1. Introduction
Successful reforestation following harvest is an integral component of sustainable forest management, as the degree to which stands become established will influence the entire lifecycle of a forest [
1]. Given that regenerating stands are harvested approximately 80 years after planting, the reforestation phase represents a significant financial and ecological investment, the success of which rests heavily upon the ability to monitor and manage these young, regenerating stands [
1,
2]. The frequency at which stands are monitored, however, is currently limited by the cost of field-based assessment methods, which can result in poorly performing stands being assessed with insufficient frequency [
3].
The pronounced size, abundance, and remoteness of Alberta’s forested land base relative to other jurisdictions increases the cost and difficulty of operating field-based assessments. These factors are confounded by the restricted availability of field crews as well as poor road access to many harvested sites. Moreover, licensee holders are obligated to return harvested stands to a free-to-grow status, and silvicultural surveys determining this status present significant operational challenges, including additional financial constraints [
4]. However, the increasing use of remote sensing datasets for forest inventory and assessment has seen marked improvements to the speed, scale, and cost of forest monitoring and the continued development of these applications provides new solutions to address some of the difficulties managers are faced with around regeneration assessment [
5].
One area of long-term investment in optical remote sensing applications is the extraction of information at the individual tree level at a pace that is more efficient and less expensive than field surveying or photo-interpretation. This need has driven research into tree crown detection and delineation algorithms since the 1980s, when advances in digital optical sensors made individual tree crowns (ITC) visible in imagery and increased computing power made automated crown delineation possible [
6]. The foundational concept driving these algorithms is that in nadir imagery, treetops will receive more solar illumination than other parts of the crown. Accordingly, pixels corresponding to treetops will be the brightest pixels within a tree crown. In early studies (e.g., [
7]), tree crowns were detected by identifying the brightest pixels (local maxima) within a moving window of fixed dimensions (for example, 5 by 5 pixels) as the window scanned across an image. This technique was named local maxima filtering. Subsequently, the valley-following algorithm was developed [
8], which isolated ITC by detecting local minima in shaded gaps between crowns rather than by detecting local maxima within them. An extension of the valley-following algorithm, watershed algorithms saw wide-spread adoption for ITC delineation in the early 2000s [
5]. Watershed algorithms generate a surface in which the brightest pixels correspond to the lowest elevations where water would gather if the surface were flooded, thus representing watersheds. Boundaries formed where watersheds met, which allowed for discrete tree crowns to be delineated.
While effective for stands with evenly sized and well-spaced crowns, these algorithms faced challenges with complex forest conditions where crowns varied in diameter and grew in clustered formations [
5]. The fixed window size of local maxima filtering approaches was found to be inadequate under these conditions. Large windows captured large crowns and missed small ones, while small windows captured small crowns and counted large crowns multiple times. Local maxima filtering algorithms with variable window sizes were developed; however, overlapping crowns with minimal shadow between them were still a hindrance to both local maxima filtering and watershed algorithms. These algorithms also demonstrated sensitivity to solar angle and elevation as shadow-obscured crowns tended to be missed. In addition to these challenges, spatial resolution proved to be a barrier to the detection and delineation of ITC in regenerating forests. When Gougeon and Leckie [
9] applied their valley-following methods to regeneration in multispectral aerial imagery with a 30 cm spatial resolution, the small crowns of five-year old trees were completely missed. Later inquiries by Gougeon and Leckie [
10] yielded the delineation of approximately 85% of crowns with 2.5–4.0 m diameters using a semiautomatic methodology applied to IKONOS (GSD 1 m) imagery resampled to 50 cm spatial resolution. However, clusters of multiple crowns were commonly delineated as a single entity. Leckie [
11] reported similar errors for the delineation of 20 year-old conifers in multispectral aerial imagery with a 60 cm spatial resolution, which they attributed to imagery coarseness.
Currently, high spatial resolution RGB imagery is widely available, allowing even small tree crowns to be visible from practical altitudes. A new generation of algorithms have been developed to overcome some of the aforementioned challenges in delineating ITC. Convolutional neural networks (CNN) have been increasingly utilized for automatic semantic segmentation (the prediction of class labels for each pixel in an input image) in remotely sensed images and have been proven capable of outperforming conventional techniques [
12,
13,
14,
15,
16]. CNN are comprised of layers of convolution filters that work in succession to extract increasingly nuanced object features from input imagery. By analyzing annotated reference images, a CNN learns by example to activate layers that appropriately capture aspects of its target objects, resulting in powerful image segmentation capabilities that are not based exclusively on pixel brightness. Furthermore, trained CNN are highly transferable; the layer activation patterns learned by a CNN are stored in a single file, which can either be disseminated for wide-spread use or to initialize the training of a new CNN (the latter is known as transfer learning) [
17]. The aptitude of CNN for regeneration monitoring has been demonstrated by Fromm et al. [
18] and Pearse et al. [
19] who applied CNN to automatically detect individual conifer seedlings in UAV-acquired RGB imagery (DJI Mavic Pro and DJI Phantom 4 Pro, respectively). Likewise, Fuentes-Pacheco et al. [
20] employed a CNN to detect and delineate phenotypically variable fig plants and Kentsch et al. [
21] trained a CNN to detect and classify patches of invasive and native vegetation. Both of these studies used RGB imagery collected with a DJI Phantom 4 UAV.
The proliferation of CNN applications over the last decade and their application to remotely sensed datasets has precipitated the ongoing improvement of network architectures better designed to identify features on image datasets [
14]. One key improvement has been the increased use of region-based CNN (R-CNN), which address the difficulty of detecting multiple objects of varying aspect ratios by using a selective search algorithm [
22] to generate a set amount of region proposals from which object bounding boxes may be extracted [
23]. Building on this, Mask R-CNN [
24] provides an additional step to R-CNN with an additional network branch dedicated to generating a per-pixel, binary mask for each instance of an object detected within a bounding box [
25]. This distinguishes Mask R-CNN from other R-CNN, whose processing pipelines terminate with outputting bounding boxes. While Mask R-CNN was designed for object instance segmentation in natural imagery, the advantage of per-pixel object masks has seen quick uptake in medical imaging [
26,
27,
28] and subsequently, remote sensing [
29,
30,
31,
32]. In the context of forest monitoring, Braga et al. [
32] employed Mask R-CNN to delineate ITC in a tropical forest with RGB, UAV-acquired imagery. They reported precision, recall, and F1 scores of 0.91, 0.81, and 0.86, respectively.
In addition to accurate delineation of tree crowns, a key requirement for forest management is the ability to extract information such as height from ITC. Recent advances in digital aerial photogrammetry (DAP) have enabled the construction of three-dimensional (3D) point clouds from overlapping UAV-acquired imagery [
33,
34,
35]. From these 3D point clouds, canopy height models (CHM) [
36] and fine-resolution orthomosaics can be constructed simultaneously, providing access to valuable height and spectral data.
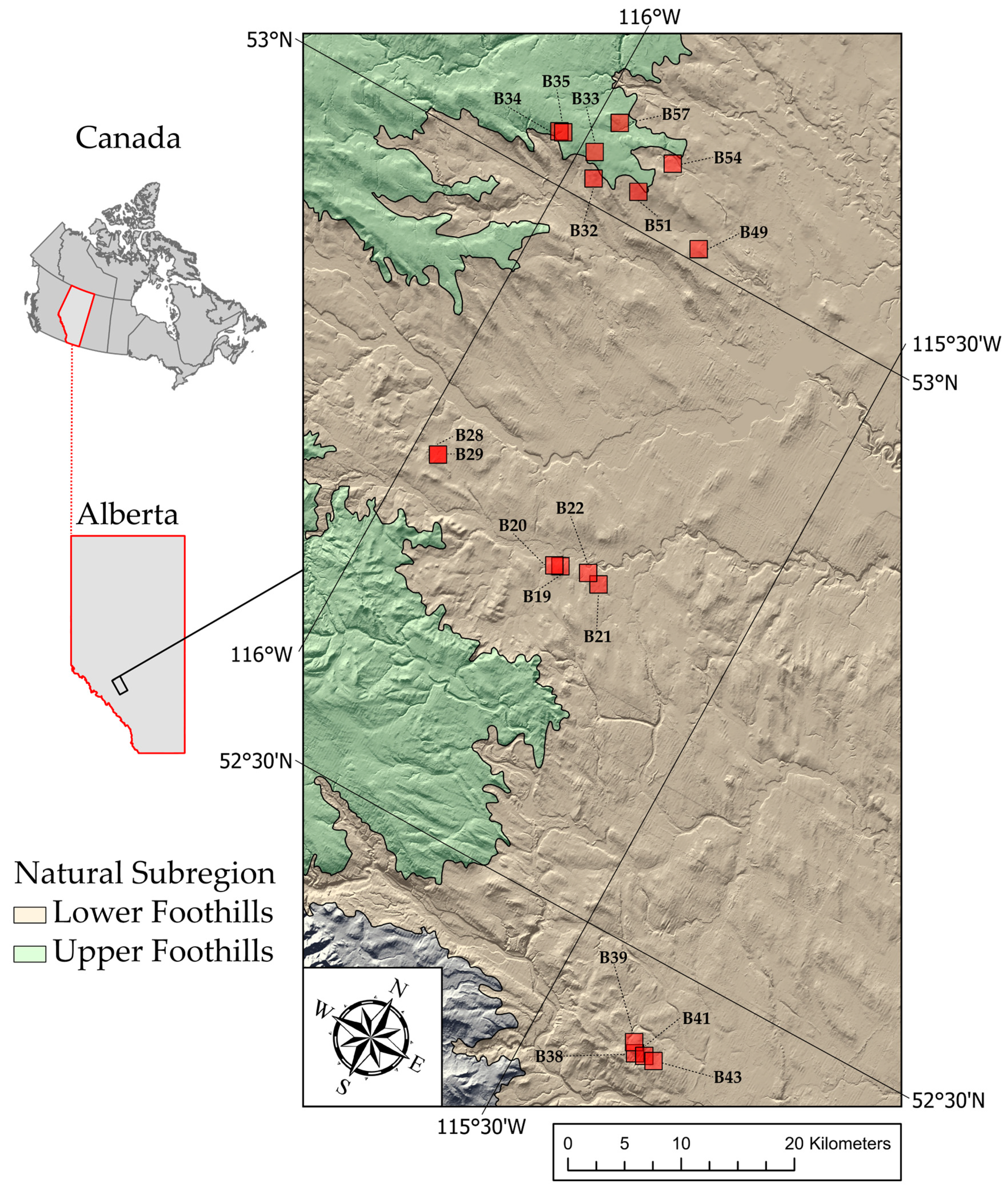
In this study, working in regenerating stands 12–14 years postharvest, we assess the ability of Mask R-CNN to delineate individual coniferous trees in imagery collected during the early spring, before deciduous green-up (leaf-off conditions). We then extract height information from DAP-derived CHM and compare the results to reference data collected in the field. To do so, we first train Mask-R-CNN to automatically delineate regeneration crowns using UAV-acquired RGB imagery. We assess the efficacy of this approach across a range of regenerating stands of different boreal forest types and growing conditions in managed forests in west-central Alberta, Canada. We then explore the potential to use predicted ITC polygons to automatically extract tree-level height information from a DAP-generated CHM. This research serves as a contribution towards the goal of automating regeneration monitoring using remote sensing techniques.
5. Conclusions
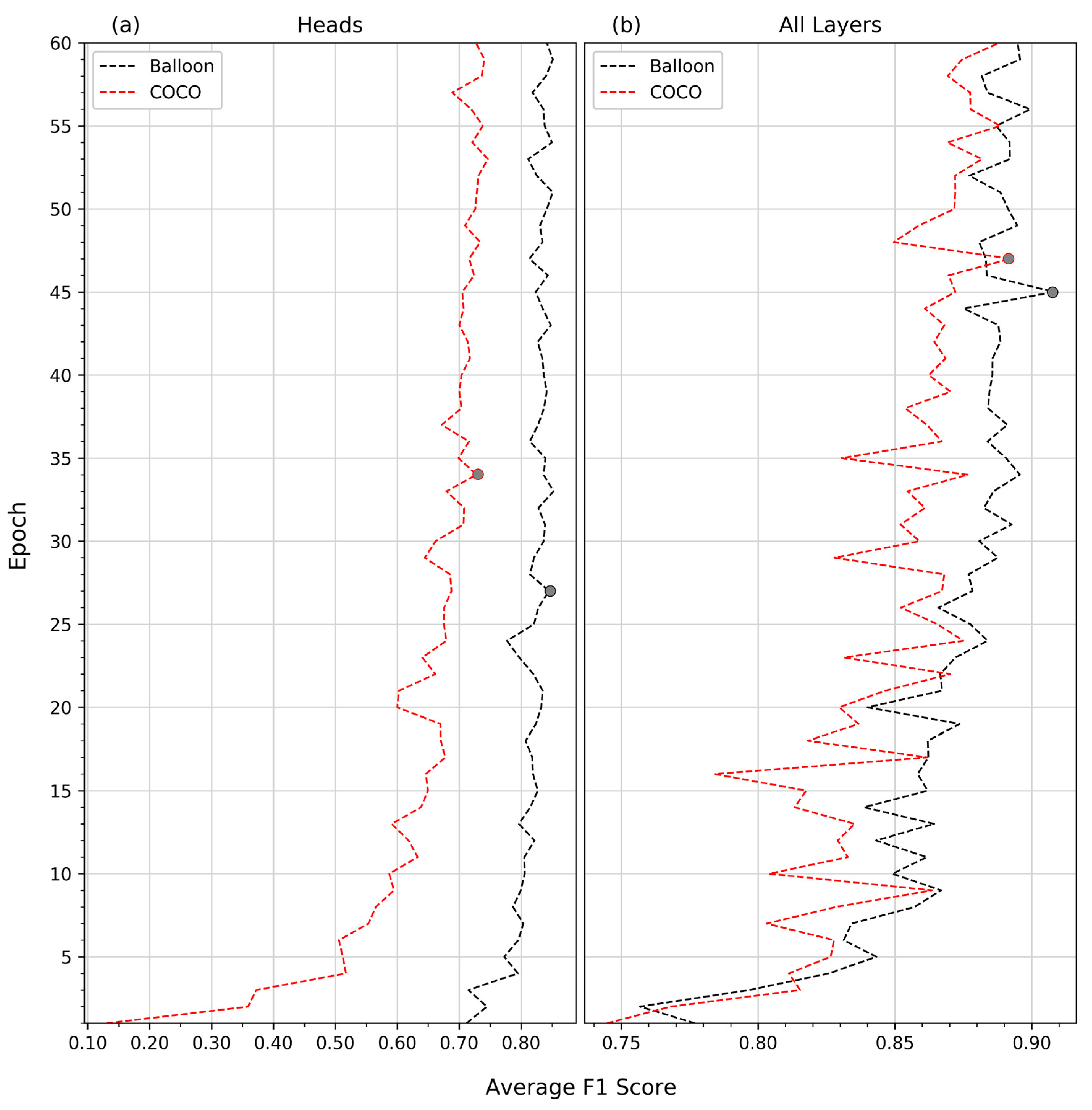
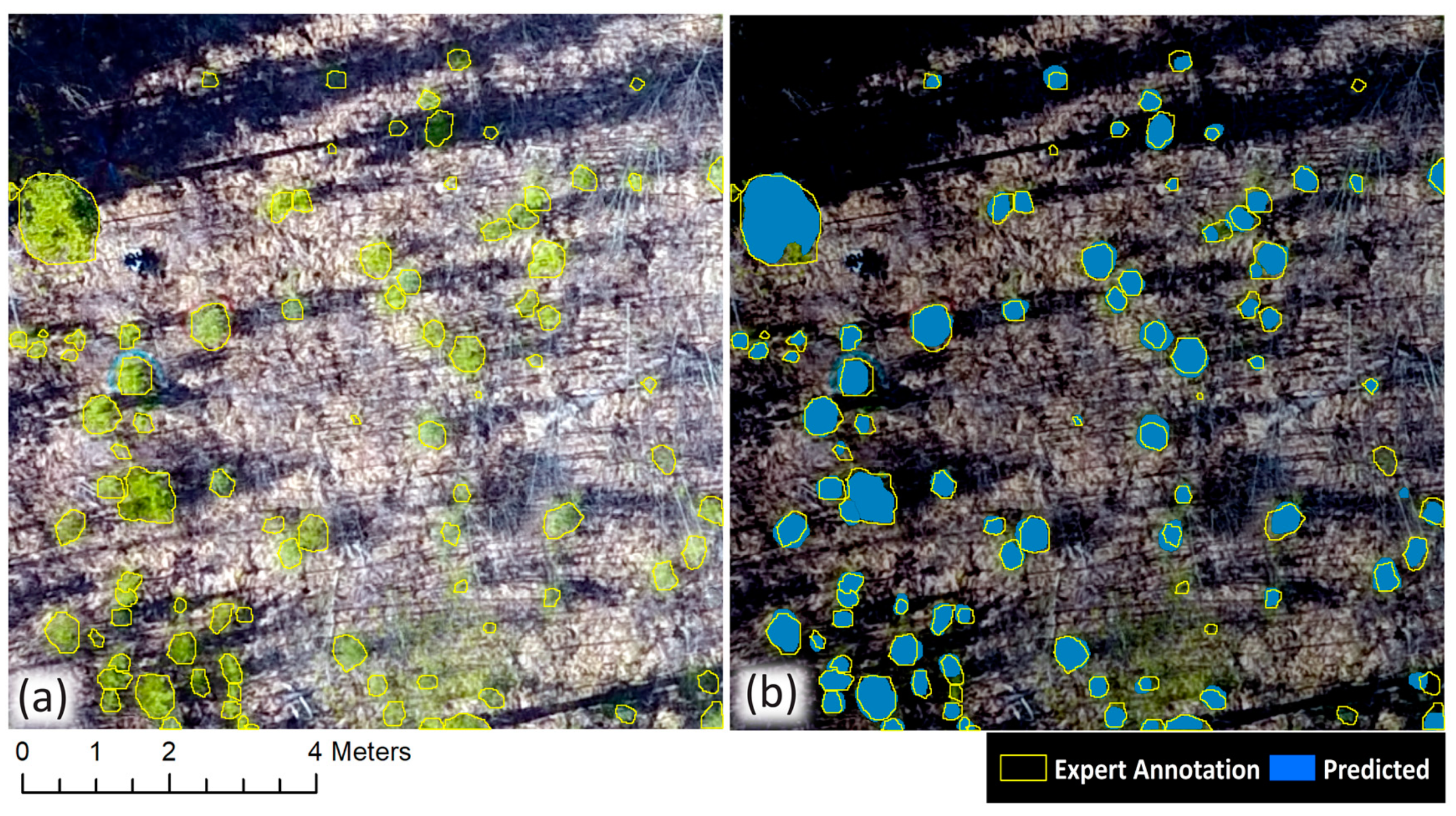
In this study, we have presented a workflow for the automatic delineation of individual conifer crowns in true colour orthomosaics using Mask R-CNN and the subsequent extraction of remote height measurements. Our best model was generated by Balloon pretrained network, where all layers of the network were adjusted during training. This achieved an average precision of 0.98, an average recall of 0.85, and an average F1 score of 0.91. Our results show that Mask R-CNN is capable of automatically delineating individual coniferous crowns in a range of conditions common to western boreal forest regeneration under leaf-off conditions and that these crown delineations can be used to extract height measurements at the individual tree level.
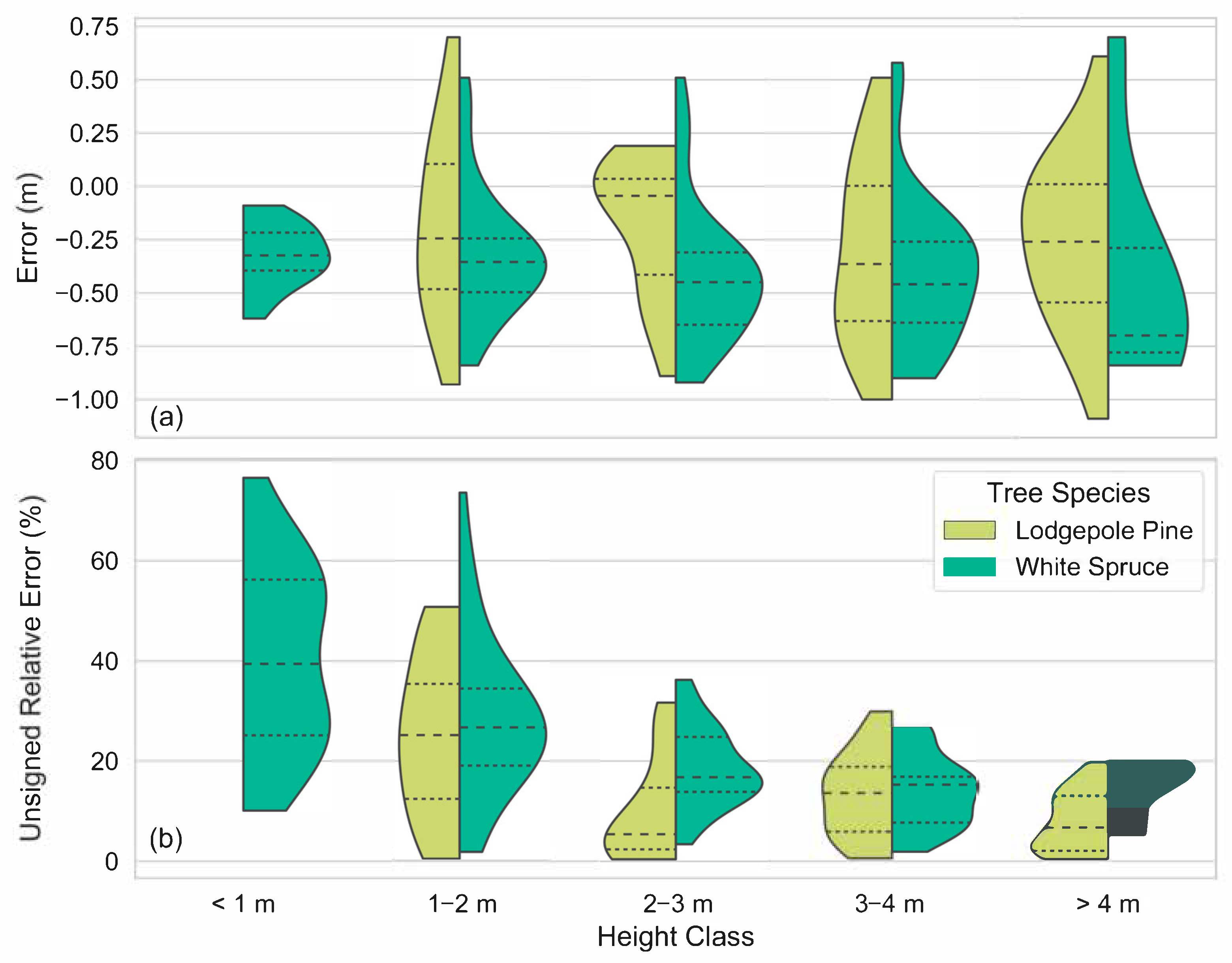
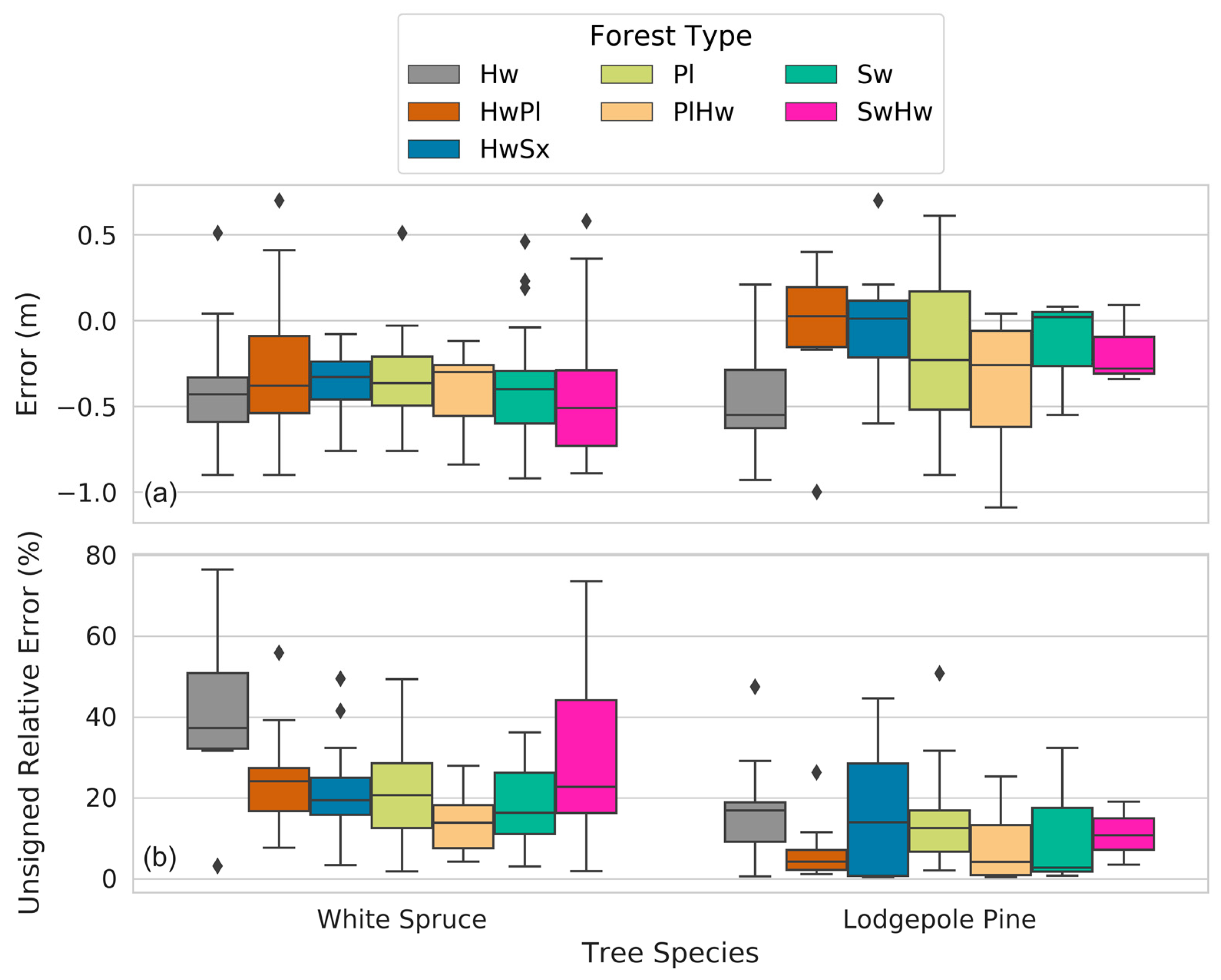
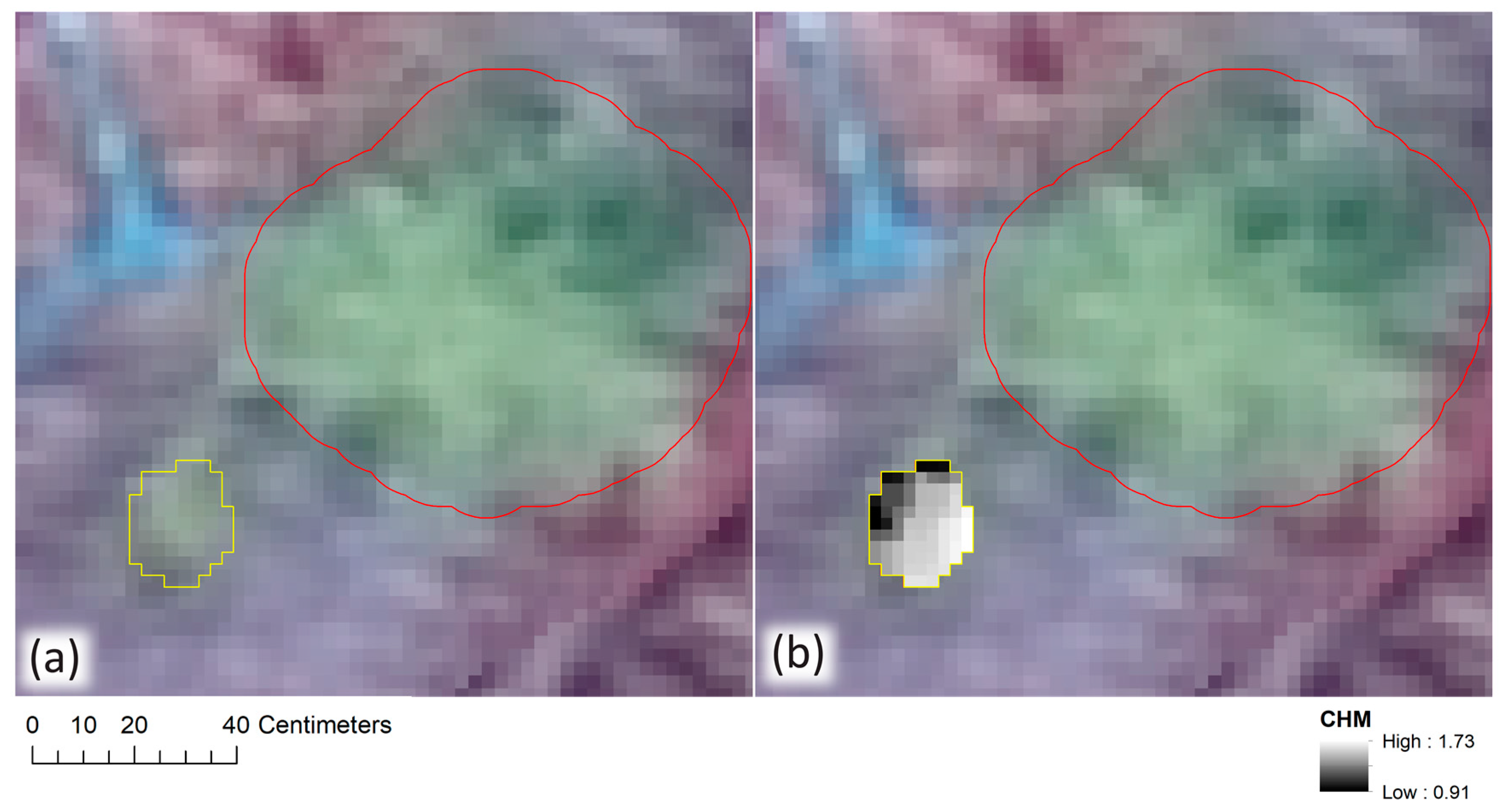
Our height measurement analysis found field and remote height measurements to be highly correlated (r2 = 0.93). Remote height measurements tended to underestimate field height measurements (162/194 samples) and mean error was greater for white spruce trees than it was for lodgepole pine (−0.37 m and −0.24 m, respectively). We speculated as to why the majority of remote height measurements underestimated their corresponding field samples as well as why lodgepole pines may be less underestimated than white spruce, and we offered suggestions for future studies to address this challenge. Furthermore, we observed that remote height measurements may overestimate their corresponding field height measurements when a delineated conifer crown contains pixels belonging to taller, adjacent coniferous or deciduous trees.
Our proposed methods to automatically delineate and extract height information for individual conifers in true colour imagery have the potential to improve the speed and scale at which forest regeneration monitoring can occur. However, techniques to classify tree species and delineate individual deciduous crowns from true colour imagery are still required if the criteria of field-based regeneration assessments are to be met by remote sensing approaches. As such, our research serves as a stepping-stone towards the remote automation of regeneration monitoring and future studies should focus on the deciduous components of boreal regeneration as well as the classification of individual tree species.