Weed Identification in Maize, Sunflower, and Potatoes with the Aid of Convolutional Neural Networks
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Field
2.2. Image Acquisition
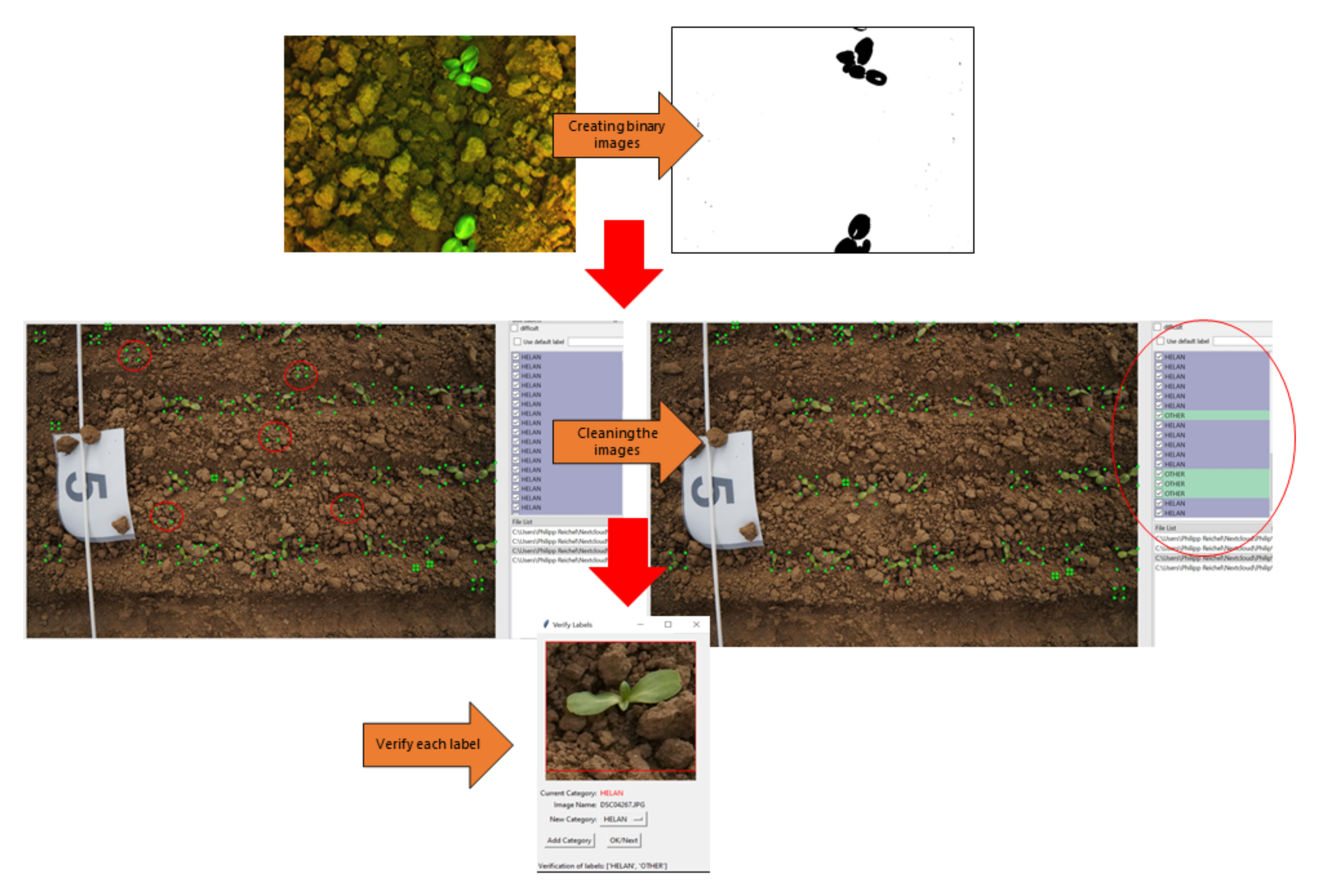
2.3. Image Preprocessing
- Pixel formations less than 400 pixels were discarded.
- Bounding boxes were expanded if needed symmetrically to the minimum size of 64 × 64 pixels. The minimum bounding box was 64 × 64 pixels.
- Regions bigger than 64 pixels were expanded only by 5 pixels in all directions. Therefore, there was no limitation to the maximum box.
- If bounding boxes overlapped, a new bounding box was created, merging all the overlapping boxes.
- Both the original and the merged bounding boxes were kept for labeling.
2.4. Neural Networks
2.4.1. VGG16
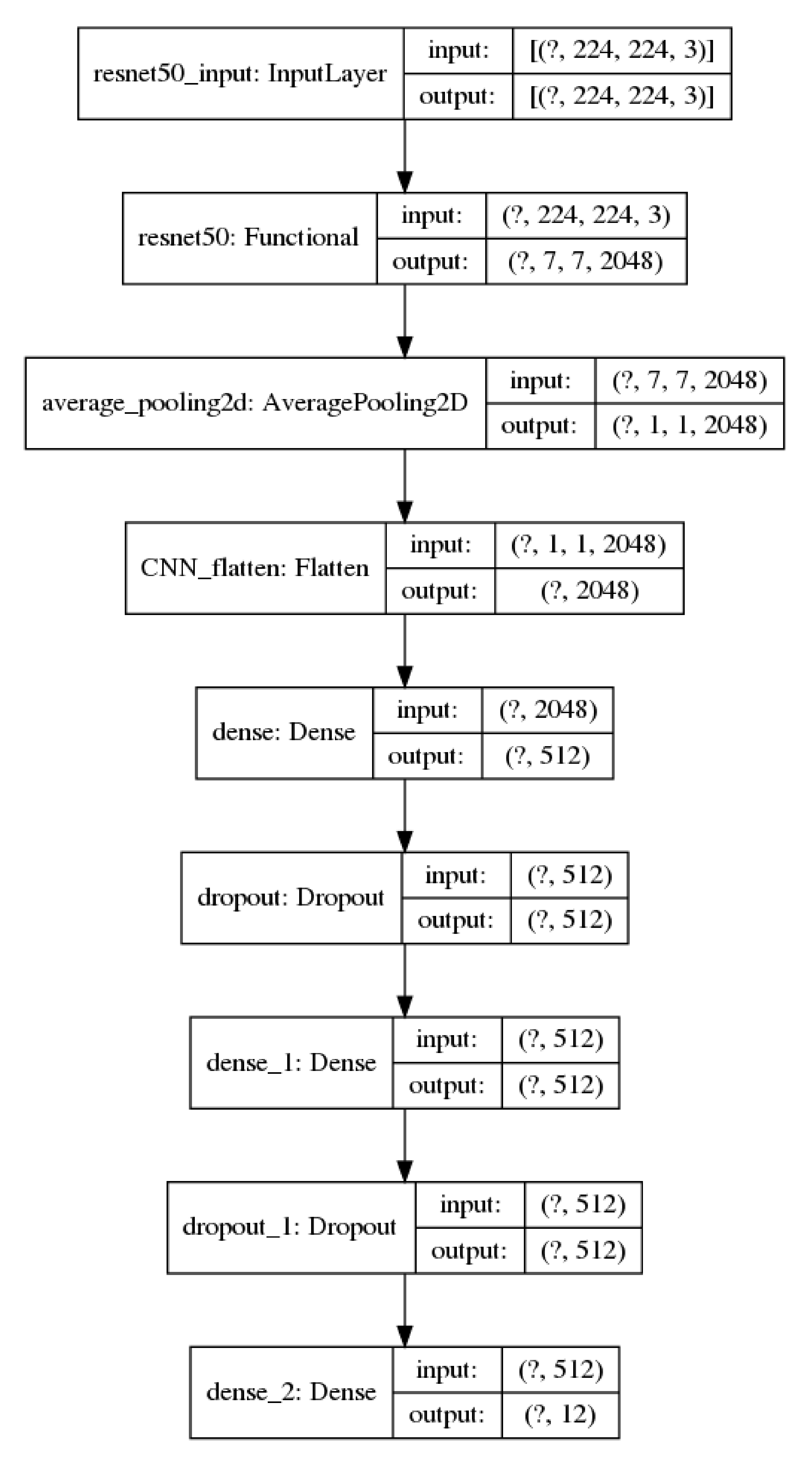
2.4.2. ResNet–50
2.4.3. Xception
2.4.4. Dataset Normalization
2.4.5. Network Training
2.5. Evaluation Metrics
3. Results
3.1. Model Accuracy/Model Loss
3.2. Classification Performance
3.3. Precision/Recall
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ANN | Artificial Neural Networks |
| CNN | Convolutional Neural Networks |
| EPPO | European and Mediterranean Plant Protection Organization |
| MDPI | Multidisciplinary Digital Publishing Institute |
| UAV | Unmanned Aerial Vehicle |
| VGG | Visual Geometry Group |
References
- Pérez-Ortiz, M.; Peña, J.M.; Gutiérrez, P.A.; Torres-Sánchez, J.; Hervás-Martínez, C.; López-Granados, F. Selecting patterns and features for between- and within- crop-row weed mapping using UAV-imagery. Expert Syst. Appl. 2016, 47, 85–94. [Google Scholar] [CrossRef]
- Oerke, E.C.; Gerhards, R.; Menz, G.; Sikora, R.A. (Eds.) Precision Crop Protection—The Challenge and Use of Heterogeneity, 1st ed.; Springer: Dordrecht, The Netherlands; Heidelberg, Germany; London, UK; New York, NY, USA, 2010; Volume 1. [Google Scholar] [CrossRef]
- Fernández-Quintanilla, C.; Peña, J.M.; Andújar, D.; Dorado, J.; Ribeiro, A.; López-Granados, F. Is the current state of the art of weed monitoring suitable for site-specific weed management in arable crops? Weed Res. 2018, 58, 259–272. [Google Scholar] [CrossRef]
- Tang, J.; Wang, D.; Zhang, Z.; He, L.; Xin, J.; Xu, Y. Weed identification based on K-means feature learning combined with convolutional neural network. Comput. Electron. Agric. 2017, 135, 63–70. [Google Scholar] [CrossRef]
- Dyrmann, M.; Christiansen, P.; Midtiby, H.S. Estimation of plant species by classifying plants and leaves in combination. J. Field Robot. 2017, 35, 202–212. [Google Scholar] [CrossRef]
- Dyrmann, M.; Karstoft, H.; Midtiby, H.S. Plant species classification using deep convolutional neural network. Biosyst. Eng. 2016, 151, 72–80. [Google Scholar] [CrossRef]
- Pantazi, X.E.; Moshou, D.; Bravo, C. Active learning system for weed species recognition based on hyperspectral sensing. Biosyst. Eng. 2016. [Google Scholar] [CrossRef]
- Sabzi, S.; Abbaspour-Gilandeh, Y.; García-Mateos, G. A fast and accurate expert system for weed identification in potato crops using metaheuristic algorithms. Comput. Ind. 2018, 98, 80–89. [Google Scholar] [CrossRef]
- European Parliament; Council of the EU. Directive 2009/128/EC of the European Parliament and of the Council of 21st October 2009 establishing a framework for Community action to achieve the sustainable use of pesticides (Text with EEA relevance). Off. J. Eur. Union 2009, L 309, 71–86. [Google Scholar]
- Machleb, J.; Peteinatos, G.G.; Kollenda, B.L.; Andújar, D.; Gerhards, R. Sensor-based mechanical weed control: Present state and prospects. Comput. Electron. Agric. 2020, 176, 105638. [Google Scholar] [CrossRef]
- Tyagi, A.C. Towards a Second Green Revolution. Irrig. Drain. 2016, 65, 388–389. [Google Scholar] [CrossRef]
- Peteinatos, G.G.; Weis, M.; Andújar, D.; Rueda Ayala, V.; Gerhards, R. Potential use of ground-based sensor technologies for weed detection. Pest Manag. Sci. 2014, 70, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Lottes, P.; Hörferlin, M.; Sander, S.; Stachniss, C. Effective Vision-based Classification for Separating Sugar Beets and Weeds for Precision Farming. J. Field Robot. 2016, 34, 1160–1178. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhu, Q.; Huang, M.; Guo, Y.; Qin, J. Maize and weed classification using color indices with support vector data description in outdoor fields. Comput. Electron. Agric. 2017, 141, 215–222. [Google Scholar] [CrossRef]
- Dos Santos Ferreira, A.; Freitas, D.M.; da Silva, G.G.; Pistori, H.; Folhes, M.T. Weed detection in soybean crops using ConvNets. Comput. Electron. Agric. 2017, 143, 314–324. [Google Scholar] [CrossRef]
- LeCun, Y.; Boser, B.; Denker, J.S.; Henderson, D.; Howard, R.E.; Hubbard, W.; Jackel, L.D. Backpropagation Applied to Handwritten Zip Code Recognition. Neural Comput. 1989, 1, 541–551. [Google Scholar] [CrossRef]
- Razavian, A.S.; Azizpour, H.; Sullivan, J.; Carlsson, S. CNN Features Off-the-Shelf: An Astounding Baseline for Recognition. In Proceedings of the 2014 IEEE Conference on Computer Vision and Pattern Recognition Workshops, Columbus, OH, USA, 23–28 June 2014; IEEE: Piscataway, NJ, USA, 2014. [Google Scholar] [CrossRef]
- Potena, C.; Nardi, D.; Pretto, A. Fast and Accurate Crop and Weed Identification with Summarized Train Sets for Precision Agriculture. In Intelligent Autonomous Systems 14; Springer International Publishing: Cham, Switzerland, 2017; pp. 105–121. [Google Scholar] [CrossRef]
- Elnemr, H.A. Convolutional Neural Network Architecture for Plant Seedling Classification. Int. J. Adv. Comput. Sci. Appl. 2019, 10. [Google Scholar] [CrossRef]
- Olsen, A.; Konovalov, D.A.; Philippa, B.; Ridd, P.; Wood, J.C.; Johns, J.; Banks, W.; Girgenti, B.; Kenny, O.; Whinney, J.; et al. DeepWeeds: A Multiclass Weed Species Image Dataset for Deep Learning. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Rawat, W.; Wang, Z. Deep Convolutional Neural Networks for Image Classification: A Comprehensive Review. Neural Comput. 2017, 29, 2352–2449. [Google Scholar] [CrossRef]
- Milioto, A.; Lottes, P.; Stachniss, C. Real-time blob-wise sugar beets vs weeds classification for monitoring fields using convolutional neural networks. ISPRS Ann. Photogramm. Remote. Sens. Spat. Inf. Sci. 2017, IV-2/W3, 41–48. [Google Scholar] [CrossRef]
- Lee, S.H.; Chan, C.S.; Mayo, S.J.; Remagnino, P. How deep learning extracts and learns leaf features for plant classification. Pattern Recognit. 2017, 71, 1–13. [Google Scholar] [CrossRef]
- Fuentes-Pacheco, J.; Torres-Olivares, J.; Roman-Rangel, E.; Cervantes, S.; Juarez-Lopez, P.; Hermosillo-Valadez, J.; Rendón-Mancha, J.M. Fig Plant Segmentation from Aerial Images Using a Deep Convolutional Encoder-Decoder Network. Remote Sens. 2019, 11, 1157. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef]
- Xinshao, W.; Cheng, C. Weed seeds classification based on PCANet deep learning baseline. In Proceedings of the 2015 Asia-Pacific Signal and Information Processing Association Annual Summit and Conference (APSIPA), Hong Kong, China, 16–19 December 2015; IEEE: Piscataway, NJ, USA, 2015. [Google Scholar] [CrossRef]
- Hoeser, T.; Kuenzer, C. Object Detection and Image Segmentation with Deep Learning on Earth Observation Data: A Review-Part I: Evolution and Recent Trends. Remote Sens. 2020, 12, 1667. [Google Scholar] [CrossRef]
- McCool, C.; Perez, T.; Upcroft, B. Mixtures of Lightweight Deep Convolutional Neural Networks: Applied to Agricultural Robotics. IEEE Robot. Autom. Lett. 2017, 2, 1344–1351. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Zhu, X.; Wu, X. Class Noise vs. Attribute Noise: A Quantitative Study. Artif. Intell. Rev. 2004, 22, 177–210. [Google Scholar] [CrossRef]
- McLaughlin, N.; Rincon, J.M.D.; Miller, P. Data-augmentation for reducing dataset bias in person re-identification. In Proceedings of the 2015 12th IEEE International Conference on Advanced Video and Signal Based Surveillance (AVSS), Karlsruhe, Germany, 25–28 August 2015; IEEE: Piscataway, NJ, USA, 2015. [Google Scholar] [CrossRef]
- Meier, U. Growth Stages of Mono- and Dicotyledonous Plants: BBCH Monograph; Open Agrar Repositorium: Göttingen, Germany, 2018. [Google Scholar] [CrossRef]
- Ge, Z.; McCool, C.; Sanderson, C.; Corke, P. Subset feature learning for fine-grained category classification. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Boston, MA, USA, 7–12 June 2015; IEEE: Piscataway, NJ, USA, 2015. [Google Scholar] [CrossRef][Green Version]
- Pan, S.J.; Yang, Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Munz, S.; Reiser, D. Approach for Image-Based Semantic Segmentation of Canopy Cover in Pea–Oat Intercropping. Agriculture 2020, 10, 354. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; IEEE: Piscataway, NJ, USA, 2016. [Google Scholar] [CrossRef]
- Sharpe, S.M.; Schumann, A.W.; Boyd, N.S. Detection of Carolina Geranium (Geranium carolinianum) Growing in Competition with Strawberry Using Convolutional Neural Networks. Weed Sci. 2018, 67, 239–245. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 1800–1807. [Google Scholar] [CrossRef]
- Keller, M.; Zecha, C.; Weis, M.; Link-Dolezal, J.; Gerhards, R.; Claupein, W. Competence center SenGIS—Exploring methods for multisensor data acquisition and handling for interdisciplinay research. In Proceedings of the 8th European Conference on Precision Agriculture 2011, Prague, Czech Republic, 11–14 July 2011; Czech Centre for Science and Society: Ampthill, UK; Prague, Czech Republic, 2011; pp. 491–500. [Google Scholar]
- Mink, R.; Dutta, A.; Peteinatos, G.; Sökefeld, M.; Engels, J.; Hahn, M.; Gerhards, R. Multi-Temporal Site-Specific Weed Control of Cirsium arvense (L.) Scop. and Rumex crispus L. in Maize and Sugar Beet Using Unmanned Aerial Vehicle Based Mapping. Agriculture 2018, 8, 65. [Google Scholar] [CrossRef]
- Meyer, G.E.; Neto, J.C.; Jones, D.D.; Hindman, T.W. Intensified fuzzy clusters for classifying plant, soil, and residue regions of interest from color images. Comput. Electron. Agric. 2004, 42, 161–180. [Google Scholar] [CrossRef]
- Theckedath, D.; Sedamkar, R.R. Detecting Affect States Using VGG16, ResNet50 and SE-ResNet50 Networks. SN Comput. Sci. 2020, 1. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. In Proceedings of the 26th Annual Conference on Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012; pp. 1097–1105. [Google Scholar]
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Chang, T.; Rasmussen, B.; Dickson, B.; Zachmann, L. Chimera: A Multi-Task Recurrent Convolutional Neural Network for Forest Classification and Structural Estimation. Remote Sens. 2019, 11, 768. [Google Scholar] [CrossRef]
- Teimouri, N.; Dyrmann, M.; Nielsen, P.; Mathiassen, S.; Somerville, G.; Jørgensen, R. Weed Growth Stage Estimator Using Deep Convolutional Neural Networks. Sensors 2018, 18, 1580. [Google Scholar] [CrossRef]
- López, V.; Fernández, A.; García, S.; Palade, V.; Herrera, F. An insight into classification with imbalanced data: Empirical results and current trends on using data intrinsic characteristics. Inf. Sci. 2013, 250, 113–141. [Google Scholar] [CrossRef]
- Batista, G.E.A.P.A.; Prati, R.C.; Monard, M.C. A study of the behavior of several methods for balancing machine learning training data. ACM SIGKDD Explor. Newsl. 2004, 6, 20–29. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. A review on the main challenges in automatic plant disease identification based on visible range images. Biosyst. Eng. 2016, 144, 52–60. [Google Scholar] [CrossRef]
- Gerhards, R.; Christensen, S. Real-time weed detection, decision making and patch spraying in maize, sugar beet, winter wheat and winter barley. Weed Res. 2003, 43, 385–392. [Google Scholar] [CrossRef]
- Tursun, N.; Datta, A.; Sakinmaz, M.S.; Kantarci, Z.; Knezevic, S.Z.; Chauhan, B.S. The critical period for weed control in three corn (Zea mays L.) types. Crop Prot. 2016, 90, 59–65. [Google Scholar] [CrossRef]
- Sökefeld, M.; Gerhards, R.; Oebel, H.; Therburg, R.D. Image acquisition for weed detection and identification by digital image analysis. In Proceedings of the 6th European Conference on Precision Agriculture (ECPA), Skiathos, Greece, 3–6 June 2007; Wageningen Academic Publishers: Wageningen, The Netherlands, 2007; Volume 6, pp. 523–529. [Google Scholar]
- Peña, J.; Torres-Sánchez, J.; Serrano-Pérez, A.; de Castro, A.; López-Granados, F. Quantifying Efficacy and Limits of Unmanned Aerial Vehicle (UAV) Technology for Weed Seedling Detection as Affected by Sensor Resolution. Sensors 2015, 15, 5609–5626. [Google Scholar] [CrossRef] [PubMed]
- Pflanz, M.; Nordmeyer, H.; Schirrmann, M. Weed Mapping with UAS Imagery and a Bag of Visual Words Based Image Classifier. Remote Sens. 2018, 10, 1530. [Google Scholar] [CrossRef]


| Plant Species | EPPO CODE | Total Images | Train Images | Validation Images | Testing Images |
|---|---|---|---|---|---|
| Alopecurus myosuroides Huds. | ALOMY | 7423 | 5196 | 1113 | 1114 |
| Amaranthus retroflexus L. | AMARE | 5274 | 3691 | 791 | 792 |
| Avena fatua L. | AVEFA | 12,409 | 8686 | 1861 | 1862 |
| Chenopodium album L. | CHEAL | 2690 | 1882 | 403 | 405 |
| Helianthus annuus L. | HELAN | 16,426 | 11,498 | 2463 | 2465 |
| Lamium purpureum L. | LAMPU | 7603 | 5322 | 1140 | 1141 |
| Matricaria chamomila L. | MATCH | 15,159 | 10,611 | 2273 | 2275 |
| Setaria spp. L. | SETSS | 2378 | 1664 | 355 | 359 |
| Solanum nigrum L. | SOLNI | 2979 | 2085 | 446 | 448 |
| Solanum tuberosum L. | SOLTU | 2742 | 1919 | 411 | 412 |
| Stellaria media Vill. | STEME | 6941 | 4858 | 1041 | 1042 |
| Zea mays L. | ZEAMX | 11,106 | 7774 | 1665 | 1667 |
| SUM | 93,130 | 65,186 | 13,962 | 13,982 |
| VGG16 | ResNet–50 | Xception | |
|---|---|---|---|
| Mean time per epoch (s) | 164 | 164 | 274 |
| Minimum epochs used | 469 | 511 | 538 |
| Maximum epochs used | 864 | 979 | 945 |
| Minimum top—1 test accuracy [%] | 81 | 97.2 | 97.5 |
| Maximum top—1 test accuracy [%] | 82.7 | 97.7 | 97.8 |
| Minimum final validation loss | 0.524 | 0.077 | 0.085 |
| Maximum final validation loss | 0.560 | 0.089 | 0.097 |
| Network Depth (Layers) | 16 | 50 | 71 |
| Total Network Parameters | 27,829,068 | 24,905,612 | 22,179,380 |
| Trained network parameters | 13,114,380 | 1,371,020 | 1,372,428 |
| Input Image Size (pixels) | 224 × 224 | 224 × 224 | 299 × 299 |
| Batch Size | 32 | ||
| Train Images per epoch | 15,600 | ||
| Validation Images per epoch | 4200 | ||
| (a) Mean Values of VGG16 | ||||||||||||
| ALOMY | AMARE | AVEFA | CHEAL | HELAN | LAMPU | MATCH | SETSS | SOLNI | SOLTU | STEME | ZEAMX | |
| ALOMY | 86.94 | 0.16 | 5.09 | 0.51 | 0.00 | 0.19 | 0.36 | 4.95 | 0.41 | 0.87 | 0.06 | 0.46 |
| AMARE | 0.74 | 92.12 | 0.06 | 0.53 | 0.00 | 0.76 | 1.26 | 3.08 | 0.47 | 0.62 | 0.16 | 0.19 |
| AVEFA | 14.46 | 0.34 | 66.57 | 1.24 | 0.17 | 0.41 | 4.26 | 2.99 | 1.83 | 1.08 | 0.19 | 6.45 |
| CHEAL | 0.00 | 4.05 | 1.51 | 55.01 | 0.47 | 6.74 | 1.80 | 2.79 | 21.60 | 3.65 | 1.78 | 0.59 |
| HELAN | 0.29 | 0.03 | 0.90 | 1.35 | 85.69 | 0.54 | 0.55 | 0.23 | 0.93 | 4.04 | 0.17 | 5.28 |
| LAMPU | 0.17 | 1.17 | 0.43 | 4.96 | 0.17 | 75.83 | 1.82 | 1.01 | 7.98 | 4.80 | 1.59 | 0.08 |
| MATCH | 1.28 | 0.89 | 0.84 | 0.15 | 0.00 | 1.01 | 93.36 | 0.88 | 0.05 | 0.44 | 0.70 | 0.40 |
| SETSS | 5.52 | 7.05 | 3.59 | 3.79 | 0.00 | 0.89 | 0.78 | 74.57 | 0.50 | 1.95 | 1.11 | 0.25 |
| SOLNI | 0.36 | 2.88 | 1.90 | 15.63 | 0.22 | 8.08 | 1.25 | 0.80 | 64.26 | 2.63 | 1.47 | 0.51 |
| SOLTU | 0.44 | 0.73 | 1.21 | 1.97 | 1.26 | 4.64 | 3.25 | 0.19 | 2.14 | 82.33 | 0.85 | 1.00 |
| STEME | 0.02 | 4.99 | 0.04 | 3.11 | 0.00 | 4.54 | 2.86 | 1.90 | 2.22 | 1.48 | 78.69 | 0.15 |
| ZEAMX | 0.56 | 0.11 | 6.66 | 0.97 | 3.88 | 0.22 | 1.33 | 0.47 | 1.13 | 1.93 | 0.52 | 82.21 |
| (b) Standard Deviation of VGG16 | ||||||||||||
| ALOMY | AMARE | AVEFA | CHEAL | HELAN | LAMPU | MATCH | SETSS | SOLNI | SOLTU | STEME | ZEAMX | |
| ALOMY | 1.23 | 0.09 | 0.91 | 0.10 | 0.00 | 0.09 | 0.17 | 0.78 | 0.13 | 0.19 | 0.06 | 0.15 |
| AMARE | 0.17 | 1.50 | 0.06 | 0.17 | 0.00 | 0.28 | 0.28 | 0.91 | 0.22 | 0.16 | 0.11 | 0.06 |
| AVEFA | 1.46 | 0.17 | 2.11 | 0.20 | 0.06 | 0.12 | 0.61 | 0.43 | 0.21 | 0.18 | 0.07 | 0.88 |
| CHEAL | 0.00 | 0.71 | 0.49 | 3.00 | 0.26 | 1.20 | 0.44 | 0.48 | 2.55 | 0.56 | 0.62 | 0.12 |
| HELAN | 0.02 | 0.02 | 0.11 | 0.17 | 0.69 | 0.11 | 0.07 | 0.04 | 0.14 | 0.45 | 0.07 | 0.51 |
| LAMPU | 0.06 | 0.30 | 0.13 | 0.69 | 0.05 | 1.46 | 0.34 | 0.17 | 1.28 | 0.37 | 0.48 | 0.03 |
| MATCH | 0.28 | 0.16 | 0.18 | 0.05 | 0.01 | 0.16 | 0.52 | 0.21 | 0.05 | 0.11 | 0.13 | 0.17 |
| SETSS | 1.50 | 1.47 | 0.70 | 0.97 | 0.00 | 0.45 | 0.41 | 2.24 | 0.24 | 0.45 | 0.51 | 0.15 |
| SOLNI | 0.15 | 0.59 | 0.36 | 2.19 | 0.00 | 1.70 | 0.20 | 0.36 | 2.94 | 0.70 | 0.29 | 0.10 |
| SOLTU | 0.18 | 0.24 | 0.24 | 0.17 | 0.26 | 1.15 | 0.36 | 0.15 | 0.40 | 1.73 | 0.29 | 0.17 |
| STEME | 0.04 | 0.94 | 0.05 | 0.93 | 0.00 | 1.14 | 0.24 | 0.26 | 0.63 | 0.46 | 2.80 | 0.05 |
| ZEAMX | 0.17 | 0.04 | 0.84 | 0.27 | 0.53 | 0.11 | 0.28 | 0.18 | 0.18 | 0.28 | 0.17 | 1.31 |
| (a) Mean Values of ResNet–50 | ||||||||||||
| ALOMY | AMARE | AVEFA | CHEAL | HELAN | LAMPU | MATCH | SETSS | SOLNI | SOLTU | STEME | ZEAMX | |
| ALOMY | 98.23 | 0.00 | 0.85 | 0.03 | 0.00 | 0.01 | 0.00 | 0.78 | 0.04 | 0.00 | 0.00 | 0.06 |
| AMARE | 0.00 | 99.66 | 0.00 | 0.08 | 0.00 | 0.00 | 0.13 | 0.01 | 0.06 | 0.00 | 0.00 | 0.06 |
| AVEFA | 3.26 | 0.01 | 95.14 | 0.15 | 0.11 | 0.13 | 0.17 | 0.01 | 0.35 | 0.22 | 0.00 | 0.45 |
| CHEAL | 0.00 | 0.44 | 0.55 | 91.52 | 0.05 | 0.69 | 0.00 | 0.74 | 4.97 | 0.82 | 0.00 | 0.22 |
| HELAN | 0.00 | 0.00 | 0.42 | 0.17 | 97.09 | 0.20 | 0.21 | 0.00 | 0.01 | 0.64 | 0.00 | 1.25 |
| LAMPU | 0.00 | 0.00 | 0.00 | 1.11 | 0.01 | 97.20 | 0.00 | 0.01 | 0.99 | 0.55 | 0.14 | 0.00 |
| MATCH | 0.04 | 0.42 | 0.00 | 0.00 | 0.00 | 0.00 | 99.54 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| SETSS | 2.91 | 0.09 | 0.03 | 0.90 | 0.00 | 0.06 | 0.06 | 95.54 | 0.00 | 0.00 | 0.34 | 0.06 |
| SOLNI | 0.02 | 0.37 | 0.92 | 5.58 | 0.02 | 2.01 | 0.02 | 0.10 | 90.33 | 0.62 | 0.00 | 0.00 |
| SOLTU | 0.19 | 0.00 | 0.08 | 0.11 | 0.00 | 0.57 | 0.05 | 0.00 | 0.00 | 98.84 | 0.11 | 0.05 |
| STEME | 0.00 | 0.17 | 0.00 | 0.15 | 0.00 | 0.00 | 0.03 | 0.15 | 0.09 | 0.00 | 99.41 | 0.00 |
| ZEAMX | 0.07 | 0.00 | 0.87 | 0.04 | 0.33 | 0.00 | 0.00 | 0.05 | 0.02 | 0.09 | 0.01 | 98.51 |
| (b) Standard Deviation of ResNet–50 | ||||||||||||
| ALOMY | AMARE | AVEFA | CHEAL | HELAN | LAMPU | MATCH | SETSS | SOLNI | SOLTU | STEME | ZEAMX | |
| ALOMY | 0.32 | 0.00 | 0.39 | 0.04 | 0.00 | 0.03 | 0.00 | 0.15 | 0.04 | 0.00 | 0.00 | 0.04 |
| AMARE | 0.00 | 0.20 | 0.00 | 0.13 | 0.00 | 0.00 | 0.10 | 0.04 | 0.06 | 0.00 | 0.00 | 0.06 |
| AVEFA | 0.76 | 0.02 | 0.87 | 0.07 | 0.04 | 0.03 | 0.08 | 0.02 | 0.07 | 0.07 | 0.00 | 0.14 |
| CHEAL | 0.00 | 0.16 | 0.19 | 0.86 | 0.10 | 0.16 | 0.00 | 0.23 | 0.61 | 0.35 | 0.00 | 0.14 |
| HELAN | 0.00 | 0.01 | 0.08 | 0.05 | 0.30 | 0.04 | 0.02 | 0.01 | 0.02 | 0.10 | 0.00 | 0.18 |
| LAMPU | 0.00 | 0.00 | 0.00 | 0.14 | 0.03 | 0.26 | 0.00 | 0.03 | 0.19 | 0.12 | 0.07 | 0.00 |
| MATCH | 0.04 | 0.10 | 0.01 | 0.00 | 0.00 | 0.00 | 0.10 | 0.00 | 0.00 | 0.00 | 0.01 | 0.00 |
| SETSS | 0.48 | 0.19 | 0.09 | 0.26 | 0.00 | 0.12 | 0.12 | 0.64 | 0.00 | 0.00 | 0.18 | 0.12 |
| SOLNI | 0.07 | 0.15 | 0.45 | 0.41 | 0.07 | 0.54 | 0.07 | 0.11 | 1.00 | 0.31 | 0.00 | 0.00 |
| SOLTU | 0.10 | 0.00 | 0.16 | 0.12 | 0.00 | 0.11 | 0.10 | 0.00 | 0.00 | 0.25 | 0.12 | 0.10 |
| STEME | 0.00 | 0.06 | 0.00 | 0.13 | 0.00 | 0.00 | 0.05 | 0.05 | 0.07 | 0.00 | 0.15 | 0.00 |
| ZEAMX | 0.02 | 0.00 | 0.12 | 0.05 | 0.12 | 0.00 | 0.00 | 0.03 | 0.03 | 0.05 | 0.02 | 0.19 |
| (a) Mean Values of Xception | ||||||||||||
| ALOMY | AMARE | AVEFA | CHEAL | HELAN | LAMPU | MATCH | SETSS | SOLNI | SOLTU | STEME | ZEAMX | |
| ALOMY | 97.63 | 0.03 | 1.21 | 0.00 | 0.00 | 0.00 | 0.01 | 1.11 | 0.01 | 0.00 | 0.00 | 0.01 |
| AMARE | 0.01 | 99.26 | 0.00 | 0.01 | 0.00 | 0.00 | 0.51 | 0.10 | 0.11 | 0.00 | 0.00 | 0.00 |
| AVEFA | 1.67 | 0.00 | 96.84 | 0.05 | 0.11 | 0.11 | 0.27 | 0.05 | 0.26 | 0.15 | 0.00 | 0.48 |
| CHEAL | 0.00 | 0.33 | 0.22 | 92.46 | 0.33 | 0.71 | 0.25 | 0.96 | 4.36 | 0.05 | 0.11 | 0.22 |
| HELAN | 0.00 | 0.00 | 0.48 | 0.09 | 97.37 | 0.19 | 0.12 | 0.00 | 0.03 | 0.37 | 0.00 | 1.34 |
| LAMPU | 0.00 | 0.04 | 0.02 | 0.59 | 0.13 | 98.15 | 0.00 | 0.01 | 0.61 | 0.38 | 0.07 | 0.00 |
| MATCH | 0.01 | 0.16 | 0.00 | 0.00 | 0.01 | 0.00 | 99.74 | 0.03 | 0.00 | 0.00 | 0.04 | 0.00 |
| SETSS | 2.10 | 0.06 | 0.15 | 0.56 | 0.00 | 0.00 | 0.09 | 96.69 | 0.00 | 0.06 | 0.25 | 0.03 |
| SOLNI | 0.15 | 0.25 | 0.97 | 4.64 | 0.07 | 2.08 | 0.05 | 0.00 | 91.49 | 0.17 | 0.02 | 0.10 |
| SOLTU | 0.22 | 0.00 | 0.00 | 0.30 | 0.59 | 0.43 | 0.08 | 0.08 | 0.00 | 98.14 | 0.11 | 0.05 |
| STEME | 0.00 | 0.07 | 0.00 | 0.05 | 0.00 | 0.00 | 0.03 | 0.14 | 0.09 | 0.01 | 99.61 | 0.00 |
| ZEAMX | 0.09 | 0.00 | 0.62 | 0.09 | 0.40 | 0.00 | 0.00 | 0.03 | 0.00 | 0.01 | 0.02 | 98.75 |
| (b) Standard Deviation of Xception | ||||||||||||
| ALOMY | AMARE | AVEFA | CHEAL | HELAN | LAMPU | MATCH | SETSS | SOLNI | SOLTU | STEME | ZEAMX | |
| ALOMY | 0.33 | 0.04 | 0.26 | 0.00 | 0.00 | 0.00 | 0.03 | 0.16 | 0.03 | 0.00 | 0.00 | 0.03 |
| AMARE | 0.04 | 0.24 | 0.00 | 0.04 | 0.00 | 0.00 | 0.16 | 0.12 | 0.11 | 0.00 | 0.00 | 0.00 |
| AVEFA | 0.64 | 0.00 | 0.65 | 0.05 | 0.05 | 0.04 | 0.06 | 0.05 | 0.04 | 0.04 | 0.00 | 0.10 |
| CHEAL | 0.00 | 0.12 | 0.18 | 0.89 | 0.23 | 0.27 | 0.00 | 0.36 | 0.87 | 0.10 | 0.17 | 0.18 |
| HELAN | 0.01 | 0.00 | 0.06 | 0.03 | 0.24 | 0.04 | 0.06 | 0.01 | 0.03 | 0.08 | 0.00 | 0.24 |
| LAMPU | 0.00 | 0.04 | 0.04 | 0.20 | 0.08 | 0.21 | 0.00 | 0.03 | 0.17 | 0.21 | 0.07 | 0.00 |
| MATCH | 0.02 | 0.06 | 0.00 | 0.00 | 0.02 | 0.00 | 0.08 | 0.04 | 0.00 | 0.01 | 0.04 | 0.00 |
| SETSS | 0.59 | 0.12 | 0.19 | 0.35 | 0.00 | 0.00 | 0.19 | 0.53 | 0.00 | 0.12 | 0.24 | 0.09 |
| SOLNI | 0.11 | 0.16 | 0.26 | 0.63 | 0.11 | 0.45 | 0.09 | 0.00 | 0.84 | 0.18 | 0.07 | 0.15 |
| SOLTU | 0.08 | 0.00 | 0.00 | 0.22 | 0.36 | 0.10 | 0.11 | 0.11 | 0.00 | 0.51 | 0.12 | 0.10 |
| STEME | 0.00 | 0.06 | 0.00 | 0.07 | 0.00 | 0.00 | 0.05 | 0.05 | 0.03 | 0.03 | 0.12 | 0.00 |
| ZEAMX | 0.04 | 0.00 | 0.15 | 0.06 | 0.12 | 0.00 | 0.00 | 0.03 | 0.00 | 0.02 | 0.06 | 0.31 |
| Plants Per | VGG 16 | ResNet–50 | Xception | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Category | precision | recall | f1-score | precision | recall | f1-score | precision | recall | f1-score | |
| ALOMY | 1114 | 0.77 | 0.86 | 0.81 | 0.93 | 0.99 | 0.96 | 0.94 | 0.98 | 0.96 |
| AMARE | 792 | 0.81 | 0.93 | 0.86 | 0.98 | 1.00 | 0.99 | 0.99 | 0.99 | 0.99 |
| AVEFA | 1862 | 0.85 | 0.69 | 0.76 | 0.98 | 0.95 | 0.97 | 0.98 | 0.95 | 0.97 |
| CHEAL | 405 | 0.47 | 0.57 | 0.51 | 0.87 | 0.92 | 0.89 | 0.90 | 0.93 | 0.92 |
| HELAN | 2465 | 0.97 | 0.86 | 0.91 | 1.00 | 0.97 | 0.98 | 0.99 | 0.97 | 0.98 |
| LAMPU | 1141 | 0.80 | 0.78 | 0.79 | 0.98 | 0.97 | 0.98 | 0.98 | 0.99 | 0.98 |
| MATCH | 2275 | 0.91 | 0.94 | 0.93 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 1.00 |
| SETSS | 359 | 0.53 | 0.77 | 0.63 | 0.96 | 0.95 | 0.96 | 0.93 | 0.96 | 0.95 |
| SOLNI | 448 | 0.51 | 0.63 | 0.57 | 0.92 | 0.90 | 0.91 | 0.93 | 0.92 | 0.92 |
| SOLTU | 412 | 0.57 | 0.82 | 0.67 | 0.93 | 0.99 | 0.96 | 0.95 | 0.98 | 0.96 |
| STEME | 1042 | 0.94 | 0.77 | 0.84 | 1.00 | 0.99 | 0.99 | 1.00 | 0.99 | 1.00 |
| ZEAMX | 1667 | 0.83 | 0.84 | 0.83 | 0.97 | 0.99 | 0.98 | 0.97 | 0.99 | 0.98 |
| accuracy | 13,982 | 0.82 | 0.97 | 0.98 | ||||||
| macro avg | 13,982 | 0.75 | 0.79 | 0.76 | 0.96 | 0.97 | 0.96 | 0.96 | 0.97 | 0.97 |
| weighted avg | 13,982 | 0.83 | 0.82 | 0.82 | 0.98 | 0.97 | 0.97 | 0.98 | 0.98 | 0.98 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peteinatos, G.G.; Reichel, P.; Karouta, J.; Andújar, D.; Gerhards, R. Weed Identification in Maize, Sunflower, and Potatoes with the Aid of Convolutional Neural Networks. Remote Sens. 2020, 12, 4185. https://doi.org/10.3390/rs12244185
Peteinatos GG, Reichel P, Karouta J, Andújar D, Gerhards R. Weed Identification in Maize, Sunflower, and Potatoes with the Aid of Convolutional Neural Networks. Remote Sensing. 2020; 12(24):4185. https://doi.org/10.3390/rs12244185
Chicago/Turabian StylePeteinatos, Gerassimos G., Philipp Reichel, Jeremy Karouta, Dionisio Andújar, and Roland Gerhards. 2020. "Weed Identification in Maize, Sunflower, and Potatoes with the Aid of Convolutional Neural Networks" Remote Sensing 12, no. 24: 4185. https://doi.org/10.3390/rs12244185
APA StylePeteinatos, G. G., Reichel, P., Karouta, J., Andújar, D., & Gerhards, R. (2020). Weed Identification in Maize, Sunflower, and Potatoes with the Aid of Convolutional Neural Networks. Remote Sensing, 12(24), 4185. https://doi.org/10.3390/rs12244185









