Ensemble Modelling of Skipjack Tuna (Katsuwonus pelamis) Habitats in the Western North Pacific Using Satellite Remotely Sensed Data; a Comparative Analysis Using Machine-Learning Models
Abstract
:1. Introduction
2. Materials and Methods
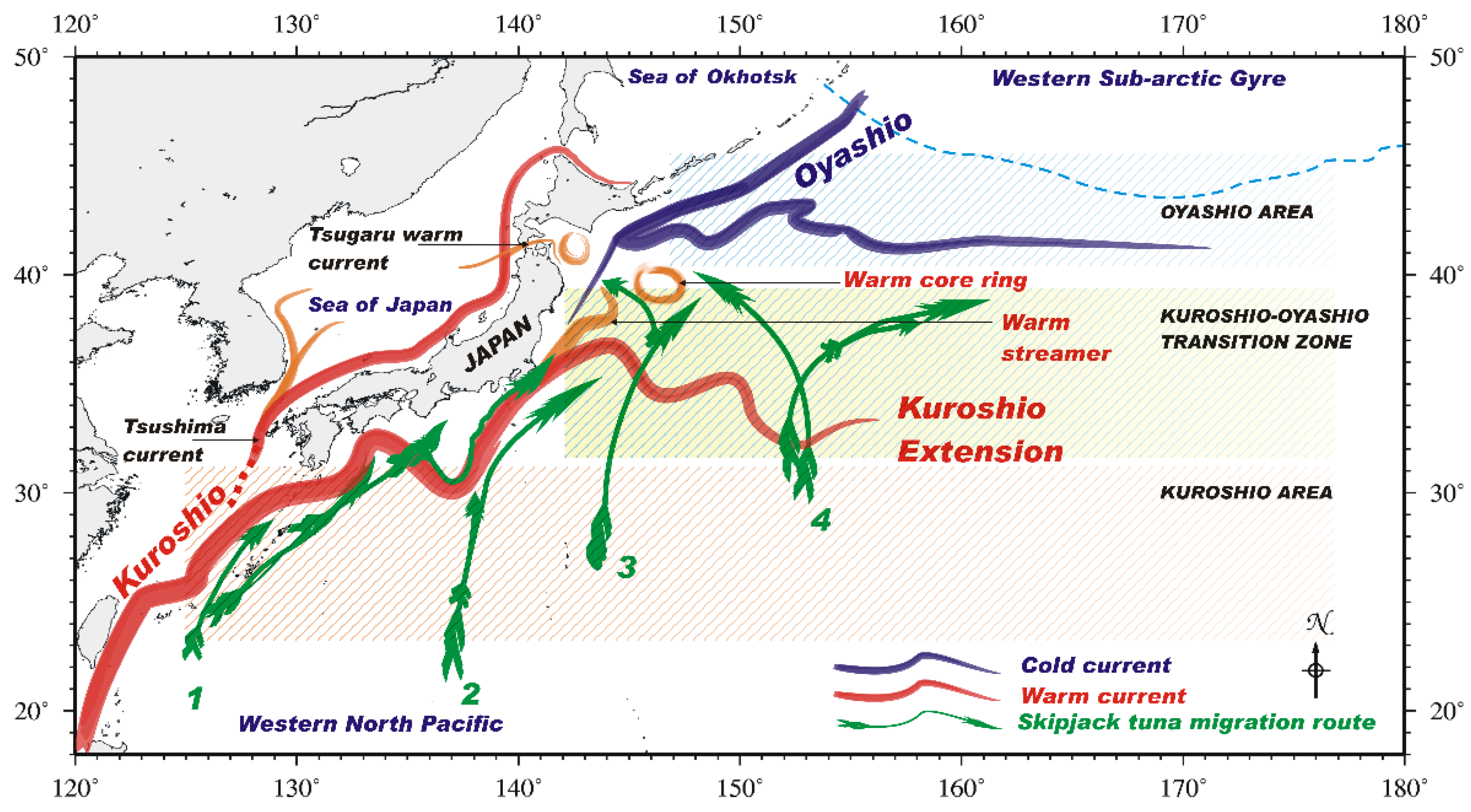
2.1. Study Area
2.2. Fishery Data
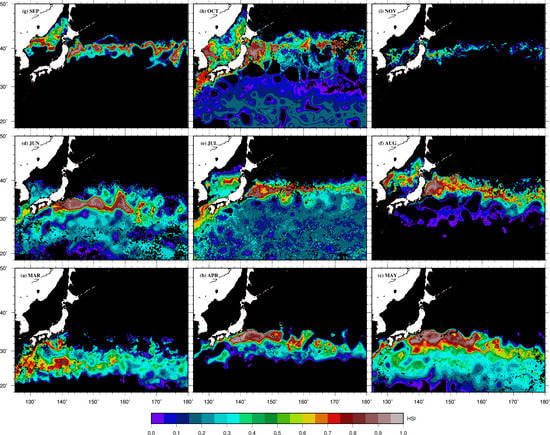
2.3. Environmental Data
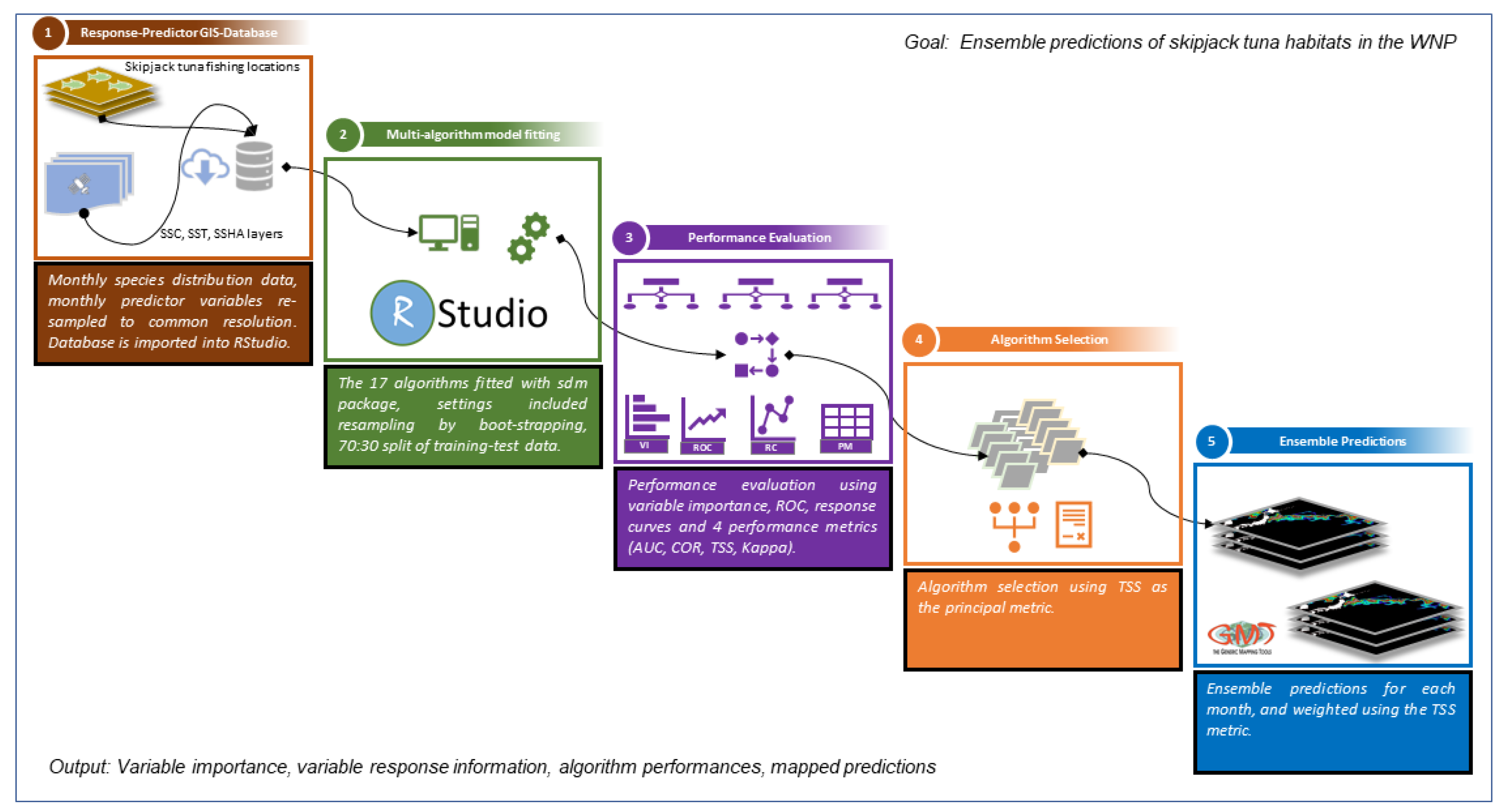
2.4. Habitat Modeling Using sdm Package
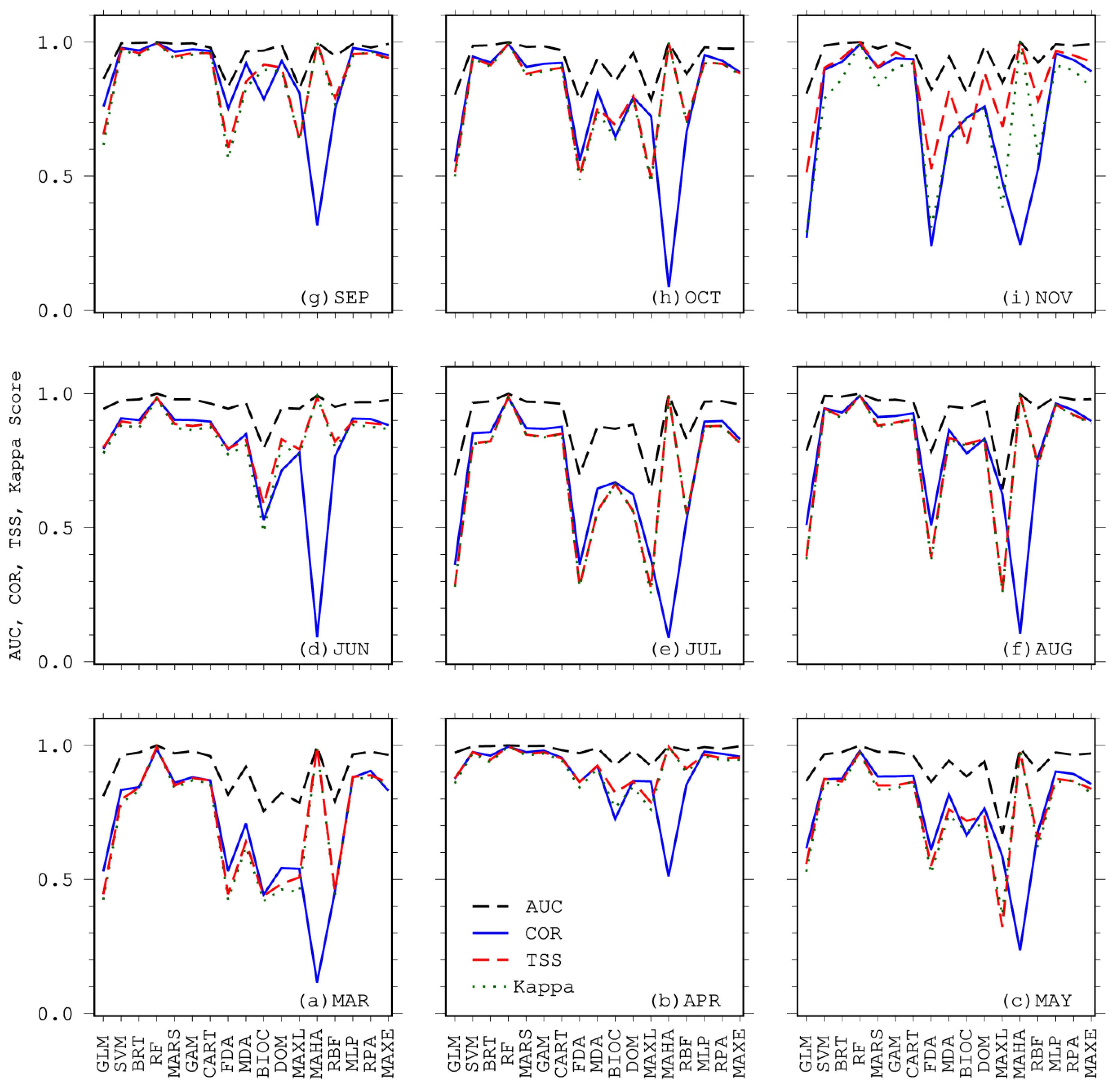
2.5. Evaluation of Model Performance
2.6. Ensemble Model Development
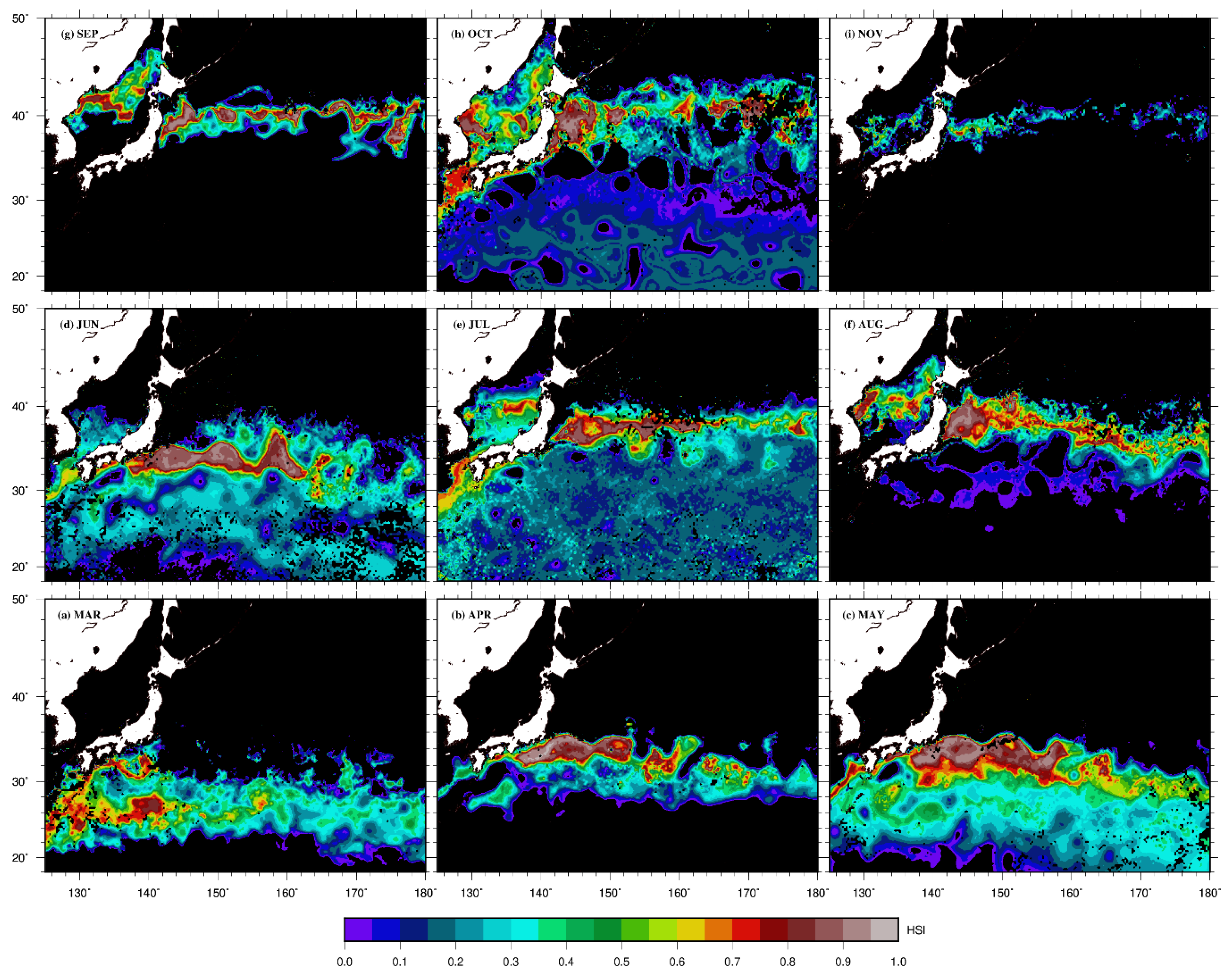
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hutchinson, G.E. Concluding Remarks. Cold Spring Harb. Symp. Quant. Biol. 1957, 22, 415–427. [Google Scholar] [CrossRef]
- Vandermeer, J.H. Niche Theory. Annu. Rev. Ecol. Syst. 1972, 3, 107–132. [Google Scholar] [CrossRef]
- Hirzel, A.H.; Le Lay, G. Habitat suitability modelling and niche theory. J. Appl. Ecol. 2008, 45, 1372–1381. [Google Scholar] [CrossRef]
- Kobayashi, D.R.; Farman, R.; Polovina, J.J.; Parker, D.M.; Rice, M.; Balazs, G.H. ‘Going with the Flow’ or Not: Evidence of Positive Rheotaxis in Oceanic Juvenile Loggerhead Turtles (Caretta caretta) in the South Pacific Ocean Using Satellite Tags and Ocean Circulation Data. PLoS ONE 2014, 9, e103701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mugo, R.; Saitoh, S.-I.; Nihira, A.; Kuroyama, T. Habitat characteristics of skipjack tuna (Katsuwonus pelamis) in the western North Pacific: A remote sensing perspective. Fish. Oceanogr. 2010, 19, 382–396. [Google Scholar] [CrossRef]
- Zainuddin, M.; Kiyofuji, H.; Saitoh, K.; Saitoh, S.-I. Using multi-sensor satellite remote sensing and catch data to detect ocean hot spots for albacore (Thunnus alalunga) in the northwestern North Pacific. Deep Sea Res. Part II Top. Stud. Oceanogr. 2006, 53, 419–431. [Google Scholar] [CrossRef]
- Zainuddin, M.; Saitoh, K.; Saitoh, S.-I. Albacore (Thunnus alalunga) fishing ground in relation to oceanographic conditions in the western North Pacific Ocean using remotely sensed satellite data. Fish. Oceanogr. 2008, 17, 61–73. [Google Scholar] [CrossRef] [Green Version]
- Mugo, R.M.; Saitoh, S.-I.; Takahashi, F.; Nihira, A.; Kuroyama, T. Evaluating the role of fronts in habitat overlaps between cold and warm water species in the western North Pacific: A proof of concept. Deep Sea Res. Part II Top. Stud. Oceanogr. 2014, 107, 29–39. [Google Scholar] [CrossRef]
- Robinson, N.M.; Nelson, W.A.; Costello, M.J.; Sutherland, J.E.; Lundquist, C.J. A Systematic Review of Marine-Based Species Distribution Models (SDMs) with Recommendations for Best Practice. Front. Mar. Sci. 2017, 4, 421. [Google Scholar] [CrossRef] [Green Version]
- Thuiller, W.; Lafourcade, B.; Engler, R.; Araújo, M.B. BIOMOD—A platform for ensemble forecasting of species distributions. Ecography 2009, 32, 369–373. [Google Scholar] [CrossRef]
- Muñoz, M.E.D.S.; De Giovanni, R.; De Siqueira, M.F.; Sutton, T.; Brewer, P.; Pereira, R.S.; Canhos, D.A.L.; Canhos, V.P. openModeller: A generic approach to species’ potential distribution modelling. Geoinformatica 2009, 15, 111–135. [Google Scholar] [CrossRef]
- Guo, Q.; Liu, Y. ModEco: An integrated software package for ecological niche modeling. Ecography 2010, 33, 637–642. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Elith, J. Species Distribution Modeling. 2016. Available online: https://rspatial.org/raster/sdm/index.html (accessed on 21 May 2020).
- Iturbide, M.; Bedia, J.; Herrera, S.; Del Hierro, O.; Pinto, M.; Gutiérrez, J.M. A framework for species distribution modelling with improved pseudo-absence generation. Ecol. Model. 2015, 312, 166–174. [Google Scholar] [CrossRef] [Green Version]
- Naimi, B.; Araújo, M.B. sdm: A reproducible and extensible R platform for species distribution modelling. Ecography 2016, 39, 368–375. [Google Scholar] [CrossRef] [Green Version]
- Kaschner, K.; Rius-Barile, J.; Kesner-Reyes, K.; Garilao, C.; Kullander, S.O.; Rees, T.; Froese, R. AquaMaps: Predicted Range Maps for Aquatic Species; World Wide Web Electronic Publication. 2019. Available online: https://www.aquamaps.org/ (accessed on 21 May 2020).
- Kachelriess, D.; Wegmann, M.; Gollock, M.; Pettorelli, N. The application of remote sensing for marine protected area management. Ecol. Indic. 2014, 36, 169–177. [Google Scholar] [CrossRef]
- Jones, M.C.; Cheung, W.W.L. Multi-model ensemble projections of climate change effects on global marine biodiversity. ICES J. Mar. Sci. 2014, 72, 741–752. [Google Scholar] [CrossRef] [Green Version]
- Byrne, M.; Gall, M.; Wolfe, K.; Agüera, A. From pole to pole: The potential for the Arctic seastar Asterias amurensis to invade a warming Southern Ocean. Glob. Chang. Biol. 2016, 22, 3874–3887. [Google Scholar] [CrossRef]
- Melo-Merino, S.M.; Reyes-Bonilla, H.; Lira-Noriega, A. Ecological niche models and species distribution models in marine environments: A literature review and spatial analysis of evidence. Ecol. Model. 2020, 415, 108837. [Google Scholar] [CrossRef]
- Zainuddin, M.; Farhum, A.; Safruddin, S.; Selamat, M.B.; Sudirman, S.; Nurdin, N.; Syamsuddin, M.; Ridwan, M.; Saitoh, S.-I. Detection of pelagic habitat hotspots for skipjack tuna in the Gulf of Bone-Flores Sea, southwestern Coral Triangle tuna, Indonesia. PLoS ONE 2017, 12, e0185601. [Google Scholar] [CrossRef] [Green Version]
- Alabia, I.D.; Saitoh, S.-I.; Igarashi, H.; Ishikawa, Y.; Usui, N.; Kamachi, M.; Awaji, T.; Seito, M. Ensemble squid habitat model using three-dimensional ocean data. ICES J. Mar. Sci. 2016, 73, 1863–1874. [Google Scholar] [CrossRef] [Green Version]
- Erauskin-Extramiana, M.; Arrizabalaga, H.; Hobday, A.J.; Cabré, A.; Ibaibarriaga, L.; Arregui, I.; Murua, H.; Chust, G. Large-scale distribution of tuna species in a warming ocean. Glob. Chang. Biol. 2019, 25, 2043–2060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Báez, J.; Barbosa, A.M.; Pascual, P.; Ramos, M.L.; Abascal, F. Ensemble modeling of the potential distribution of the whale shark in the Atlantic Ocean. Ecol. Evol. 2019, 10, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Shabani, F.; Kumar, L.; Ahmadi, M. A comparison of absolute performance of different correlative and mechanistic species distribution models in an independent area. Ecol. Evol. 2016, 6, 5973–5986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, K.S.; Bradley, B.A.; Cord, A.F.; Rocchini, D.; Tuanmu, M.-N.; Schmidtlein, S.; Turner, W.; Wegmann, M.; Pettorelli, N. Will remote sensing shape the next generation of species distribution models? Remote Sens. Ecol. Conserv. 2015, 1, 4–18. [Google Scholar] [CrossRef] [Green Version]
- Turner, W. Sensing biodiversity. Science 2014, 346, 301–302. [Google Scholar] [CrossRef]
- Wild, A.; Hampton, J. A review of the biology and fisheries for skipjack tuna, Katsuwonus pelamis, in the Pacific Ocean. FAO Fish. Tech. Pap. 1994, 336, 1–151. [Google Scholar]
- Nihira, A. Studies on the behavioral ecology and physiology of migratory fish schools of skipjack tuna (Katsuwonus pelamis) in the oceanic frontal area [Japan]. Bull. Tohoku Natl. Fish. Res. Inst. 1996, 58, 137–233. [Google Scholar]
- Saitoh, S.; Kosaka, S.; Iisaka, J. Satellite infrared observations of Kuroshio warm-core rings and their application to study of Pacific saury migration. Deep Sea Res. Part A Oceanogr. Res. Pap. 1986, 33, 1601–1615. [Google Scholar] [CrossRef]
- Sugimoto, T.; Tameishi, H. Warm-core rings, streamers and their role on the fishing ground formation around Japan. Deep Sea Res. Part A Oceanogr. Res. Pap. 1992, 39, S183–S201. [Google Scholar] [CrossRef]
- Sund, P.N.; Blackburn, M.; Williams, F. Tunas and their environment in the Pacific Ocean: A review. Oceanogr. Mar. Biol. Ann. Rev. 1981, 19, 443–512. [Google Scholar]
- Madureira, L.S.P.; Coletto, J.L.; Pinho, M.P.; Weigert, S.C.; Varela, C.M.; Campello, M.E.S.; Llopart, A. Skipjack (Katsuwonus pelamis) fishery improvement project: From satellite and 3D oceanographic models to acoustics, towards predator-prey landscapes. In Proceedings of the 2017 IEEE/OES Acoustics in Underwater Geosciences Symposium (RIO Acoustics), Rio de Janeiro, Brazil, 25–27 July 2017; pp. 1–7. [Google Scholar] [CrossRef]
- Nakamura, E.L. Food and Feeding Habits of Skipjack Tuna (Katsuwonus pelamis) from the Marquesas and Tuamotu Islands. Trans. Am. Fish. Soc. 1965, 94, 236–242. [Google Scholar] [CrossRef]
- Iizuka, K.; Asano, M.; Naganuma, A. Feeding habits of skipjack tuna (Katsuwonus pelamis Linnaeus) caught by pole and line and the state of young skipjack tuna distribution in the tropical seas of the Western Pacific Ocean. Bull. Tohoku Reg. Fish. Res. Lab. 1989, 51, 107–116. [Google Scholar]
- Kiyofuji, H.; Aoki, Y.; Kinoshita, J.; Okamoto, S.; Masujima, M.; Matsumoto, T.; Fujioka, K.; Ogata, R.; Nakao, T.; Sugimoto, N.; et al. Northward migration dynamics of skipjack tuna (Katsuwonus pelamis) associated with the lower thermal limit in the western Pacific Ocean. Prog. Oceanogr. 2019, 175, 55–67. [Google Scholar] [CrossRef]
- Ogura, M. Swimming Behavior of Skipjack, Katsuwonus pelamis, Observed by the Data Storage Tag at the Northwestern Pacific, Off Northern Japan, in Summer of 2001 and 2002; SCTB16 Working Paper; National Research Institute of Far Seas Fisheries: Shizuoka, Japan, 2003; Volume 16, pp. 1–10. Available online: http://wwwx.spc.int/coastfish/Sections/reef/Library/Meetings/SCTB/16/SKJ_7.pdf (accessed on 24 April 2020).
- Schaefer, K.M.; Fuller, D.W. Vertical movement patterns of skipjack tuna (Katsuwonus pelamis) in the eastern equatorial Pacific Ocean, as revealed with archival tags. Fish. Bull. 2007, 105, 379–389. [Google Scholar]
- Saitoh, S.; Chassot, E.; Dwivedi, R.; Fonteneau, A.; Kiyofuji, H.; Kumari, B. Remote Sensing Applications to Fish Harvesting. In Remote Sensing in Fisheries and Aquaculture; IOCCG: Dartmouth, NS, Canada, 2009; Volume 8. [Google Scholar]
- Fujino, K. Range of the skipjack tuna sub-population in the western Pacific Ocean. In Proceedings of the Second Symposium on the Results of the Cooperative Study of the Kuroshio and Adjacent Region, Tokyo, Japan, 28 September–1 October 1972; pp. 373–384. [Google Scholar]
- Matsumoto, W.M. Distribution, Relative Abundance and Movement of Skipjack Tuna, Katsuwonus pelamis, in the Pacific Ocean Based on Japanese Tuna Longline Catches 1964–67; NOAA Technical Report, NMFS SSRF; National Marine Fisheries Service: Seattle, Washington, USA, 1975; Volume 695, pp. 1–30.
- Akiyama, H.; Hidaka, K.; Hirai, M.; Ishida, Y.; Moku, M.; Sugimoto, S. Oyashio and Kuroshio. In Marine Ecosystems of the North Pacific; Perry, R.I., Mckinnell, S.M., Eds.; PICES: Sidney, BC, Canada, 2004; Volume 1, pp. 113–127. [Google Scholar]
- Yasuda, I. Hydrographic Structure and Variability in the Kuroshio-Oyashio Transition Area. J. Oceanogr. 2003, 59, 389–402. [Google Scholar] [CrossRef]
- Sakurai, Y. An overview of the Oyashio ecosystem. Deep Sea Res. Part II Top. Stud. Oceanogr. 2007, 54, 2526–2542. [Google Scholar] [CrossRef] [Green Version]
- Kawai, H. Hydrography of the Kuroshio Extension. In Kuroshio, Its Physical Aspects; University of Tokyo Press: Tokyo, Japan, 1972; pp. 235–352. [Google Scholar]
- Talley, L.D.; Nagata, Y.; Fujimura, M.; Iwao, T.; Kono, T.; Inagake, D.; Hirai, M.; Okuda, K. North Pacific Intermediate Water in the Kuroshio/Oyashio Mixed Water Region. J. Phys. Oceanogr. 1995, 25, 475–501. [Google Scholar] [CrossRef] [Green Version]
- Yasuda, I.; Okuda, K.; Hirai, M. Evolution of a Kuroshio warm-core ring—Variability of the hydrographic structure. Deep Sea Res. Part A Oceanogr. Res. Pap. 1992, 39, S131–S161. [Google Scholar] [CrossRef]
- Seki, M.P.; Flint, E.N.; Howell, E.; Ichii, T.; Polovina, J.J.; Yatsu, A. Transition Zone. In Marine Ecosystems of the North Pacific; PICES: Sidney, BC, Canada, 2004; Volume 1, pp. 201–209. [Google Scholar]
- Tameishi, H. Understanding Japanese sardine migrations using acoustic and other aids. ICES J. Mar. Sci. 1996, 53, 167–171. [Google Scholar] [CrossRef]
- Hirzel, A.H.; Hausser, J.; Chessel, D.; Perrin, N. Ecological Niche Factor Analysis: How to compute habitat suitability maps without absence data? Ecology 2002, 83, 2027–2036. [Google Scholar] [CrossRef]
- Wilson, C.; Morales, J.; Nayak, S.; Asanuma, I.; Feldman, G. Ocean-color radiometry and fisheries. In Why Ocean Colour? The Societal Benefits of Ocean-Colour Technology; Reports of the International Ocean-Colour Coordinating Group; IOCCG: Dartmouth, NS, Canada, 2008; pp. 47–57. [Google Scholar]
- Mueller, J.L. SeaWiFS algorithm for the diffuse attenuation coefficient, K(490), using water-leaving radiances at 490 and 555 nm. In SeaWiFS Postlaunch Technical Report Series: Volume 11; Technical Report; NASA Goddard Space Flight Center: Greenbelt, MD, USA, 2000; Volume 11, pp. 24–27. [Google Scholar]
- Takahashi, W.; Kawamura, H. Detection method of the Kuroshio front using the satellite-derived chlorophyll-a images. Remote Sens. Environ. 2005, 97, 83–91. [Google Scholar] [CrossRef]
- Ayers, J.M.; Lozier, M.S. Physical controls on the seasonal migration of the North Pacific transition zone chlorophyll front. J. Geophys. Res. 2010, 115, 05001. [Google Scholar] [CrossRef] [Green Version]
- Baith, K.; Lindsay, R.; Fu, G.; McClain, C.R. Data analysis system developed for ocean color satellite sensors. Eos Trans. AGU 2001, 82, 202. [Google Scholar] [CrossRef]
- Wessel, P.; Luís, J.F.; Uieda, L.; Scharroo, R.; Wobbe, F.; Smith, W.; Tian, D. The Generic Mapping Tools Version 6. Geochem. Geophys. Geosyst. 2019, 20, 5556–5564. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Pourghasemi, H.R.; Zhang, S.; Wang, J.A. Comparative Study of Functional Data Analysis and Generalized Linear Model Data-Mining Techniques for Landslide Spatial Modeling. In Spatial Modeling in GIS and R for Earth and Environmental Sciences; Elsevier: Amsterdam, The Netherlands, 2019; pp. 467–484. [Google Scholar]
- De La Hoz, C.F.; Ramos, E.; Puente, A.; Juanes, J.A. Climate change induced range shifts in seaweeds distributions in Europe. Mar. Environ. Res. 2019, 148, 1–11. [Google Scholar] [CrossRef]
- Shabani, F.; Kumar, L.; Ahmadi, M. Assessing Accuracy Methods of Species Distribution Models: AUC, Specificity, Sensitivity and the True Skill Statistic. Acta Sci. Hum. Soc. Sci. 2018, 18, 7–18. [Google Scholar]
- Hanley, J.A.; McNeil, B.J. The meaning and use of the area under a Receiver Operating Characteristic (ROC) curve. Radiology 1982, 143, 29–36. [Google Scholar] [CrossRef] [Green Version]
- Elith, J.; Graham, C.; Anderson, R.P.; Dudík, M.; Ferrier, S.; Guisan, A.; Hijmans, R.; Huettmann, F.; Leathwick, J.R.; Lehmann, A.; et al. Novel methods improve prediction of species’ distributions from occurrence data. Ecography 2006, 29, 129–151. [Google Scholar] [CrossRef] [Green Version]
- Allouche, O.; Tsoar, A.; Kadmon, R. Kadmon Assessing the accuracy of species distribution models: Prevalence, kappa and the true skill statistic (TSS): Assessing the accuracy of distribution models. J. Appl. Ecol. 2006, 43, 1223–1232. [Google Scholar] [CrossRef]
- Cohen, J. A Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Hao, T.; Elith, J.; Guillera-Arroita, G.; Lahoz-Monfort, J.J. A review of evidence about use and performance of species distribution modelling ensembles like BIOMOD. Divers. Distrib. 2019, 25, 839–852. [Google Scholar] [CrossRef]
- Zhang, Z.; Mammola, S.; Zhang, H. Does weighting presence records improve the performance of species distribution models? A test using fish larval stages in the Yangtze Estuary. Sci. Total Environ. 2020, 741, 140393. [Google Scholar] [CrossRef] [PubMed]
- Araujo, M.; New, M. Ensemble forecasting of species distributions. Trends Ecol. Evol. 2007, 22, 42–47. [Google Scholar] [CrossRef]
- Elith, J.; Graham, C. Do they? How do they? Why do they differ? On finding reasons for differing performances of species distribution models. Ecography 2009, 32, 66–77. [Google Scholar] [CrossRef]
- Aguirre-Gutiérrez, J.; Carvalheiro, L.G.; Polce, C.; Van Loon, E.E.; Raes, N.; Reemer, M.; Biesmeijer, J.C. Fit-for-Purpose: Species Distribution Model Performance Depends on Evaluation Criteria—Dutch Hoverflies as a Case Study. PLoS ONE 2013, 8, e63708. [Google Scholar] [CrossRef] [Green Version]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef] [Green Version]
- Phillips, S.J.; Dudík, M.; Elith, J.; Graham, C.; Lehmann, A.; Leathwick, J.; Ferrier, S. Sample selection bias and presence-only distribution models: Implications for background and pseudo-absence data. Ecol. Appl. 2009, 19, 181–197. [Google Scholar] [CrossRef] [Green Version]
- Druon, J.-N.; Chassot, E.; Murua, H.; Lopez, J. Skipjack Tuna Availability for Purse Seine Fisheries Is Driven by Suitable Feeding Habitat Dynamics in the Atlantic and Indian Oceans. Front. Mar. Sci. 2017, 4, 315. [Google Scholar] [CrossRef]
- Howell, E.A.; Hawn, D.R.; Polovina, J.J. Spatiotemporal variability in bigeye tuna (Thunnus obesus) dive behavior in the central North Pacific Ocean. Prog. Oceanogr. 2010, 86, 81–93. [Google Scholar] [CrossRef]
| Data | Resolution | Source | Agency |
|---|---|---|---|
| SST-v2, AVHRR-AMSR-E | 0.25 | https://eclipse.ncdc.noaa.gov/pub/OI-daily-v2/IEEE | NOAA |
| SSC | 0.05 | https://oceancolor.gsfc.nasa.gov/l3/ | NASA |
| SSHA-global | 0.25 | https://coastwatch.pfeg.noaa.gov/coastwatch/CWBrowserWW180.jsp | NOAA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mugo, R.; Saitoh, S.-I. Ensemble Modelling of Skipjack Tuna (Katsuwonus pelamis) Habitats in the Western North Pacific Using Satellite Remotely Sensed Data; a Comparative Analysis Using Machine-Learning Models. Remote Sens. 2020, 12, 2591. https://doi.org/10.3390/rs12162591
Mugo R, Saitoh S-I. Ensemble Modelling of Skipjack Tuna (Katsuwonus pelamis) Habitats in the Western North Pacific Using Satellite Remotely Sensed Data; a Comparative Analysis Using Machine-Learning Models. Remote Sensing. 2020; 12(16):2591. https://doi.org/10.3390/rs12162591
Chicago/Turabian StyleMugo, Robinson, and Sei-Ichi Saitoh. 2020. "Ensemble Modelling of Skipjack Tuna (Katsuwonus pelamis) Habitats in the Western North Pacific Using Satellite Remotely Sensed Data; a Comparative Analysis Using Machine-Learning Models" Remote Sensing 12, no. 16: 2591. https://doi.org/10.3390/rs12162591
APA StyleMugo, R., & Saitoh, S.-I. (2020). Ensemble Modelling of Skipjack Tuna (Katsuwonus pelamis) Habitats in the Western North Pacific Using Satellite Remotely Sensed Data; a Comparative Analysis Using Machine-Learning Models. Remote Sensing, 12(16), 2591. https://doi.org/10.3390/rs12162591