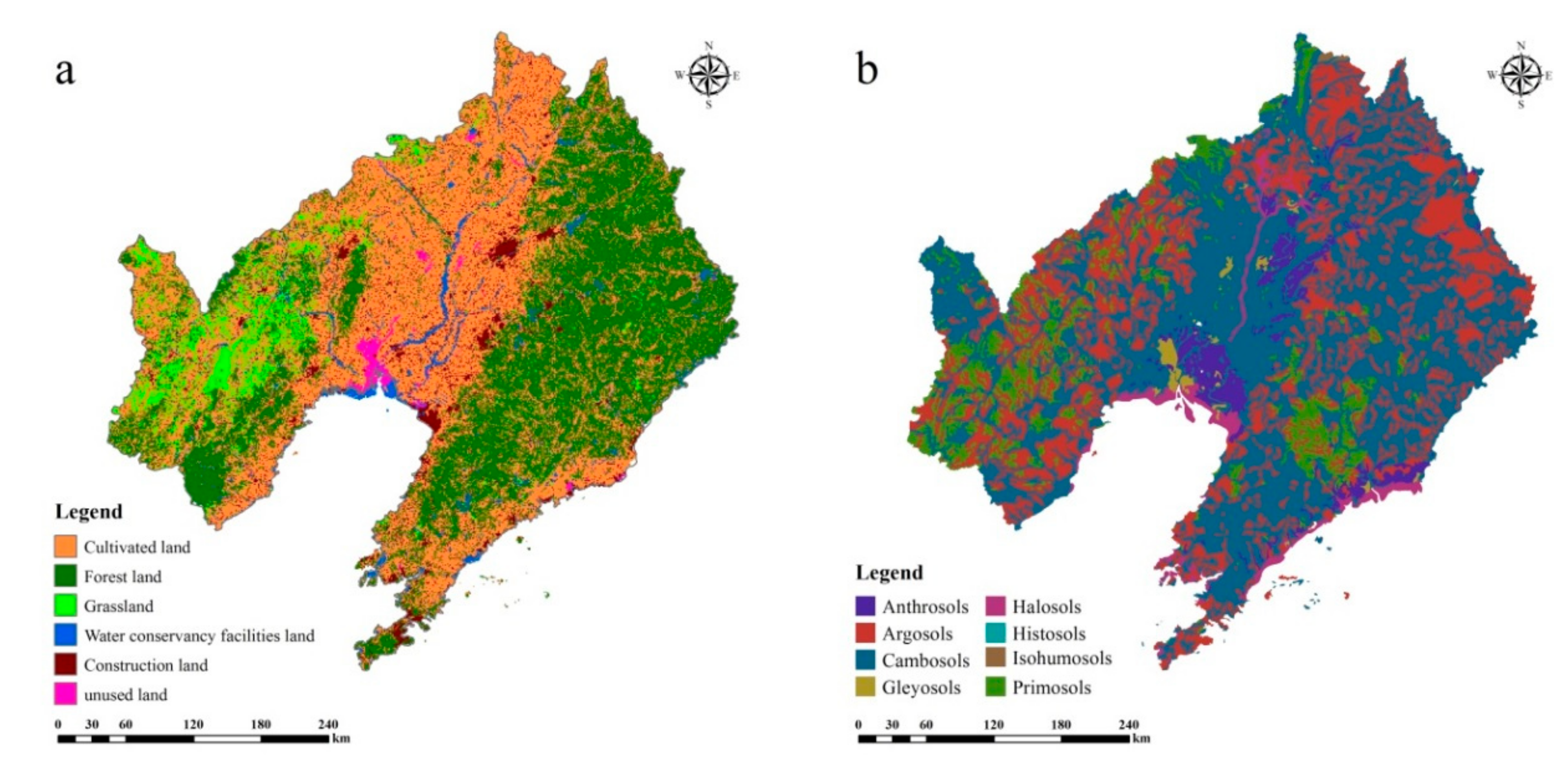
2.1. Study Area
2.2. Data Sources
2.2.1. Soil Sampling Collection and Measurement
2.2.2. Remote Sensing Related Data
2.3. Prediction Models
2.3.1. Geographically Weighted Regression
2.3.2. Multiple Stepwise Linear Regressions
2.3.3. Boosted Regression Trees
2.4. Prediction Accuracy
3. Results
3.1. Exploratory Data Analysis
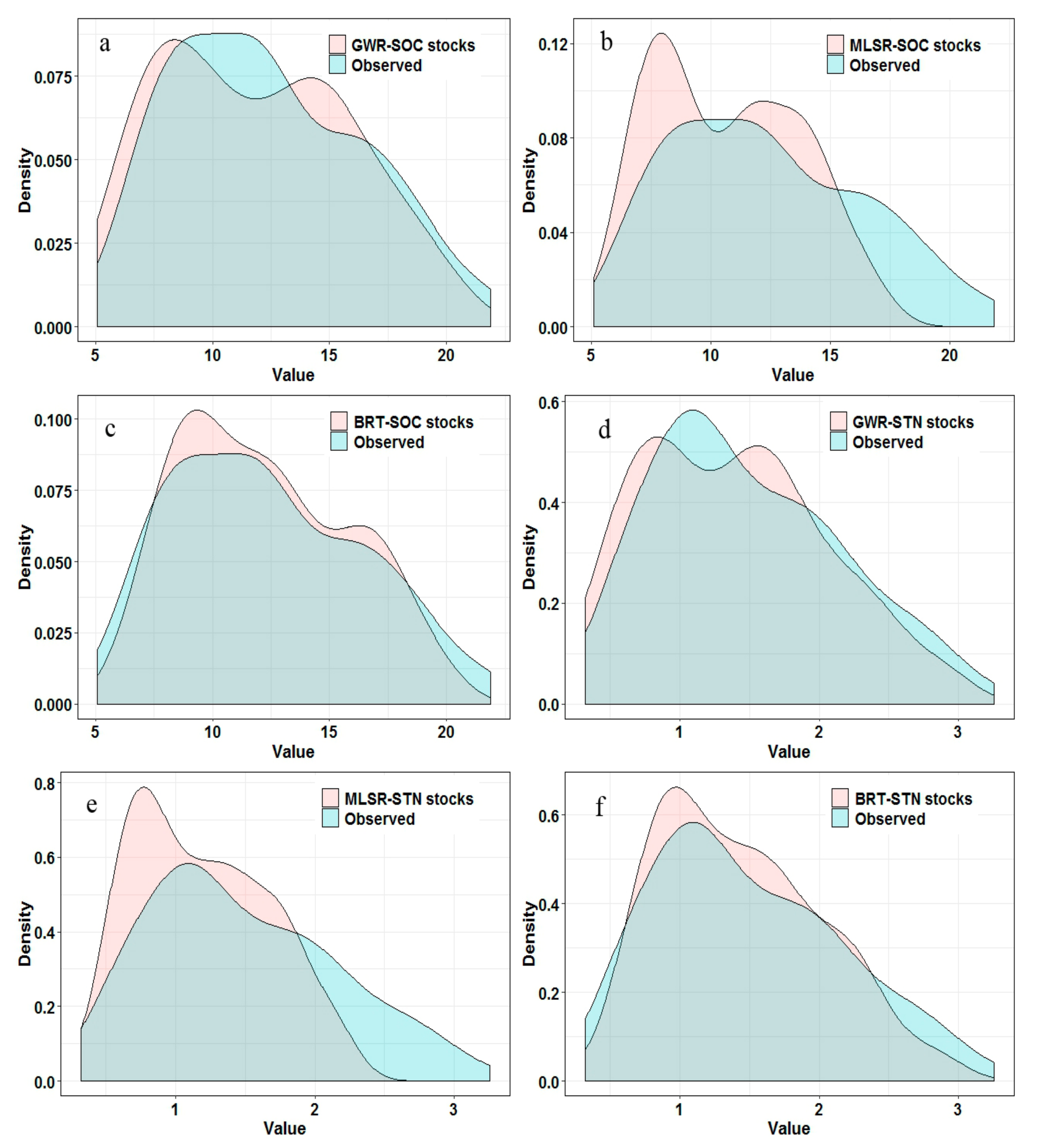
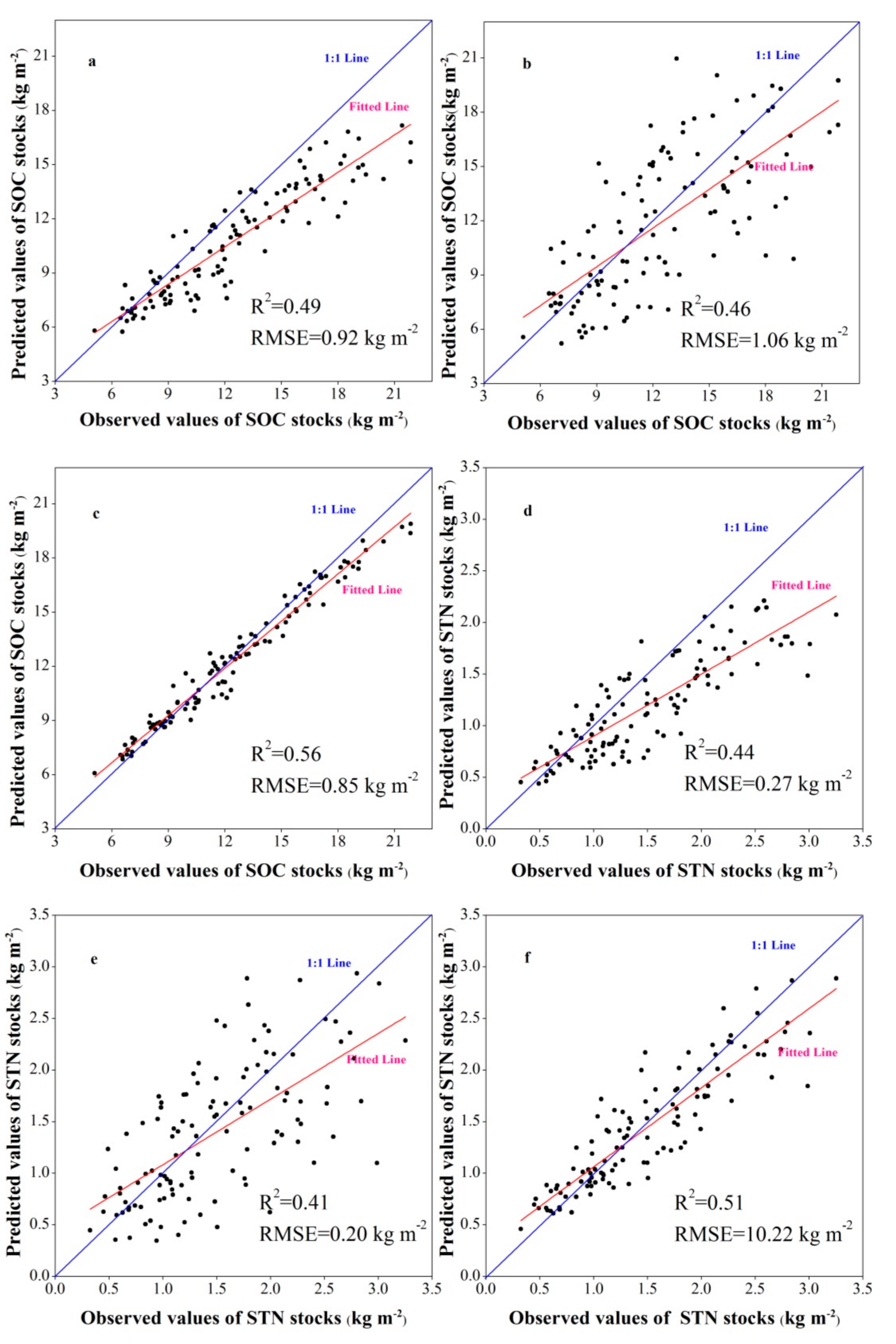
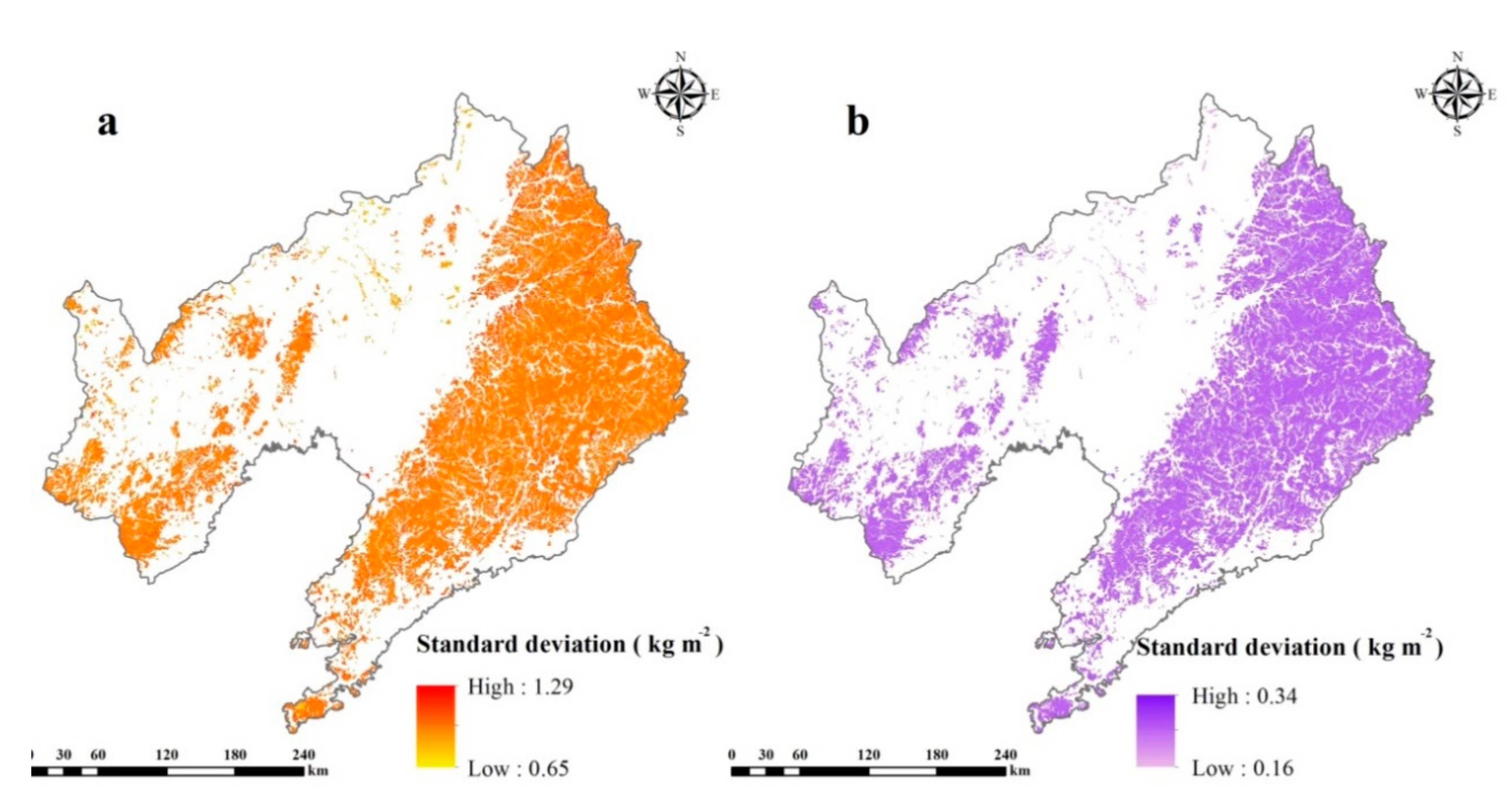
3.2. Model Performance and Uncertainty
3.3. Importance of Remotely Sensed Environmental Variables
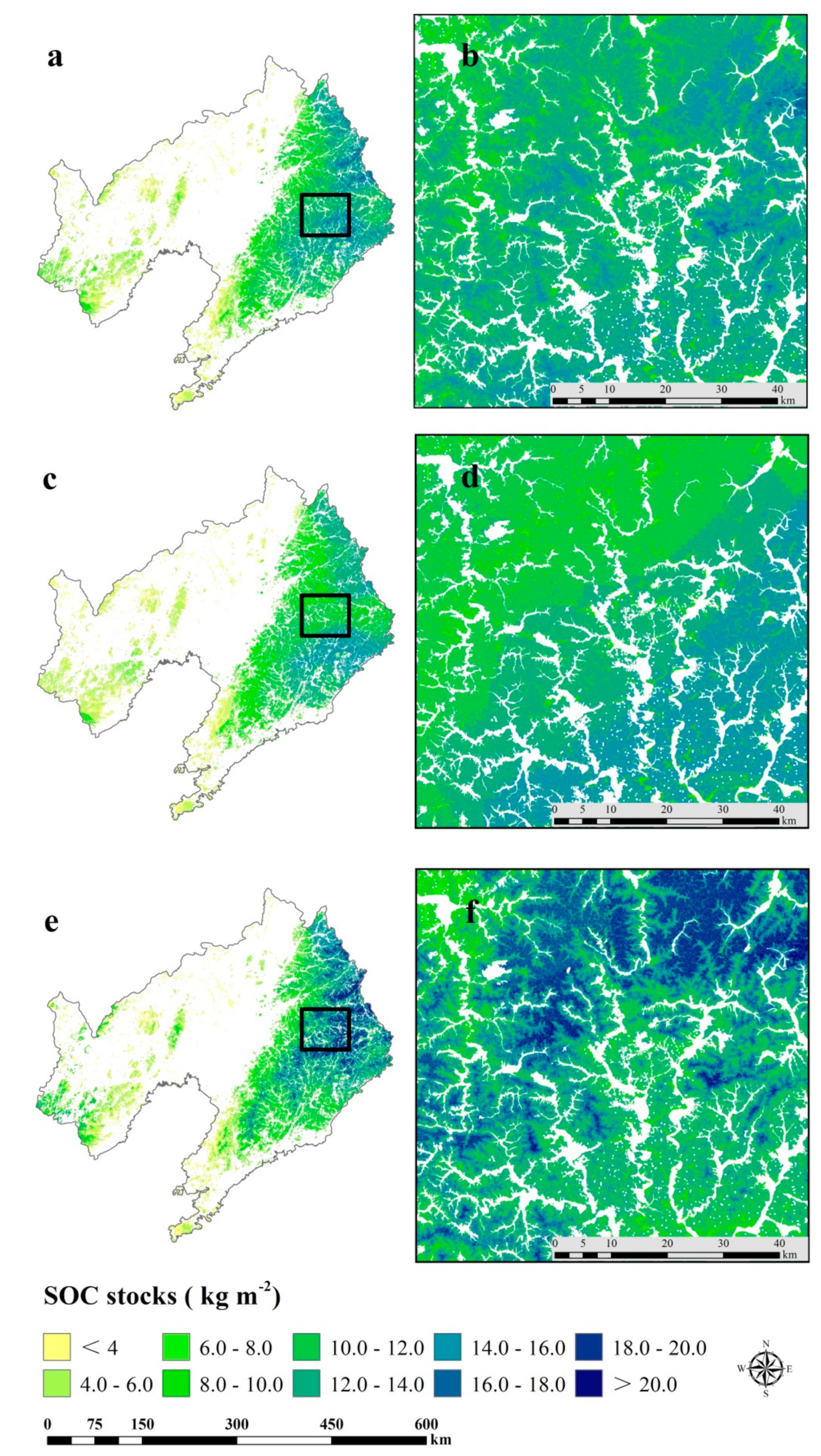
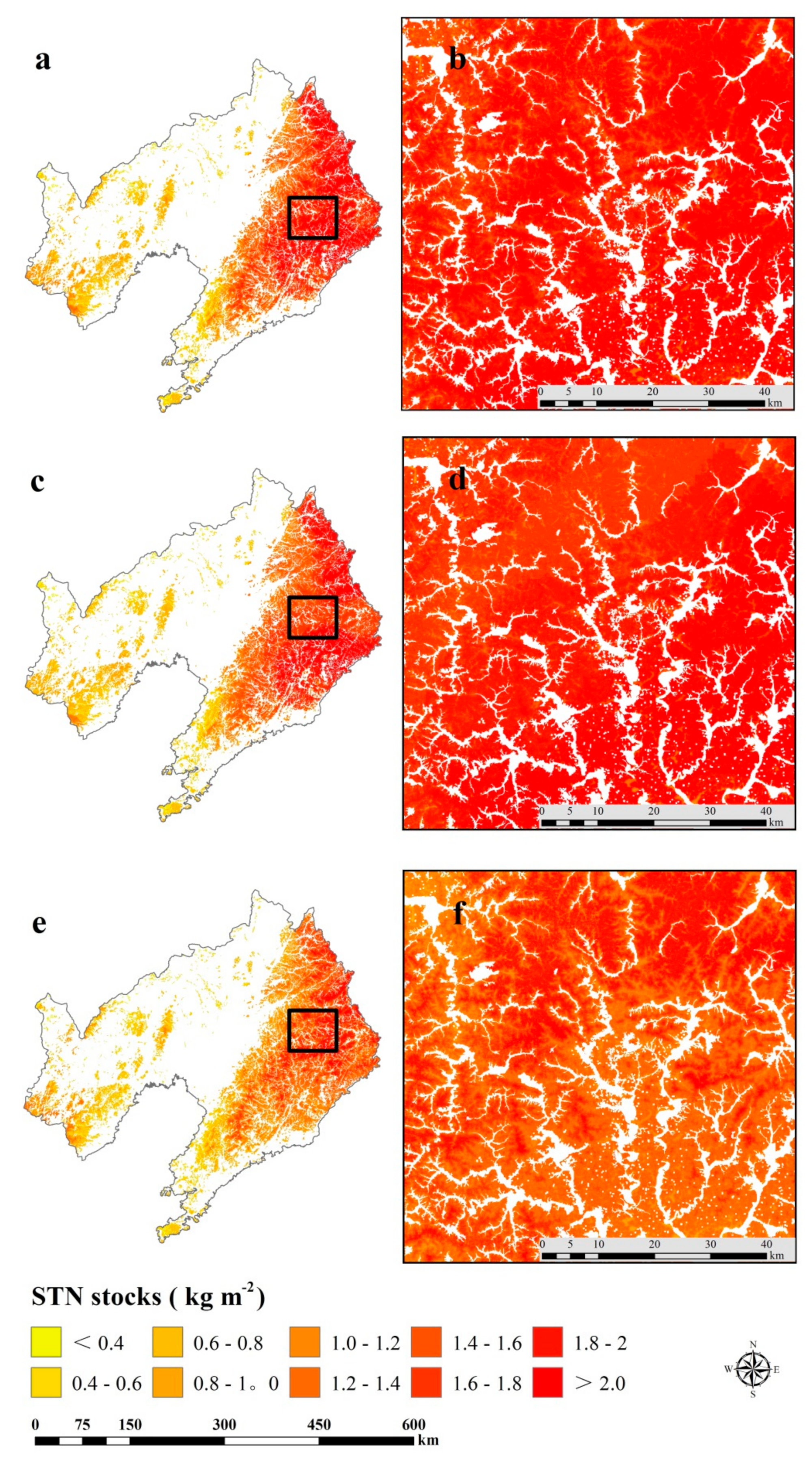
3.4. Spatial Prediction of SOC and STN Stocks
4. Discussion
4.1. Role of Remotely-Sensed Enviroment Variables in Predicting Topsoil SOC and STN Stocks
4.2. Uncertainty in Current Research
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Batjes, N.H. Total carbon and nitrogen in the soils of the world. Eur. J. Soil Sci. 1996, 47, 151–163. [Google Scholar] [CrossRef]
- Hessen, D.O.; Ågren, G.I.; Anderson, T.R.; Elser, J.J.; De Ruiter, P.C. Carbon sequestration in ecosystems: The role of stoichiometry. Ecology 2004, 85, 1179–1192. [Google Scholar] [CrossRef]
- Melillo, J.M. Carbon and nitrogen interactions in the terrestrial biosphere: Anthropogenic effects. Glob. Chang. Terr. Ecosyst. 1996, 2, 431. [Google Scholar]
- Vejre, H.; Callesen, I.; Vesterdal, L.; Raulund-Rasmussen, K. Carbon and nitrogen in Danish forest soils—Contents and distribution determined by soil order. Soil Sci. Soc. Am. J. 2003, 67, 335–343. [Google Scholar] [CrossRef]
- Lal, R. Forest soils and carbon sequestration. For. Ecol. Manag. 2005, 220, 242–258. [Google Scholar] [CrossRef]
- Gao, Q.; Guo, Y.; Xu, H.; Ganjurjav, H.; Li, Y.; Wan, Y.; Liu, S. Climate change and its impacts on vegetation distribution and net primary productivity of the alpine ecosystem in the Qinghai-Tibetan Plateau. Sci. Total Environ. 2016, 554, 34–41. [Google Scholar] [CrossRef]
- Xu, S.; Liu, L.; Sayer, E.J. Variability of above-ground litter inputs alters soil physicochemical and biological processes: A meta-analysis of litterfall-manipulation experiments. Biogeosciences 2013, 10, 7423–7433. [Google Scholar] [CrossRef]
- Li, C. Quantifying greenhouse gas emissions from soils: Scientific basis and modeling approach. Soil Sci. Plant Nutr. 2007, 53, 344–352. [Google Scholar] [CrossRef]
- Wang, S.; Zhuang, Q.; Wang, Q.; Jin, X.; Han, C. Mapping stocks of soil organic carbon and soil total nitrogen in Liaoning Province of China. Geoderma 2017, 305, 250–263. [Google Scholar] [CrossRef]
- McBratney, A.B.; Santos, M.M.; Minasny, B. On digital soil mapping. Geoderma 2003, 117, 3–52. [Google Scholar] [CrossRef]
- Jenny, H. Factors of Soil Formation; McGraw-Hill: New York, NY, USA, 1941. [Google Scholar]
- Vermote, E.; Justice, C.; Claverie, M.; Franch, B. Preliminary analysis of the performance of the Landsat 8/OLI land surface reflectance product. Remote Sens. Environ. 2016, 185, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.P.; Wattenbach, M.; Smith, P.; Meersmans, J.; Jolivet, C.; Boulonne, L.; Arrouays, D. Spatial distribution of soil organic carbon stocks in France. Biogeosciences 2011, 8, 1053–1065. [Google Scholar] [CrossRef]
- Liddicoat, C.; Maschmedt, D.; Clifford, D.; Searle, R.; Herrmann, T.; Macdonald, L.M.; Baldock, J. Predictive mapping of soil organic carbon stocks in South Australia’s agricultural zone. Soil Res. 2015, 53, 956–973. [Google Scholar] [CrossRef]
- Wang, S.; Jin, X.; Adhikari, K.; Li, W.; Yu, M.; Bian, Z.; Wang, Q. Mapping total soil nitrogen from a site in northeastern China. Catena 2018, 166, 134–146. [Google Scholar] [CrossRef]
- Duane, M.V.; Cohen, W.B.; Campbell, J.L.; Hudiburg, T.; Turner, D.P.; Weyermann, D.L. Implications of alternative field-sampling designs on Landsat-based mapping of stand age and carbon stocks in Oregon forests. For. Sci. 2010, 56, 405–416. [Google Scholar]
- Niwa, K.; Yokobori, J.; Hongo, C.; Nagata, O. Estimating soil carbon stocks in an upland area of Tokachi District, Hokkaido, Japan, by satellite remote sensing. Soil Sci. Plant Nutr. 2011, 57, 283–293. [Google Scholar] [CrossRef]
- Yigini, Y.; Panagos, P. Assessment of soil organic carbon stocks under future climate and land cover changes in Europe. Sci. Total Environ. 2016, 557, 838–850. [Google Scholar] [CrossRef] [PubMed]
- Mishra, U.; Lal, R.; Slater, B.; Calhoun, F.; Liu, D.; Van Meirvenne, M. Predicting soil organic carbon stock using profile depth distribution functions and ordinary kriging. Soil Sci. Soc. Am. J. 2009, 73, 614–621. [Google Scholar] [CrossRef]
- Grimm, R.; Behrens, T.; Märker, M.; Elsenbeer, H. Soil organic carbon concentrations and stocks on Barro Colorado Island—Digital soil mapping using Random Forests analysis. Geoderma 2008, 146, 102–113. [Google Scholar] [CrossRef]
- Wang, S.; Gao, J.; Zhuang, Q.; Lu, Y.; Gu, H.; Jin, X. Multispectral Remote Sensing Data Are Effective and Robust in Mapping Regional Forest Soil Organic Carbon Stocks in a Northeast Forest Region in China. Remote Sens. 2020, 12, 393. [Google Scholar] [CrossRef]
- Kumar, S.; Lal, R.; Liu, D. A geographically weighted regression kriging approach for mapping soil organic carbon stock. Geoderma 2012, 189, 627–634. [Google Scholar] [CrossRef]
- Nocita, M.; Stevens, A.; Noon, C.; van Wesemael, B. Prediction of soil organic carbon for different levels of soil moisture using Vis-NIR spectroscopy. Geoderma 2013, 199, 37–42. [Google Scholar] [CrossRef]
- Yu, D.S.; Shi, X.Z.; Wang, H.J.; Sun, W.X.; Chen, J.M.; Liu, Q.H.; Zhao, Y.C. Regional patterns of soil organic carbon stocks in China. J. Environ. Manag. 2007, 85, 680–689. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.P.; Wulder, M.A.; Loveland, T.R.; Woodcock, C.E.; Allen, R.G.; Anderson, M.C.; Scambos, T.A. Landsat-8: Science and product vision for terrestrial global change research. Remote Sens. Environ. 2014, 145, 154–172. [Google Scholar] [CrossRef]
- Zhu, A.X.; Yang, L.; Li, B.; Qin, C.; English, E.; Burt, J.E.; Zhou, C. Purposive sampling for digital soil mapping for areas with limited data. In Digital Soil Mapping with Limited Data; Hartemink, A.E., Ed.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 33–245. [Google Scholar]
- Liu, Z.K.; Hunt, B.R. A new approach to removing cloud cover from satellite imagery. Comput. Vis. Graph. Image Process. 1984, 25, 252–256. [Google Scholar] [CrossRef]
- Odebiri, O.; Mutanga, O.; Odindi, J.; Peerbhay, K.; Dovey, S. Predicting soil organic carbon stocks under commercial forest plantations in KwaZulu-Natal province, South Africa using remotely sensed data. GIScience Remote Sens. 2020, 1–14. [Google Scholar] [CrossRef]
- Perkins, T.; Adlergolden, S.; Matthew, M.; Berk, A.; Anderson, G.; Gardner, J. Retrieval of atmospheric properties from hyper and multispectral imagery with the FLAASH atmospheric correction algorithm. In Remote Sensing of Clouds & the Atmosphere X; International Society for Optics and Photonics: Orlando, FL, USA, 2005. [Google Scholar]
- Pimple, U.; Sitthi, A.; Simonetti, D.; Pungkul, S.; Leadprathom, K.; Chidthaisong, A. Topographic Correction of Landsat TM-5 and Landsat OLI-8 Imagery to Improve the Performance of Forest Classification in the Mountainous Terrain of Northeast Thailand. Sustainability 2017, 9, 258. [Google Scholar] [CrossRef]
- Goward, S.N.; Markham, B.; Dye, D.G.; Dulaney, W.; Yang, J. Normalized difference vegetation index measurements from the Advanced Very High Resolution Radiometer. Remote Sens. Environ. 1991, 35, 257–277. [Google Scholar] [CrossRef]
- Gilabert, M.A.; González-Piqueras, J.; Garcıa-Haro, F.J.; Meliá, J. A generalized soil-adjusted vegetation index. Remote Sens. Environ. 2002, 82, 303–310. [Google Scholar] [CrossRef]
- Huete, A.R. A soil-adjusted vegetation index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Richardson, A.J.; Wiegand, C.L. Distinguishing vegetation from soil background information. Photogramm. Eng. Remote Sens. 1977, 43, 1541–1552. [Google Scholar]
- Major, D.J.; Baret, F.; Guyot, G. A ratio vegetation index adjusted for soil brightness. Int. J. Remote Sens. 1990, 11, 727–740. [Google Scholar] [CrossRef]
- Payero, J.O.; Neale, C.M.U.; Wright, J.L. Comparison of eleven vegetation indices for estimating plant height of alfalfa and grass. Appl. Eng. Agric. 2004, 20, 385. [Google Scholar] [CrossRef]
- Brunsdon, C.; Fotheringham, S.; Charlton, M. Geographically weighted regression. R. Stat. Soc. 1998, 47, 431–443. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, C.; Li, W. Predictive mapping of soil total nitrogen at a regional scale: A comparison between geographically weighted regression and cokriging. Appl. Geogr. 2013, 42, 73–85. [Google Scholar] [CrossRef]
- Foster, S.A.; Gorr, W.L. An adaptive filter for estimating spatially-varying parameters: Application to modeling police hours spent in response to calls for service. Manag. Sci. 1986, 32, 878–889. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.; Huang, C. Spatial variability of soil total nitrogen and soil total phosphorus under different land uses in a small watershed on the Loess Plateau, China. Geoderma 2009, 150, 141–149. [Google Scholar] [CrossRef]
- Lv, J.; Liu, Y.; Zhang, Z.; Dai, J. Factorial kriging and stepwise regression approach to identify environmental factors influencing spatial multi-scale variability of heavy metals in soils. J. Hazard. Mater. 2013, 261, 387–397. [Google Scholar] [CrossRef]
- Ishii, Y.; Murakami, J.; Sasaki, K.; Tsukahara, M.; Wakamatsu, K. Efficient folding/assembly in Chinese hamster ovary cells is critical for high quality (low aggregate content) of secreted trastuzumab as well as for high production: Stepwise multivariate regression analyses. J. Biosci. Bioeng. 2014, 118, 223–230. [Google Scholar] [CrossRef]
- Hong, S.Y.; Minasny, H.K.H.; Kim, Y.; Lee, K. Predicting and mapping soil available water capacity in Korea. PeerJ 2013, 1, e71. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Additive logistic regression: A statistical view of boosting. Ann. Stat. 2000, 28, 337–407. [Google Scholar] [CrossRef]
- Elith, J.; Leathwick, J.R.; Hastie, T. A working guide to boosted regression trees. J. Anim. Ecol. 2008, 77, 802–813. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhuang, Q.; Yang, Z.; Yu, N.; Jin, X. Temporal and Spatial Changes of Soil Organic Carbon Stocks in the Forest Area of Northeastern China. Forests 2019, 10, 1023. [Google Scholar] [CrossRef]
- Lin, L. A concordance correlation coefficient to evaluate reproducibility. Biometrics 1989, 45, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Kissel, D.E.; West, L.T.; Adkins, W. Field-scale mapping of surface soil organic carbon using remotely sensed imagery. Soil Sci. Soc. Am. J. 2000, 64, 746–753. [Google Scholar] [CrossRef]
- Wang, B.; Waters, C.; Orgill, S.; Cowie, A.; Clark, A.; Li Liu, D.; Sides, T. Estimating soil organic carbon stocks using different modelling techniques in the semi-arid rangelands of eastern Australia. Ecol. Indic. 2018, 88, 425–438. [Google Scholar] [CrossRef]
- Yimer, F.; Ledin, S.; Abdelkadir, A. Soil property variations in relation to topographic aspect and vegetation community in the south-eastern highlands of Ethiopia. For. Ecol. Manag. 2006, 232, 90–99. [Google Scholar] [CrossRef]
- Gong, Y.; Guo, J.; Li, J.; Zhu, K.; Liao, M.; Liu, X.; Zhang, D. Experimental realization of an intrinsic magnetic topological insulator. Chin. Phys. Lett. 2019, 36, 076801. [Google Scholar] [CrossRef]
- Qi, L.; Wang, S.; Zhuang, Q.; Yang, Z.; Bai, S.; Jin, X.; Lei, G. Spatial-temporal changes in soil organic carbon and pH in the Liaoning Province of China: A modeling analysis based on observational data. Sustainability 2019, 11, 3569. [Google Scholar] [CrossRef]
- Yang, R.; Rossiter, D.G.; Liu, F.; Lu, Y.; Yang, F.; Yang, F.; Zhang, G. Predictive mapping of topsoil organic carbon in an alpine environment aided by Landsat TM. PLoS ONE 2015, 10, e0139042. [Google Scholar] [CrossRef]
- Lu, W.; Lu, D.; Wang, G.; Wu, J.; Huang, J.; Li, G. Examining soil organic carbon distribution and dynamic change in a hickory plantation region with Landsat and ancillary data. Catena 2018, 165, 576–589. [Google Scholar] [CrossRef]
- Gomez, C.; Rossel, R.A.V.; McBratney, A.B. Soil organic carbon prediction by hyperspectral remote sensing and field vis-NIR spectroscopy: An Australian case study. Geoderma 2008, 146, 403–411. [Google Scholar] [CrossRef]
- Pouladi, N.; Møller, A.B.; Tabatabai, S.; Greve, M.H. Mapping soil organic matter contents at field level with Cubist, Random Forest and kriging. Geoderma 2019, 342, 85–92. [Google Scholar] [CrossRef]

| Property | Min. | Median | Mean | Max. | SD | Skewness | Kurtosis |
|---|---|---|---|---|---|---|---|
| SOC stocks | 0.23 | 9.32 | 10.32 | 30.23 | 0.53 | 0.54 | 2.31 |
| STN stocks | 0.13 | 1.15 | 1.21 | 2.96 | 0.32 | 0.63 | 3.12 |
| BGREEN | 0.09 | 0.14 | 0.16 | 0.37 | 0.17 | 0.54 | 1.65 |
| BRED | 0.18 | 0.34 | 0.35 | 0.50 | 0.21 | 0.32 | 1.72 |
| BNIR | 0.18 | 0.40 | 0.41 | 0.63 | 0.24 | 0.48 | 1.83 |
| SAVI | 0.16 | 0.41 | 0.44 | 0.76 | 0.24 | −0.41 | 1.87 |
| NDVI | 0.13 | 0.39 | 0.41 | 0.72 | 0.22 | −0.38 | 1.75 |
| RVI | 1.65 | 2.29 | 2.78 | 4.12 | 1.16 | −1.13 | 1.23 |
| DVI | 65.31 | 135.33 | 136.21 | 193.11 | 24.51 | 0.04 | 2.24 |
| EVI | 0.18 | 0.59 | 0.60 | 0.95 | 0.33 | −0.58 | 2.51 |
| RDVI | 72.44 | 138.61 | 143.21 | 201.43 | 26.41 | 0.62 | 3.13 |
| Property | Soil Groups | Number | Min. | Median | Mean | Max. | SD | Skewness | Kurtosis |
|---|---|---|---|---|---|---|---|---|---|
| SOC stocks | Argosols | 154 | 0.49 | 9.86 | 10.38 | 26.16 | 0.54 | 0.57 | 2.41 |
| Cambosols | 148 | 0.44 | 8.84 | 9.30 | 22.41 | 0.47 | 0.47 | 2.21 | |
| Gleyosols | 23 | 0.26 | 5.36 | 5.58 | 13.67 | 0.43 | 0.52 | 2.14 | |
| Halosols | 43 | 0.23 | 4.62 | 4.86 | 11.23 | 0.37 | 0.43 | 1.95 | |
| Histosols | 27 | 0.62 | 12.48 | 13.14 | 30.23 | 0.49 | 0.55 | 2.17 | |
| Isohumosols | 29 | 0.5 | 10.39 | 10.71 | 28.38 | 0.38 | 0.49 | 1.87 | |
| Primosols | 89 | 0.23 | 4.1 | 4.32 | 10.02 | 0.36 | 0.41 | 1.72 | |
| STN stocks | Argosols | 154 | 0.26 | 1.32 | 1.35 | 2.47 | 0.35 | 0.68 | 3.23 |
| Cambosols | 148 | 0.23 | 1.17 | 1.23 | 2.25 | 0.27 | 0.59 | 2.75 | |
| Gleyosols | 23 | 0.22 | 1.06 | 1.14 | 1.97 | 0.25 | 0.52 | 2.17 | |
| Halosols | 43 | 0.19 | 0.97 | 1.02 | 1.66 | 0.22 | 0.47 | 2.83 | |
| Histosols | 27 | 0.27 | 1.39 | 1.43 | 2.96 | 0.32 | 0.63 | 2.95 | |
| Isohumosols | 29 | 0.26 | 1.3 | 1.37 | 2.53 | 0.29 | 0.54 | 1.98 | |
| Primosols | 89 | 0.13 | 0.89 | 0.93 | 1.27 | 0.23 | 0.52 | 2.03 |
| Property | SOC Stocks | STN Stocks | BGREEN | BRED | BNIR | SAVI | NDVI | RVI | DVI | EVI |
|---|---|---|---|---|---|---|---|---|---|---|
| STN stocks | 0.65 ** | |||||||||
| BGREEN | −0.53 ** | −0.43 ** | ||||||||
| BRED | 0.21 * | 0.13 | −0.39 ** | |||||||
| BNIR | −0.36 ** | −0.29 ** | 0.61 ** | 0.14 | ||||||
| SAVI | 0.45 ** | 0.34 ** | −0.53 ** | 0.33 ** | −0.41 ** | |||||
| NDVI | 0.56 ** | 0.45 ** | −0.65 ** | 0.57 ** | −0.39 ** | 0.73 ** | ||||
| RVI | −0.18 * | −0.23 * | −0.22 * | 0.39 ** | −0.29 ** | −0.22 ** | −0.26 ** | |||
| DVI | 0.26 * | 0.34 ** | 0.16 * | −0.28 ** | 0.43 ** | 0.17 * | 0.29 ** | −0.33 ** | ||
| EVI | 0.39 ** | 0.32 ** | 0.15 * | −0.09 | 0.32 ** | −0.25 ** | −0.35 ** | −0.24 ** | 0.37 ** | |
| RDVI | −0.35 ** | −0.25 * | −0.07 | 0.17 | −0.18 * | 0.43 ** | 0.46 ** | −0.08 | 0.41 ** | 0.26 ** |
| Property | Model | Index | Min. | Median | Mean | Max. |
|---|---|---|---|---|---|---|
| SOC stocks | GWR | MAE | 0.82 | 0.84 | 0.84 | 0.90 |
| RMSE | 0.89 | 0.91 | 0.92 | 0.95 | ||
| R2 | 0.48 | 0.49 | 0.49 | 0.51 | ||
| LCCC | 0.60 | 0.61 | 0.61 | 0.66 | ||
| MLSR | MAE | 0.82 | 0.84 | 0.84 | 0.89 | |
| RMSE | 1.02 | 1.04 | 1.06 | 1.08 | ||
| R2 | 0.45 | 0.45 | 0.46 | 0.48 | ||
| LCCC | 0.58 | 0.62 | 0.64 | 0.68 | ||
| BRT | MAE | 0.66 | 0.70 | 0.71 | 0.75 | |
| RMSE | 0.82 | 0.84 | 0.85 | 0.88 | ||
| R2 | 0.53 | 0.56 | 0.56 | 0.58 | ||
| LCCC | 0.77 | 0.78 | 0.80 | 0.82 | ||
| STN stocks | GWR | MAE | 0.15 | 0.16 | 0.16 | 0.19 |
| RMSE | 0.26 | 0.27 | 0.27 | 0.31 | ||
| R2 | 0.39 | 0.42 | 0.44 | 0.45 | ||
| LCCC | 0.47 | 0.47 | 0.47 | 0.50 | ||
| MLSR | MAE | 0.18 | 0.19 | 0.19 | 0.20 | |
| RMSE | 0.17 | 0.19 | 0.20 | 0.22 | ||
| R2 | 0.39 | 0.40 | 0.41 | 0.43 | ||
| LCCC | 0.42 | 0.44 | 0.45 | 0.47 | ||
| BRT | MAE | 0.16 | 0.16 | 0.16 | 0.17 | |
| RMSE | 0.20 | 0.22 | 0.22 | 0.25 | ||
| R2 | 0.47 | 0.50 | 0.51 | 0.54 | ||
| LCCC | 0.63 | 0.64 | 0.65 | 0.65 |
| Soil Groups | Area (km2) | Average SOC Stocks (kg m−2) | Average STN Stocks (kg m−2) | SOC Stock (Tg) | STN Stock (Tg) |
|---|---|---|---|---|---|
| Argosols | 5486.00 | 3.46 | 0.45 | 19.01 | 2.46 |
| Cambosols | 43959.00 | 2.92 | 0.49 | 128.35 | 21.54 |
| Gleyosols | 233.00 | 1.72 | 0.37 | 0.40 | 0.09 |
| Halosols | 774.00 | 1.51 | 0.32 | 1.17 | 0.25 |
| Histosols | 13.00 | 4.13 | 0.35 | 0.05 | 0.01 |
| Isohumosols | 12.00 | 3.57 | 0.40 | 0.04 | 0.01 |
| Primosols | 12275.00 | 1.36 | 0.29 | 16.68 | 3.61 |
| Sum | 62752.00 | 165.69 | 27.96 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Zhuang, Q.; Jin, X.; Yang, Z.; Liu, H. Predicting Soil Organic Carbon and Soil Nitrogen Stocks in Topsoil of Forest Ecosystems in Northeastern China Using Remote Sensing Data. Remote Sens. 2020, 12, 1115. https://doi.org/10.3390/rs12071115
Wang S, Zhuang Q, Jin X, Yang Z, Liu H. Predicting Soil Organic Carbon and Soil Nitrogen Stocks in Topsoil of Forest Ecosystems in Northeastern China Using Remote Sensing Data. Remote Sensing. 2020; 12(7):1115. https://doi.org/10.3390/rs12071115
Chicago/Turabian StyleWang, Shuai, Qianlai Zhuang, Xinxin Jin, Zijiao Yang, and Hongbin Liu. 2020. "Predicting Soil Organic Carbon and Soil Nitrogen Stocks in Topsoil of Forest Ecosystems in Northeastern China Using Remote Sensing Data" Remote Sensing 12, no. 7: 1115. https://doi.org/10.3390/rs12071115
APA StyleWang, S., Zhuang, Q., Jin, X., Yang, Z., & Liu, H. (2020). Predicting Soil Organic Carbon and Soil Nitrogen Stocks in Topsoil of Forest Ecosystems in Northeastern China Using Remote Sensing Data. Remote Sensing, 12(7), 1115. https://doi.org/10.3390/rs12071115