Quantifying Leaf Chlorophyll Concentration of Sorghum from Hyperspectral Data Using Derivative Calculus and Machine Learning
Abstract
1. Introduction
2. Materials and Methods
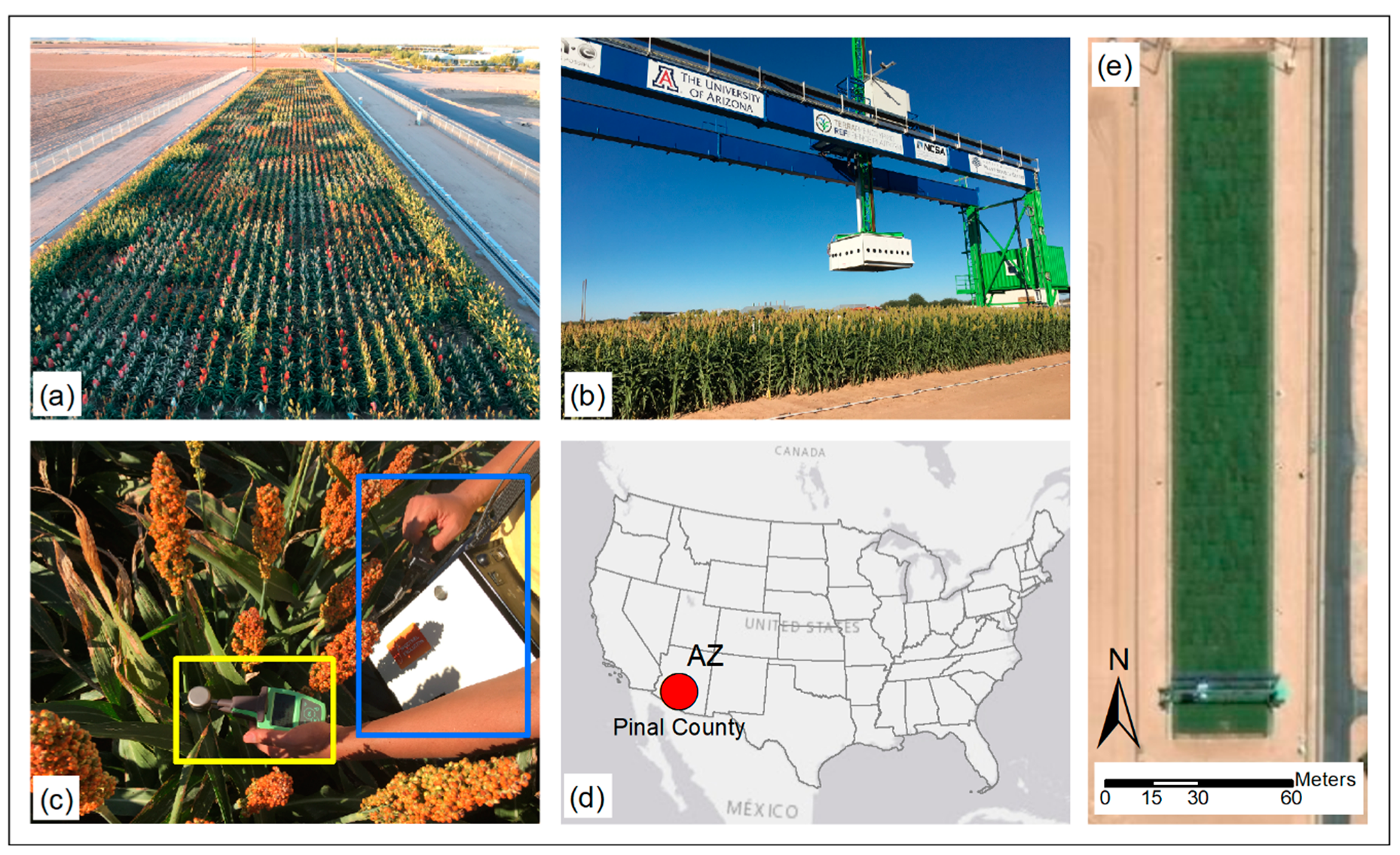
2.1. Study Site and Plant Material
2.2. Data Collection
2.2.1. Leaf Chlorophyll Concentration Measurements
2.2.2. Hyperspectral Reflectance Measurements
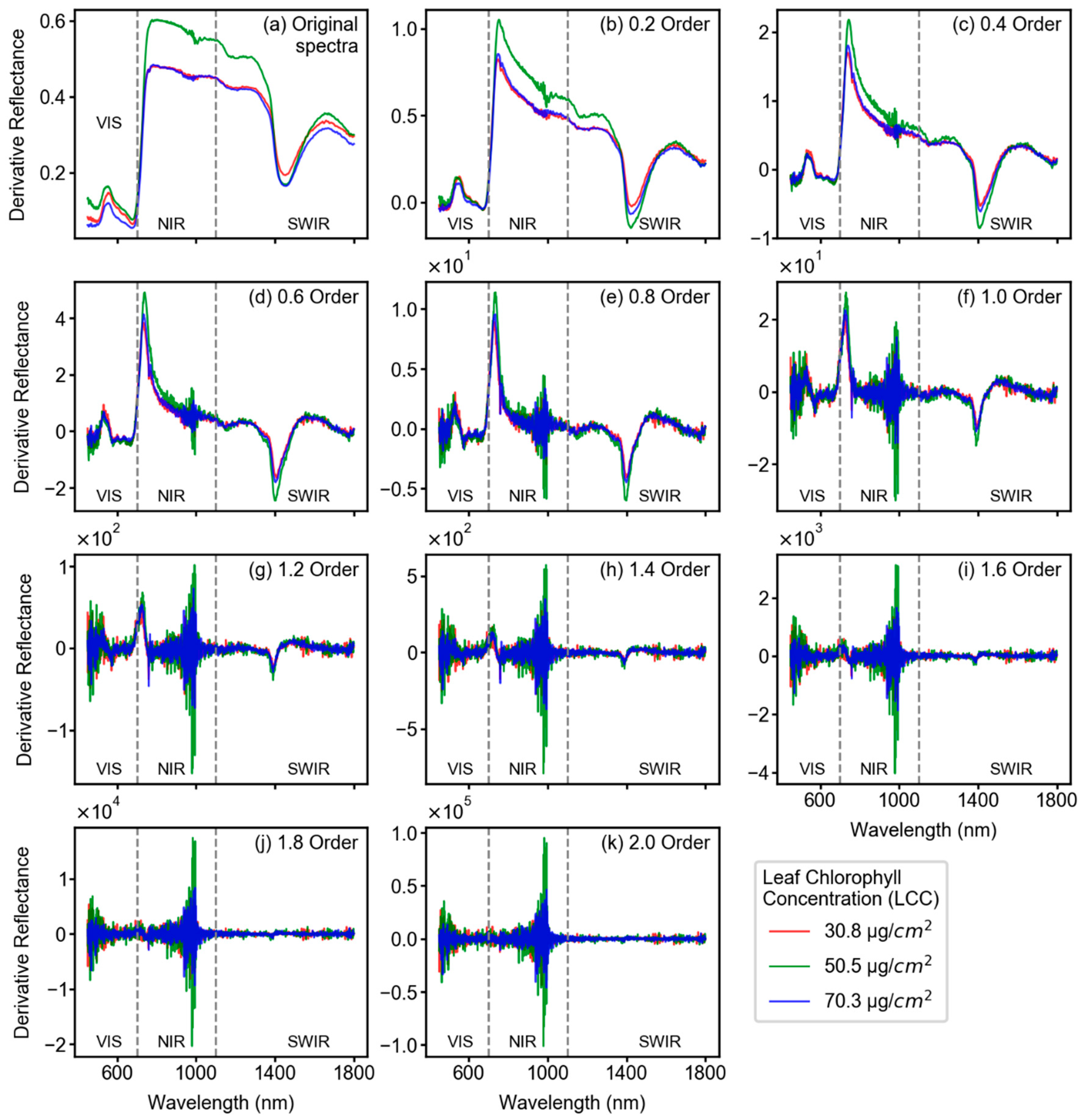
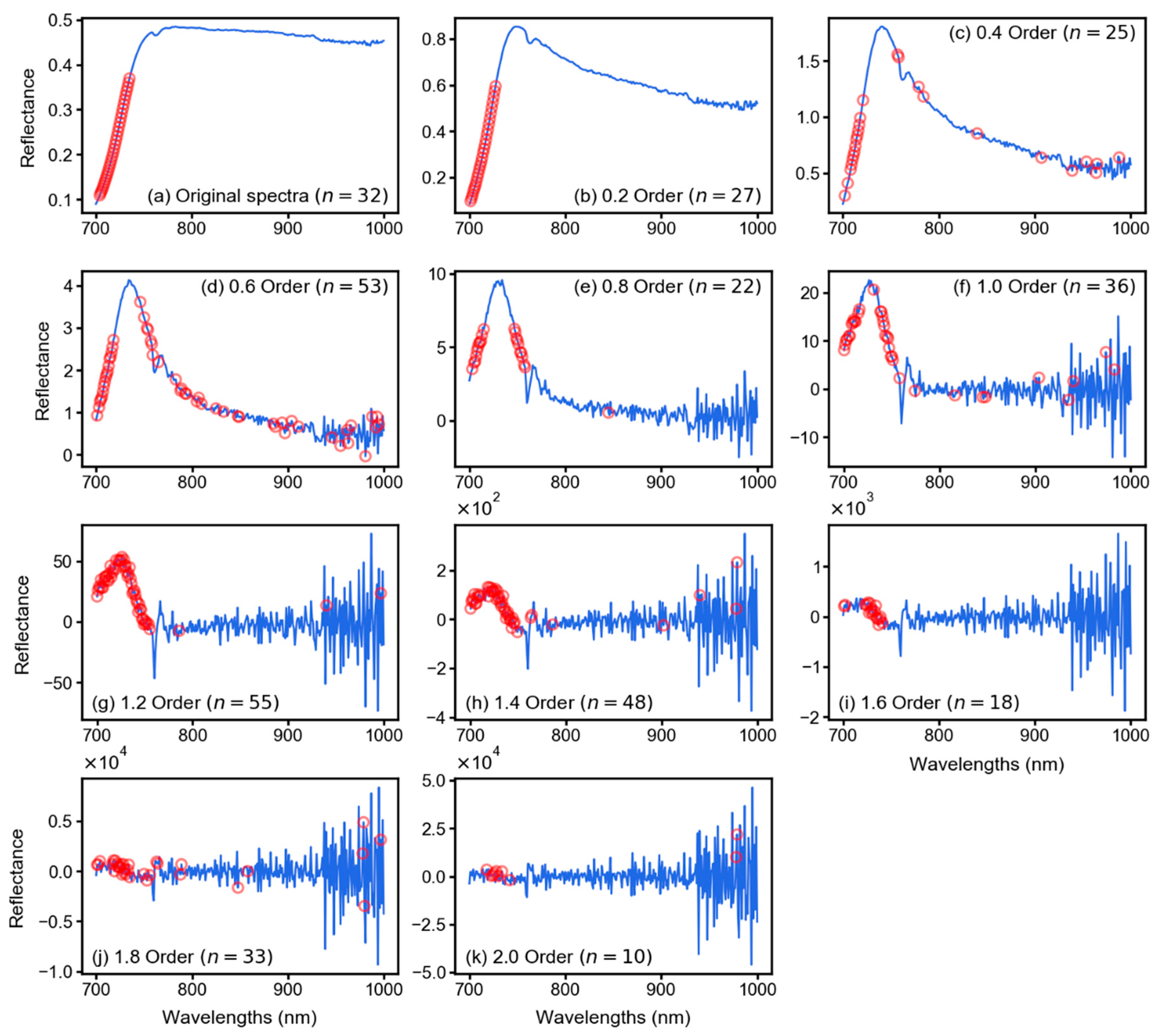
2.3. Fractional Derivative Calculation
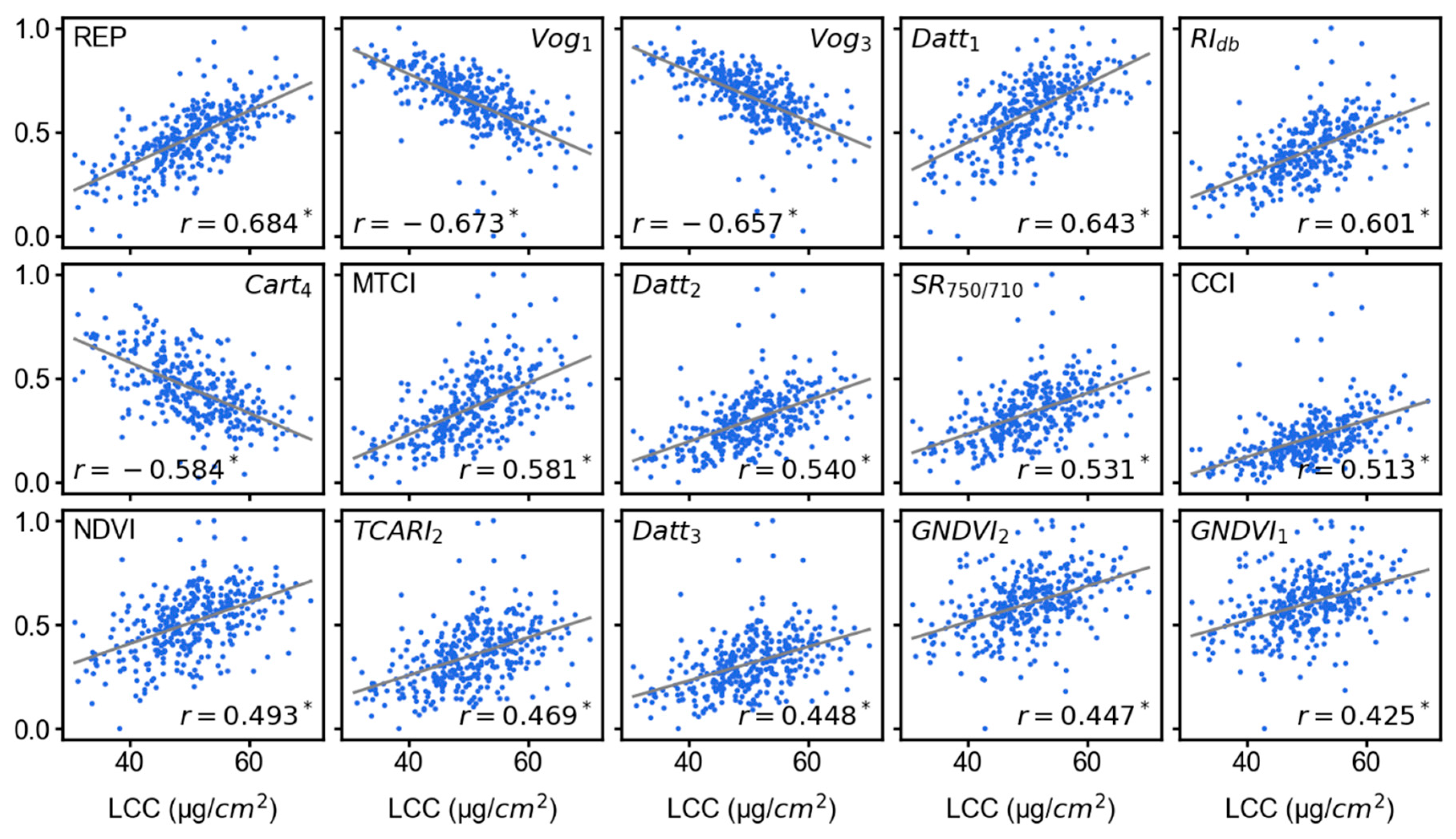
2.4. Calculation of Vegetation Indices
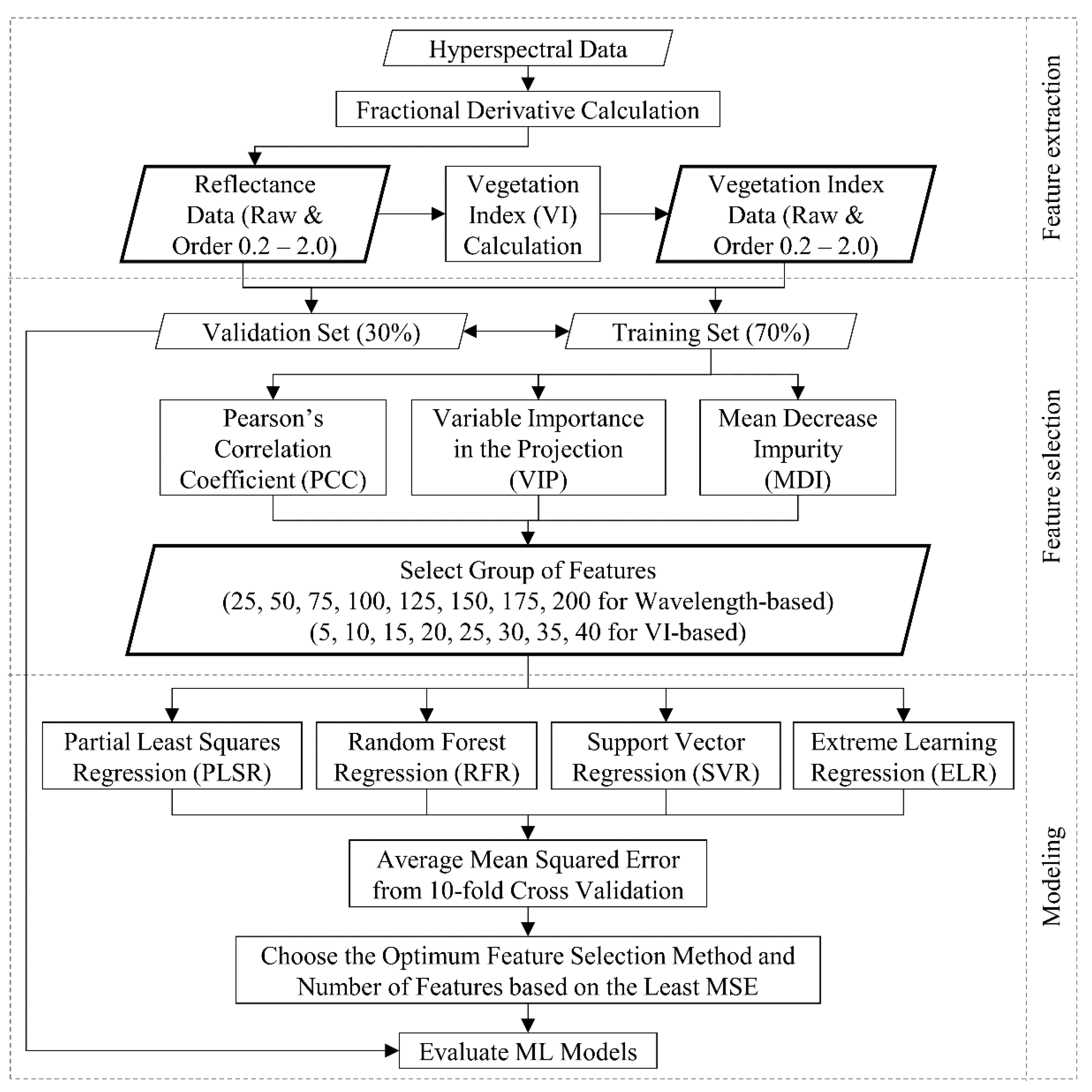
2.5. Feature Selection Methods
2.5.1. Pearson’s Correlation Coefficient (PCC)
2.5.2. Variable Importance in the Projection (VIP)
2.5.3. Mean Decrease Impurity (MDI)
2.6. Machine Learning Algorithms
2.7. Modeling Pipeline and Evaluation
3. Results
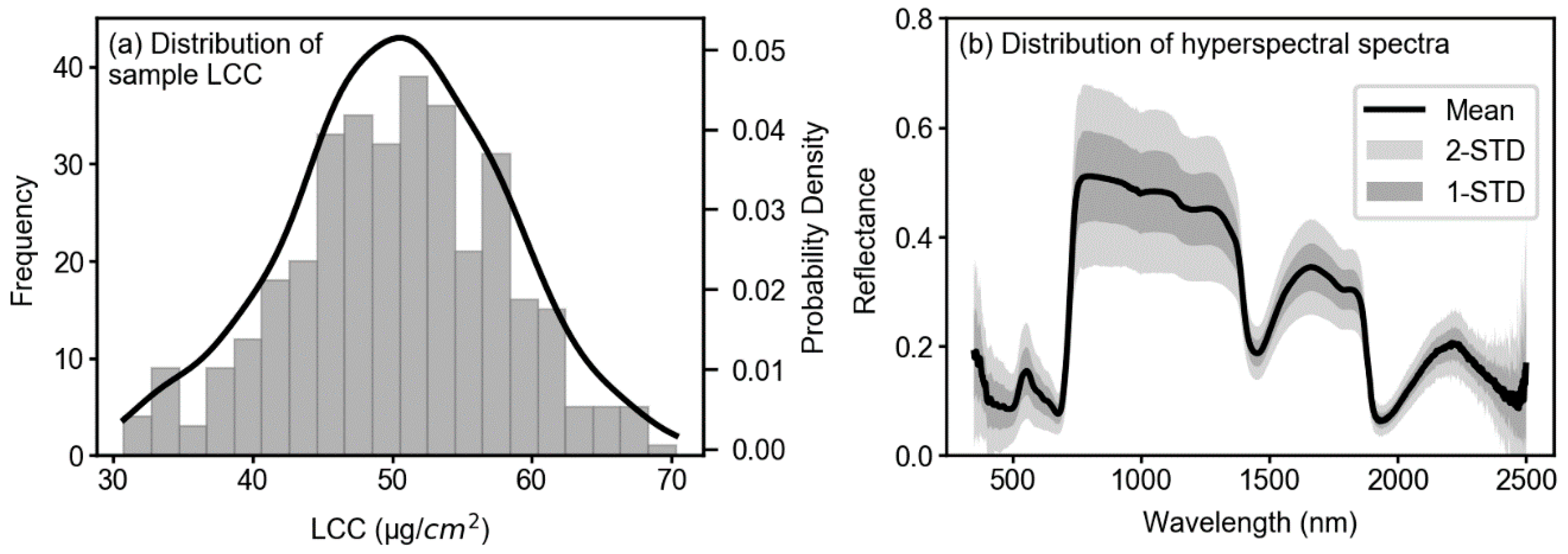
3.1. Descriptive Statistics of Collected Samples
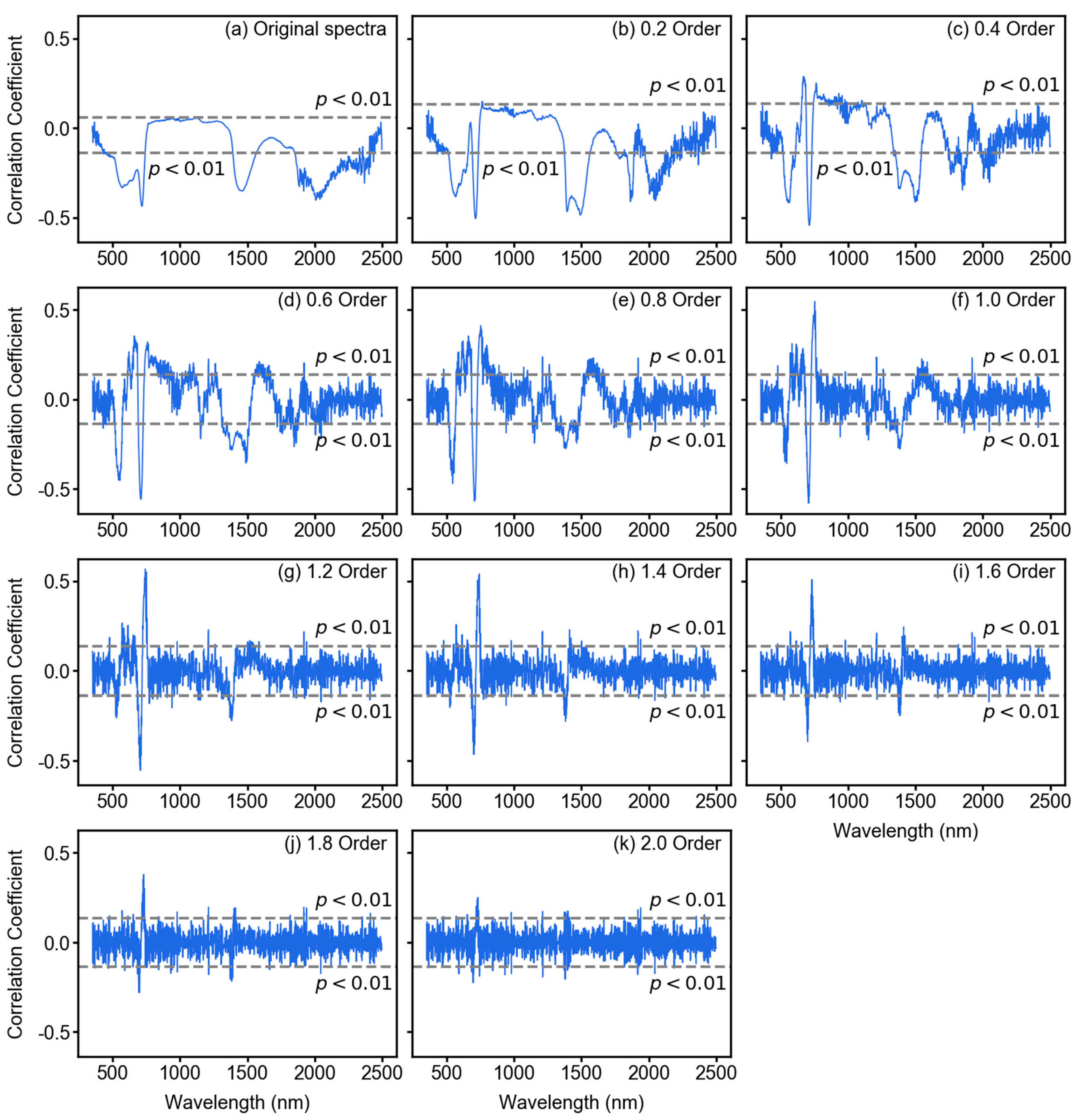
3.2. Spectral Features After Fractional Derivative Analysis
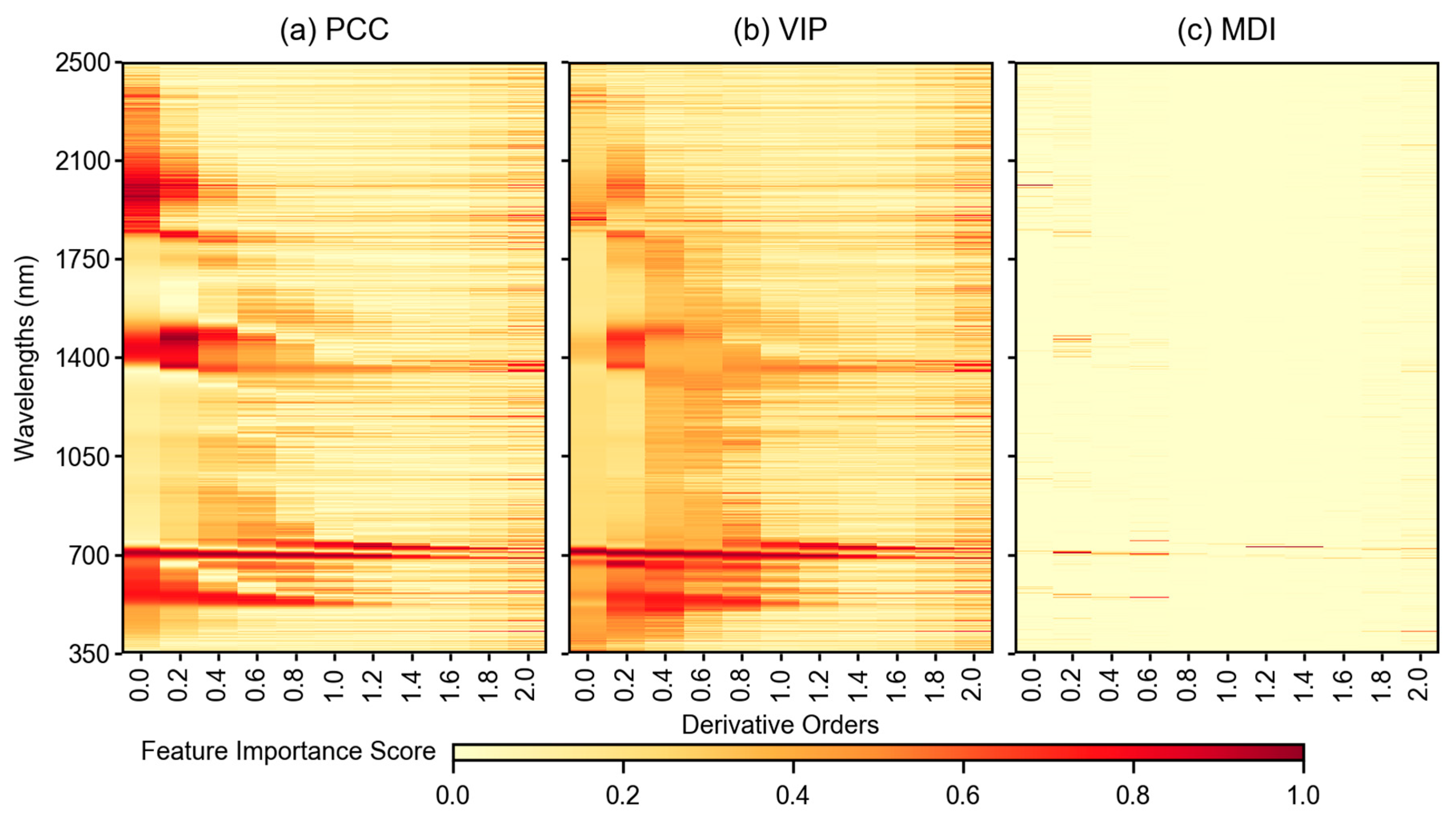
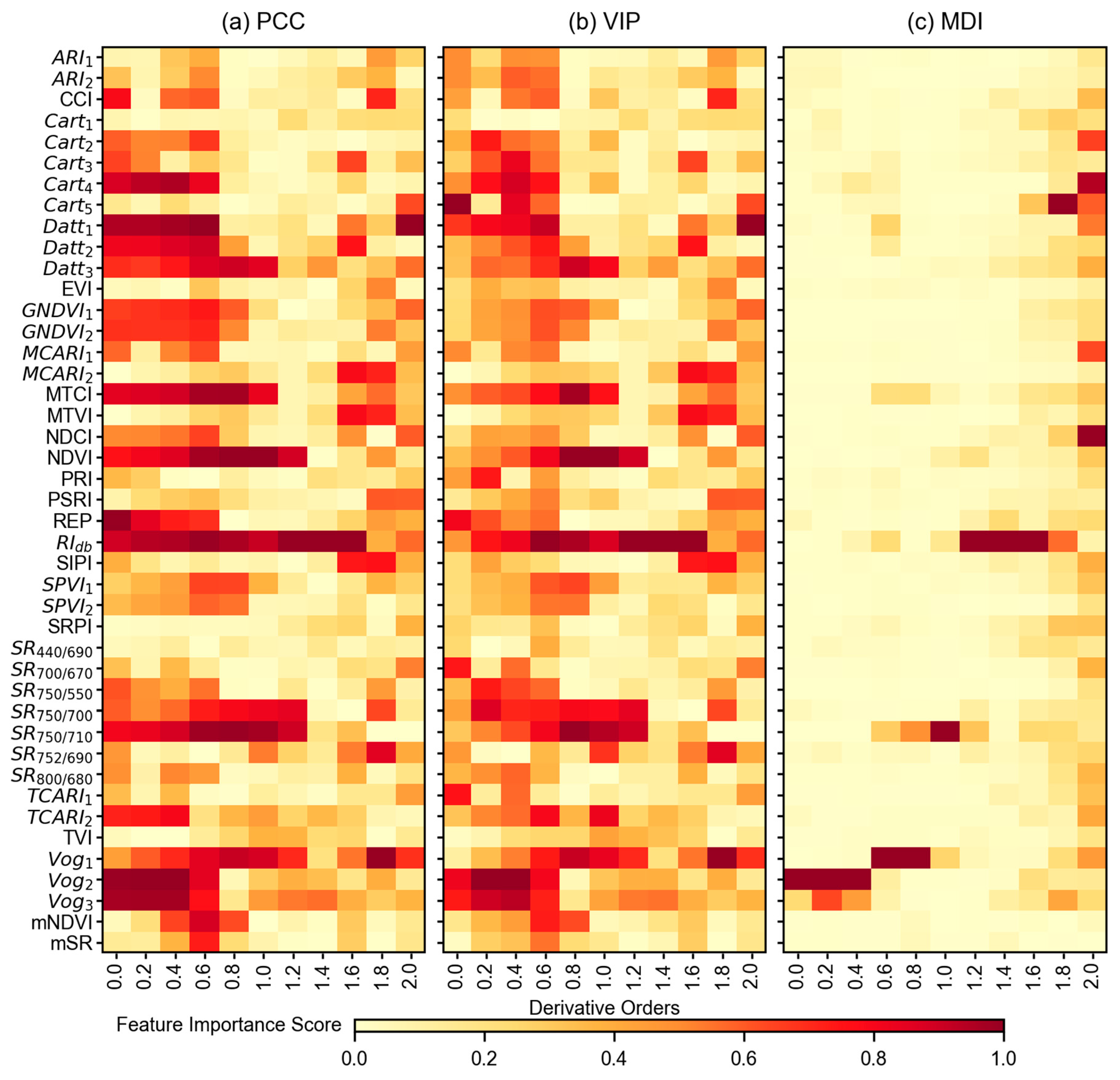
3.3. Feature Importance Scores
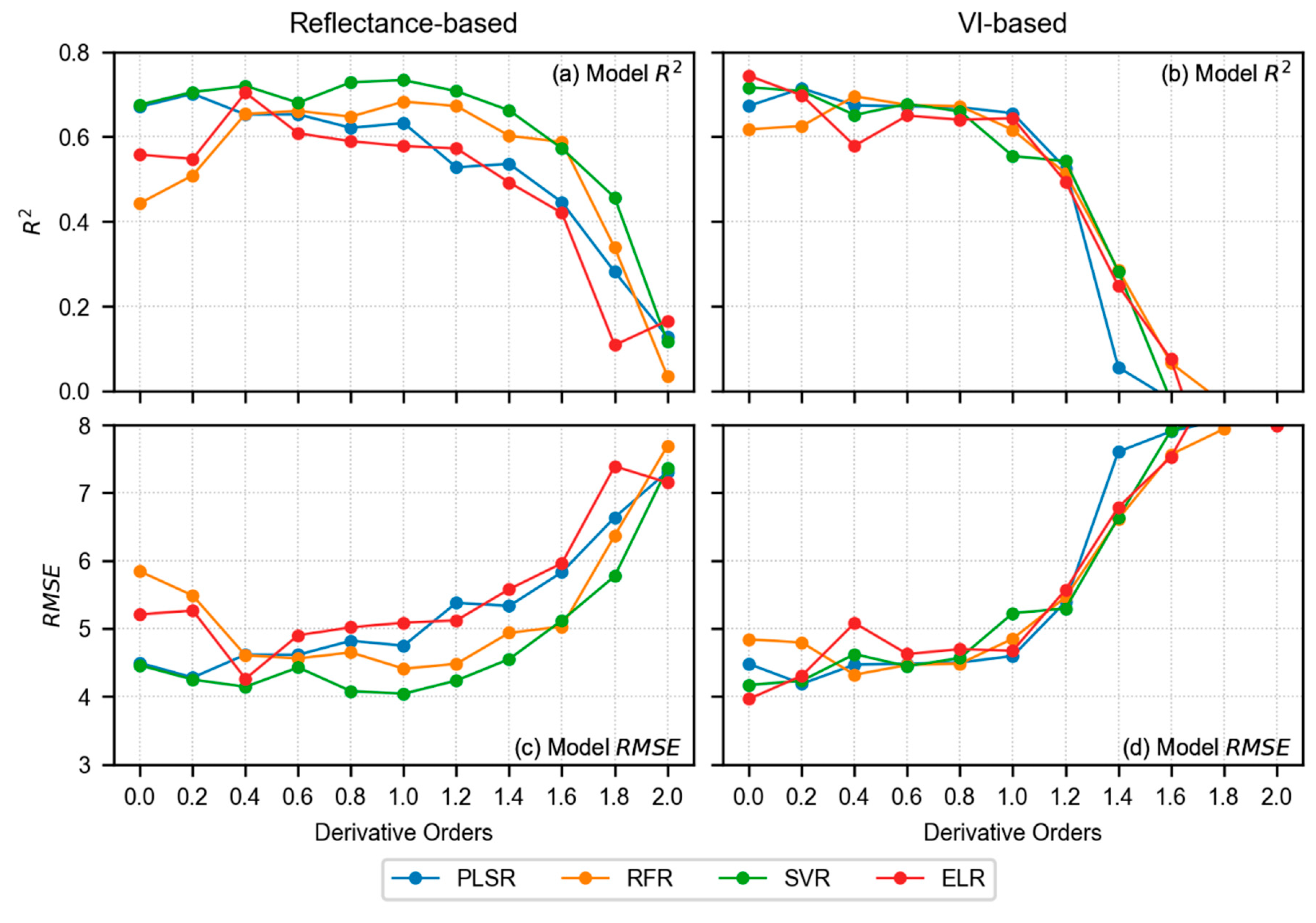
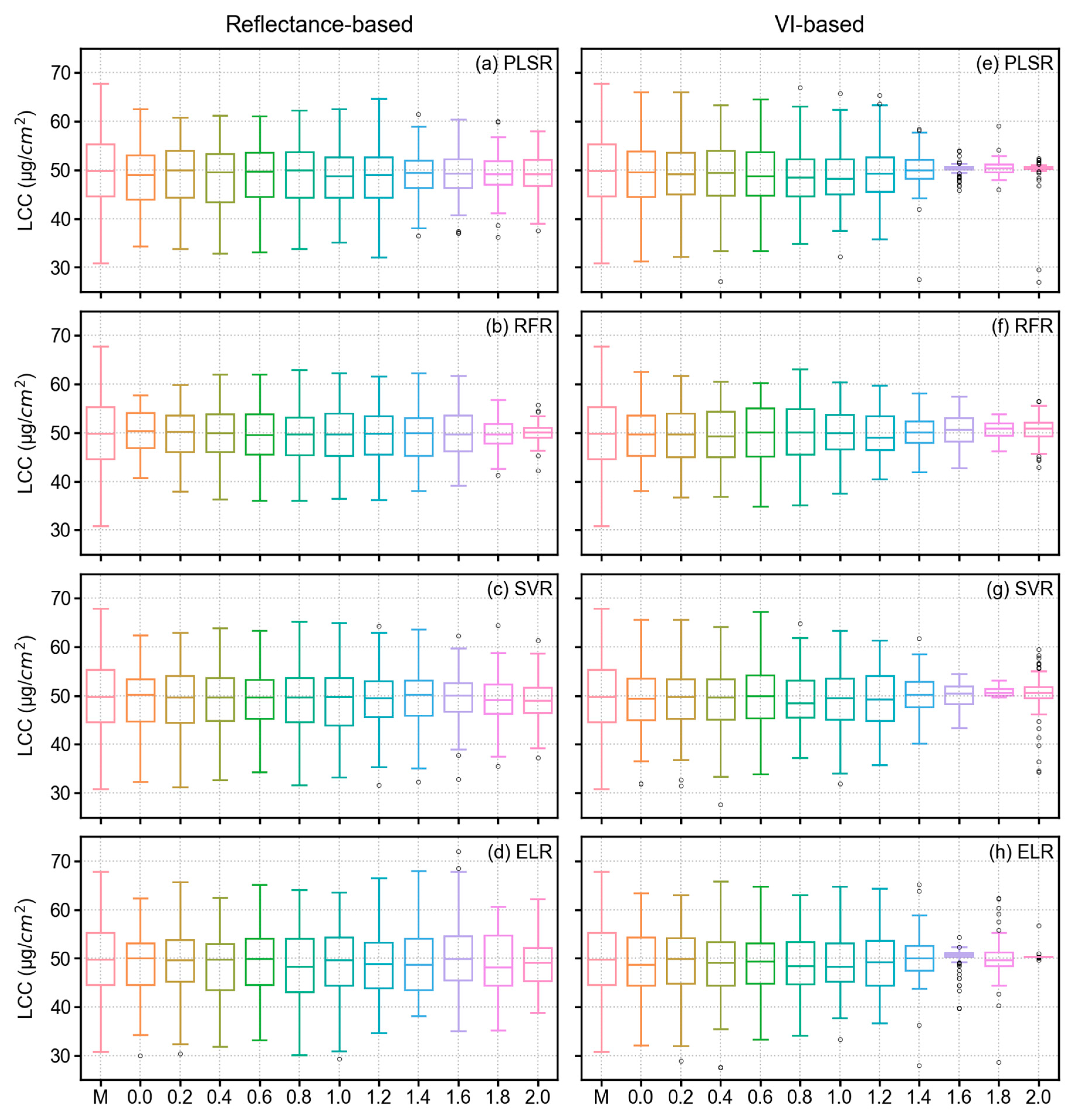
3.4. Model Results of LCC Estimation
4. Discussion
4.1. Performance Analysis of Derivative Spectra and VIs in LCC Estimation
4.2. Impact of Feature Selection Methods in Modeling Pipeline
4.3. Performance of Machine Learning Models in LCC Estimation
5. Conclusions
- In terms of reflectance-based analysis, increasing derivative order can show improved model performance until a certain order; however, it is inconclusive to state that a particular derivative order is optimal for estimating LCC of sorghum. Further assessment with data from multiple study sites and growth stages is required to make such an inference.
- VI-based modeling with original spectra outperformed reflectance-based modeling with derivative-augmented spectra.
- Sensitive feature selection is a crucial step in any machine learning pipeline. MDI score was found effective in selecting sensitive features from a large feature space (reflectance-based analysis), whereas PCC worked better with a smaller feature space (VI-based analysis).
- When single wavelengths were used in the analysis from different FD orders, SVR outperformed all other models. However, PLSR and ELR required fewer model parameters and computational time, which can be advantageous in model training. Alternatively, ELR with VIs from original spectra yielded slightly better results compared to all other models. Therefore, ELR worked better when hand-crafted features (VIs) were used.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stringer, R. Food Security Global Overview. In Food Poverty and Insecurity: International Food Inequalities; Caraher, M., Coveney, J., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 11–18. [Google Scholar]
- Myers, S.S.; Smith, M.R.; Guth, S.; Golden, C.D.; Vaitla, B.; Mueller, N.D.; Dangour, A.D.; Huybers, P. Climate Change and Global Food Systems: Potential Impacts on Food Security and Undernutrition. Annu. Rev. Publ. Health 2017, 38, 259–277. [Google Scholar] [CrossRef] [PubMed]
- Stephens, E.C.; Jones, A.D.; Parsons, D. Agricultural systems research and global food security in the 21st century: An overview and roadmap for future opportunities. Agric. Syst. 2018, 163, 1–6. [Google Scholar] [CrossRef]
- FAO; IFAD; WFP. The State of Food Insecurity in the World: Strengthening the Enabling Environment for Food Security and Nutrition; Food and Agriculture Organization of the United Nations: Rome, Italy, 2014; p. 57. [Google Scholar]
- Monk, R.; Franks, C.; Dahlberg, J. Sorghum. In Yield Gains in Major U.S. Field Crops; Smith, S., Diers, B., Specht, J., Carver, B., Eds.; Wiley: Hoboken, NJ, USA, 2014; pp. 293–310. [Google Scholar]
- Hadebe, S.T.; Modi, A.T.; Mabhaudhi, T. Drought Tolerance and Water Use of Cereal Crops: A Focus on Sorghum as a Food Security Crop in Sub-Saharan Africa. J. Agric. Crop. Sci 2017, 203, 177–191. [Google Scholar] [CrossRef]
- Morris, G.P.; Ramu, P.; Deshpande, S.P.; Hash, C.T.; Shah, T.; Upadhyaya, H.D.; Riera-Lizarazu, O.; Brown, P.J.; Acharya, C.B.; Mitchell, S.E.; et al. Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc. Natl. Acad. Sci. USA 2013, 110, 453–458. [Google Scholar] [CrossRef]
- Maimaitijiang, M.; Ghulam, A.; Sidike, P.; Hartling, S.; Maimaitiyiming, M.; Peterson, K.; Shavers, E.; Fishman, J.; Peterson, J.; Kadam, S.; et al. Unmanned Aerial System (UAS)-based phenotyping of soybean using multi-sensor data fusion and extreme learning machine. ISPRS J. Photogramm. Remote Sens. 2017, 134, 43–58. [Google Scholar] [CrossRef]
- Furbank, R.T.; Tester, M. Phenomics—Technologies to relieve the phenotyping bottleneck. Trends Plant. Sci. 2011, 16, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Chapman, S.C.; Merz, T.; Chan, A.; Jackway, P.; Hrabar, S.; Dreccer, M.F.; Holland, E.; Zheng, B.; Ling, T.J.; Jimenez-Berni, J. Pheno-Copter: A Low-Altitude, Autonomous Remote-Sensing Robotic Helicopter for High-Throughput Field-Based Phenotyping. Agronomy 2014, 4, 279–301. [Google Scholar] [CrossRef]
- Watanabe, K.; Guo, W.; Arai, K.; Takanashi, H.; Kajiya-Kanegae, H.; Kobayashi, M.; Yano, K.; Tokunaga, T.; Fujiwara, T.; Tsutsumi, N.; et al. High-Throughput Phenotyping of Sorghum Plant Height Using an Unmanned Aerial Vehicle and Its Application to Genomic Prediction Modeling. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Malenovsky, Z.; Homolova, L.; Zurita-Milla, R.; Lukes, P.; Kaplan, V.; Hanus, J.; Gastellu-Etchegorry, J.P.; Schaepman, M.E. Retrieval of spruce leaf chlorophyll content from airborne image data using continuum removal and radiative transfer. Remote Sens. Environ. 2013, 131, 85–102. [Google Scholar] [CrossRef]
- Houborg, R.; Fisher, J.B.; Skidmore, A.K. Advances in remote sensing of vegetation function and traits. Int. J. Appl. Earth Obs. 2015, 43, 1–6. [Google Scholar] [CrossRef]
- Sun, J.; Yang, J.; Shi, S.; Chen, B.W.; Du, L.; Gong, W.; Song, S.L. Estimating Rice Leaf Nitrogen Concentration: Influence of Regression Algorithms Based on Passive and Active Leaf Reflectance. Remote Sens. 2017, 9, 951. [Google Scholar] [CrossRef]
- Curran, P.J.; Dungan, J.L.; Peterson, D.L. Estimating the foliar biochemical concentration of leaves with reflectance spectrometry testing the Kokaly and Clark methodologies. Remote Sens. Environ. 2001, 76, 349–359. [Google Scholar] [CrossRef]
- Blackburn, G.A. Hyperspectral remote sensing of plant pigments. J. Exp. Bot. 2007, 58, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Golhani, K.; Balasundram, S.K.; Vadamalai, G.; Pradhan, B. Estimating chlorophyll content at leaf scale in viroid-inoculated oil palm seedlings (Elaeis guineensis Jacq.) using reflectance spectra (400 nm–1050 nm). Int. J. Remote Sens. 2019, 40, 7647–7662. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Tremblay, N.; Zarco-Tejada, P.J.; Dextraze, L. Integrated narrow-band vegetation indices for prediction of crop chlorophyll content for application to precision agriculture. Remote Sens. Environ. 2002, 81, 416–426. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Vina, A.; Ciganda, V.; Rundquist, D.C.; Arkebauer, T.J. Remote estimation of canopy chlorophyll content in crops. Geophys. Res. Lett. 2005, 32. [Google Scholar] [CrossRef]
- Wu, C.Y.; Niu, Z.; Tang, Q.; Huang, W.J. Estimating chlorophyll content from hyperspectral vegetation indices: Modeling and validation. Agric. For. Meteorol. 2008, 148, 1230–1241. [Google Scholar] [CrossRef]
- Croft, H.; Chen, J.M.; Zhang, Y. The applicability of empirical vegetation indices for determining leaf chlorophyll content over different leaf and canopy structures. Ecol. Complex. 2014, 17, 119–130. [Google Scholar] [CrossRef]
- Meroni, M.; Colombo, R.; Panigada, C. Inversion of a radiative transfer model with hyperspectral observations for LAI mapping in poplar plantations. Remote Sens. Environ. 2004, 92, 195–206. [Google Scholar] [CrossRef]
- Weiss, M.; Baret, F. Evaluation of canopy biophysical variable retrieval performances from the accumulation of large swath satellite data. Remote Sens. Environ. 1999, 70, 293–306. [Google Scholar] [CrossRef]
- Atzberger, C. Object-based retrieval of biophysical canopy variables using artificial neural nets and radiative transfer models. Remote Sens. Environ. 2004, 93, 53–67. [Google Scholar] [CrossRef]
- Tsai, F.; Philpot, W.D. A derivative-aided hyperspectral image analysis system for land-cover classification. IEEE Trans. Geosci. Remote 2002, 40, 416–425. [Google Scholar] [CrossRef]
- Stavroulakis, P.; Liatsis, P.; Tipping, N.; Craddock, P. Evaluation and Optimization of the Savitzky-Golay Smoothing Filter for Noise Reduction in Thin Film Interference Signal Analysis; SPIE: Bellingham, WA, USA, 2013; Volume 8842. [Google Scholar]
- Shafri, H.; Shafri, M.; Rozni, M.; Yusof, R. Trends and Issues in Noise Reduction for Hyperspectral Vegetation Reflectance Spectra. Eur. J. Sci. Res. 2009, 29, 404–410. [Google Scholar]
- Han, L.H. Estimating chlorophyll-a concentration using first-derivative spectra in coastal water. Int. J. Remote Sens. 2005, 26, 5235–5244. [Google Scholar] [CrossRef]
- Wiggins, K.; Palmer, R.; Hutchinson, W.; Drummond, P. An investigation into the use of calculating the first derivative of absorbance spectra as a tool for forensic fibre analysis. Sci. Justice 2007, 47, 9–18. [Google Scholar] [CrossRef]
- Zhang, X.; He, Y.; Wang, C.; Xu, F.; Li, X.; Tan, C.; Chen, D.; Wang, G.; Shi, L. Estimation of Corn Canopy Chlorophyll Content Using Derivative Spectra in the O2–A Absorption Band. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Holden, H.; LeDrew, E. Accuracy Assessment of Hyperspectral Classification of Coral Reef Features. Geocarto Int. 2000, 15, 7–14. [Google Scholar] [CrossRef]
- Pu, Y.F.; Wang, W.X.; Zhou, J.L.; Wang, Y.Y.; Jia, H.D. Fractional differential approach to detecting textural features of digital image and its fractional differential filter implementation. Sci. Chin. Ser. 2008, 51, 1319–1339. [Google Scholar] [CrossRef]
- Wang, X.P.; Zhang, F.; Ding, J.L.; Kung, H.T.; Latif, A.; Johnson, V.C. Estimation of soil salt content (SSC) in the Ebinur Lake Wetland National Nature Reserve (ELWNNR), Northwest China, based on a Bootstrap-BP neural network model and optimal spectral indices. Sci. Total Environ. 2018, 615, 918–930. [Google Scholar] [CrossRef]
- Kharintsev, S.S.; Salakhov, M.K. A simple method to extract spectral parameters using fractional derivative spectrometry. Spectrochim. Acta 2004, 60, 2125–2133. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.P.; Zhang, F.; Kung, H.T.; Johnson, V.C. New methods for improving the remote sensing estimation of soil organic matter content (SOMC) in the Ebinur Lake Wetland National Nature Reserve (ELWNNR) in northwest China. Remote Sens. Environ. 2018, 218, 104–118. [Google Scholar] [CrossRef]
- Cao, H.F.; Zhang, R.X.; Yan, F.L. Spread spectrum communication and its circuit implementation using fractional-order chaotic system via a single driving variable. Commun. Nonlinear Sci. 2013, 18, 341–350. [Google Scholar] [CrossRef]
- Duong, P.L.T.; Lee, M. Optimal design of fractional order linear system with stochastic inputs/parametric uncertainties by hybrid spectral method. J. Process. Contr. 2014, 24, 1639–1645. [Google Scholar] [CrossRef]
- Huang, X.; Sun, T.T.; Li, Y.X.; Liang, J.L. A Color Image Encryption Algorithm Based on a Fractional-Order Hyperchaotic System. Entropy 2015, 17, 28–38. [Google Scholar] [CrossRef]
- Fu, C.B.; Xiong, H.G.; Tian, A.H. Study on the Effect of Fractional Derivative on the Hyperspectral Data of Soil Organic Matter Content in Arid Region. J. Spectrosc. 2019, 7159317. [Google Scholar] [CrossRef]
- Dariusz, W.B. Comparison of Fractional Order Derivatives Computational Accuracy—Right Hand vs Left Hand Definition. Appl. Math. Nonlinear Sci. 2017, 2, 237–248. [Google Scholar]
- Schmitt, J.M. Fractional derivative analysis of diffuse reflectance spectra. Appl. Spectrosc. 1998, 52, 840–846. [Google Scholar] [CrossRef]
- Li, Y.-l.; Tang, H.-q.; Chen, H.-x. Fractional-order derivative spectroscopy for resolving simulated overlapped Lorenztian peaks. Chemometr. Intell. Lab. 2011, 107, 83–89. [Google Scholar] [CrossRef]
- Tong, P.J.; Du, Y.P.; Zheng, K.Y.; Wu, T.; Wang, J.J. Improvement of NIR model by fractional order Savitzky-Golay derivation (FOSGD) coupled with wavelength selection. Chemometr. Intell. Lab. 2015, 143, 40–48. [Google Scholar] [CrossRef]
- Wang, J.; Tashpolat, T.; Ding, J.; Zhang, D.; Liu, W. Estimation of desert soil organic carbon content based on hyperspectral data preprocessing with fractional differential. Trans. Chin. Soc. Agric. Eng. 2016, 32, 161–169. [Google Scholar]
- Xia, N.; Tiyip, T.; Kelimu, A.; Nurmemet, I.; Ding, J.L.; Zhang, F.; Zhang, D. Influence of Fractional Differential on Correlation Coefficient between EC1:5 and Reflectance Spectra of Saline Soil. J. Spectrosc. 2017. [Google Scholar] [CrossRef]
- Wang, J.; Tashpolat, T.; Zhang, D. Spectral Detection of Chromium Content in Desert Soil Based on Fractional Differential. Trans. Chin. Soc. Agric. Mach. 2017, 48, 152–158. [Google Scholar] [CrossRef]
- Chen, K.; Li, C.; Tang, R. Estimation of the nitrogen concentration of rubber tree using fractional calculus augmented NIR spectra. Ind. Crop. Prod. 2017, 108, 831–839. [Google Scholar] [CrossRef]
- Abulaiti, Y.; Sawut, M.; Maimaitiaili, B.; Chunyue, M. A possible fractional order derivative and optimized spectral indices for assessing total nitrogen content in cotton. Comput. Electron. Agric. 2020, 171, 105275. [Google Scholar] [CrossRef]
- Xia, Z.Z.; Yang, J.; Wang, J.; Wang, S.P.; Liu, Y. Optimizing Rice Near-Infrared Models Using Fractional Order Savitzky-Golay Derivation (FOSGD) Combined with Competitive Adaptive Reweighted Sampling (CARS). Appl. Spectrosc. 2020. [Google Scholar] [CrossRef]
- Chen, L.; Huang, J.F.; Wang, F.M.; Tang, Y.L. Comparison between back propagation neural network and regression models for the estimation of pigment content in rice leaves and panicles using hyperspectral data. Int. J. Remote Sens. 2007, 28, 3457–3478. [Google Scholar] [CrossRef]
- Darvishzadeh, R.; Skidmore, A.; Schlerf, M.; Atzberger, C.; Corsi, F.; Cho, M. LAI and chlorophyll estimation for a heterogeneous grassland using hyperspectral measurements. ISPRS J. Photogramm. Remote Sens. 2008, 63, 409–426. [Google Scholar] [CrossRef]
- Singh, S.K.; Hoyos-Villegas, V.; Ray, J.D.; Smith, J.R.; Fritschi, F.B. Quantification of leaf pigments in soybean (Glycine max (L.) Merr.) based on wavelet decomposition of hyperspectral features. Field Crop. Res. 2013, 149, 20–32. [Google Scholar] [CrossRef]
- Yi, Q.X.; Jiapaer, G.; Chen, J.M.; Bao, A.M.; Wang, F.M. Different units of measurement of carotenoids estimation in cotton using hyperspectral indices and partial least square regression. ISPRS J. Photogramm. Remote Sens. 2014, 91, 72–84. [Google Scholar] [CrossRef]
- Zhai, Y.F.; Cui, L.J.; Zhou, X.; Gao, Y.; Fei, T.; Gao, W.X. Estimation of nitrogen, phosphorus, and potassium contents in the leaves of different plants using laboratory-based visible and near-infrared reflectance spectroscopy: Comparison of partial least-square regression and support vector machine regression methods. Int. J. Remote Sens. 2013, 34, 2502–2518. [Google Scholar] [CrossRef]
- Kira, O.; Linker, R.; Gitelson, A. Non-destructive estimation of foliar chlorophyll and carotenoid contents: Focus on informative spectral bands. Int. J. Appl. Earth Obs. 2015, 38, 251–260. [Google Scholar] [CrossRef]
- He, Y.; Zhang, C.; Liu, F.; Kong, W.W.; Cui, P.; Zhou, W.J.; Huang, L.X. Determination of Pigments Concentration of Oilseed Rape (Brassica Napus L.) Leaves Using Hyperspectral Imaging. Appl. Eng. Agric. 2015, 31, 23–30. [Google Scholar] [CrossRef]
- Ge, Y.F.; Atefi, A.; Zhang, H.C.; Miao, C.Y.; Ramamurthy, R.K.; Sigmon, B.; Yang, J.L.; Schnable, J.C. High-throughput analysis of leaf physiological and chemical traits with VIS-NIR-SWIR spectroscopy: A case study with a maize diversity panel. Plant. Methods 2019, 15. [Google Scholar] [CrossRef] [PubMed]
- Sonobe, R.; Sano, T.; Horie, H. Using spectral reflectance to estimate leaf chlorophyll content of tea with shading treatments. Biosyst. Eng. 2018, 175, 168–182. [Google Scholar] [CrossRef]
- Shah, S.H.; Angel, Y.; Houborg, R.; Ali, S.; McCabe, M.F. A Random Forest Machine Learning Approach for the Retrieval of Leaf Chlorophyll Content in Wheat. Remote Sens. 2019, 11, 920. [Google Scholar] [CrossRef]
- Huang, G.B.; Zhu, Q.Y.; Siew, C.K. Extreme learning machine: Theory and applications. Neurocomputing 2006, 70, 489–501. [Google Scholar] [CrossRef]
- Chen, Y.; Yao, E.Y.; Basu, A. A 128-Channel Extreme Learning Machine-Based Neural Decoder for Brain Machine Interfaces. IEEE Trans. Biomed. Circ. Syst. 2016, 10, 679–692. [Google Scholar] [CrossRef]
- Sidike, P.; Krieger, E.; Alom, M.Z.; Asari, V.K.; Taha, T. A fast single-image super-resolution via directional edge-guided regularized extreme learning regression. Signal. Image Video 2017, 11, 961–968. [Google Scholar] [CrossRef]
- Maimaitiyiming, M.; Sagan, V.; Sidike, P.; Kwasniewski, M.T. Dual Activation Function-Based Extreme Learning Machine (ELM) for Estimating Grapevine Berry Yield and Quality. Remote Sens. 2019, 11, 740. [Google Scholar] [CrossRef]
- Maimaitijiang, M.; Sagan, V.; Sidike, P.; Daloye, M.A.; Erkbol, H.; Fritschi, B.F. Crop Monitoring Using Satellite/UAV Data Fusion and Machine Learning. Remote Sens. 2020, 12, 1357. [Google Scholar] [CrossRef]
- Burnette, M.; Kooper, R.; Maloney, J.D.; Rohde, G.S.; Terstriep, J.A.; Willis, C.; Fahlgren, N.; Mockler, T.; Newcomb, M.; Sagan, V.; et al. TERRA-REF Data Processing Infrastructure. In Proceedings of the Practice and Experience on Advanced Research Computing, Pittsburgh, PA, USA, 22–27 July 2018. Article 27. [Google Scholar]
- Cerovic, Z.G.; Masdoumier, G.; Ghozlen, N.B.; Latouche, G. A new optical leaf-clip meter for simultaneous non-destructive assessment of leaf chlorophyll and epidermal flavonoids. Physiol. Plant. 2012, 146, 251–260. [Google Scholar] [CrossRef]
- Schaepman-Strub, G.; Schaepman, M.E.; Painter, T.H.; Dangel, S.; Martonchik, J.V. Reflectance quantities in optical remote sensing—definitions and case studies. Remote Sens. Environ. 2006, 103, 27–42. [Google Scholar] [CrossRef]
- Atangana, A.; Secer, A. A Note on Fractional Order Derivatives and Table of Fractional Derivatives of Some Special Functions. Abstr. Appl. Anal. 2013. [Google Scholar] [CrossRef]
- Pu, Y.F. Fractional-Order Euler-Lagrange Equation for Fractional-Order Variational Method: A Necessary Condition for Fractional-Order Fixed Boundary Optimization Problems in Signal Processing and Image Processing. IEEE Access 2016, 4, 10110–10135. [Google Scholar] [CrossRef]
- Salinas, M.; Salas, R.; Mellado, D.; Glaria, A.; Saavedra, C. A Computational Fractional Signal Derivative Method. Mod. Simul. Eng. 2018. [Google Scholar] [CrossRef]
- Salahshour, S.; Ahmadian, A.; Senu, N.; Baleanu, D.; Agarwal, P. On Analytical Solutions of the Fractional Differential Equation with Uncertainty: Application to the Basset Problem. Entropy 2015, 17, 885–902. [Google Scholar] [CrossRef]
- Tariboon, J.; Ntouyas, S.K.; Agarwal, P. New concepts of fractional quantum calculus and applications to impulsive fractional q-difference equations. Adv. Differ. Equ. 2015. [Google Scholar] [CrossRef]
- Chen, Y.Q.; Wei, Y.H.; Zhong, H.; Wang, Y. Sliding mode control with a second-order switching law for a class of nonlinear fractional order systems. Nonlinear Dyn. 2016, 85, 633–643. [Google Scholar] [CrossRef]
- Agarwal, P.; Al-Mdallal, Q.; Cho, Y.J.; Jain, S. Fractional differential equations for the generalized Mittag-Leffler function. Adv. Differ. Equ. 2018. [Google Scholar] [CrossRef]
- Guan, J.L.; Ou, J.Q.; Lai, Z.H.; Lai, Y.T. Medical Image Enhancement Method Based on the Fractional Order Derivative and the Directional Derivative. Int. J. Pattern Recogn. 2018, 32. [Google Scholar] [CrossRef]
- Adams, M. differint: A Python Package for Numerical Fractional Calculus. Comput. Phys. Commun. 2019, arXiv:1912.05303. [Google Scholar]
- Gitelson, A.A.; Merzlyak, M.N.; Chivkunova, O.B. Optical properties and nondestructive estimation of anthocyanin content in plant leaves. Photochem. Photobiol. 2001, 74, 38–45. [Google Scholar] [CrossRef]
- Carter, G.A. Ratios of leaf reflectances in narrow wavebands as indicators of plant stress. Int. J. Remote Sens. 1994, 15, 697–703. [Google Scholar] [CrossRef]
- Gupta, R.K.; Vijayan, D.; Prasad, T.S. New hyperspectral vegetation characterization parameters. Adv. Space Res. 2001, 28, 201–206. [Google Scholar] [CrossRef]
- Datt, B. A New Reflectance Index for Remote Sensing of Chlorophyll Content in Higher Plants: Tests using Eucalyptus Leaves. J. Plant. Physiol 1999, 154, 30–36. [Google Scholar] [CrossRef]
- Huete, A.; Justice, C.; Liu, H. Development of vegetation and soil indices for MODIS-EOS. Remote Sens. Environ. 1994, 49, 224–234. [Google Scholar] [CrossRef]
- Huete, A.R.; Liu, H.Q.; Batchily, K.; van Leeuwen, W. A comparison of vegetation indices over a global set of TM images for EOS-MODIS. Remote Sens. Environ. 1997, 59, 440–451. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Daughtry, C.S.T.; Walthall, C.L.; Kim, M.S.; de Colstoun, E.B.; McMurtrey, J.E. Estimating Corn Leaf Chlorophyll Concentration from Leaf and Canopy Reflectance. Remote Sens Environ. 2000, 74, 229–239. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Pattey, E.; Zarco-Tejada, P.J.; Strachan, I.B. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Sims, D.A.; Gamon, J.A. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and developmental stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Dash, J.; Curran, P.J. The MERIS terrestrial chlorophyll index. Int. J. Remote Sens. 2004, 25, 5403–5413. [Google Scholar] [CrossRef]
- Marshak, A.; Knyazikhin, Y.; Davis, A.B.; Wiscombe, W.J.; Pilewskie, P. Cloud-vegetation interaction: Use of normalized difference cloud index for estimation of cloud optical thickness. Geophys. Res. Lett. 2000, 27, 1695–1698. [Google Scholar] [CrossRef]
- Rouse, J.W., Jr.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring vegetation systems in the Great Plains with ERTS. In Proceedings of the 3rd Earth Resource Technology Satellite (ERTS) Symposium, Washington, DC, USA, 10–14 December 1973; pp. 309–317. [Google Scholar]
- Gamon, J.A.; Peñuelas, J.; Field, C.B. A narrow-waveband spectral index that tracks diurnal changes in photosynthetic efficiency. Remote Sens. Environ. 1992, 41, 35–44. [Google Scholar] [CrossRef]
- Merzlyak, M.N.; Gitelson, A.A.; Chivkunova, O.B.; Rakitin, V.Y.U. Non-destructive optical detection of pigment changes during leaf senescence and fruit ripening. Physiol. Plant. 1999, 106, 135–141. [Google Scholar] [CrossRef]
- Clevers, J.G.P.W. Imaging Spectrometry in Agriculture—Plant Vitality And Yield Indicators. In Imaging Spectrometry—A Tool for Environmental Observations; Hill, J., Mégier, J., Eds.; Springer: Dordrecht, The Netherlands, 1994; pp. 193–219. [Google Scholar]
- Gupta, R.K.; Vijayan, D.; Prasad, T.S. Comparative analysis of red-edge hyperspectral indices. Adv. Space Res. 2003, 32, 2217–2222. [Google Scholar] [CrossRef]
- Penuelas, J.; Frederic, B.; Filella, I. Semi-Empirical Indices to Assess Carotenoids/Chlorophyll-a Ratio from Leaf Spectral Reflectance. Photosynthetica 1995, 31, 221–230. [Google Scholar]
- Vincini, M.; Frazzi, E.; Alessio, P. Angular dependence of maize and sugar beet VIs from directional CHRIS/Proba data. In Proceedings of the 4th ESA CHRIS PROBA Workshop, Frascati, Italy, 19–21 September 2006. [Google Scholar]
- Main, R.; Cho, M.A.; Mathieu, R.; O’Kennedy, M.M.; Ramoelo, A.; Koch, S. An investigation into robust spectral indices for leaf chlorophyll estimation. ISPRS J. Photogramm. Remote Sens. 2011, 66, 751–761. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Vegetation Stress: An Introduction to the Stress Concept in Plants. J. Plant. Physiol 1996, 148, 4–14. [Google Scholar] [CrossRef]
- McMurtrey, J.E.; Chappelle, E.W.; Kim, M.S.; Meisinger, J.J.; Corp, L.A. Distinguishing nitrogen fertilization levels in field corn (Zea mays L.) with actively induced fluorescence and passive reflectance measurements. Remote Sens. Environ. 1994, 47, 36–44. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N. Remote estimation of chlorophyll content in higher plant leaves. Int. J. Remote Sens. 1997, 18, 2691–2697. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Miller, J.R. Land cover mapping at BOREAS using red edge spectral parameters from CASI imagery. J. Geophys. Res. Atmos. 1999, 104, 27921–27933. [Google Scholar] [CrossRef]
- Penuelas, J.; Filella, I.; Lloret, P.; Munoz, F.; Vilajeliu, M. Reflectance assessment of mite effects on apple trees. Int. J. Remote Sens. 1995, 16, 2727–2733. [Google Scholar] [CrossRef]
- Broge, N.H.; Leblanc, E. Comparing prediction power and stability of broadband and hyperspectral vegetation indices for estimation of green leaf area index and canopy chlorophyll density. Remote Sens. Environ. 2001, 76, 156–172. [Google Scholar] [CrossRef]
- Vogelmann, J.E.; Rock, B.N.; Moss, D.M. Red edge spectral measurements from sugar maple leaves. Int. J. Remote Sens. 1993, 14, 1563–1575. [Google Scholar] [CrossRef]
- Gilbertson, J.K.; van Niekerk, A. Value of dimensionality reduction for crop differentiation with multi-temporal imagery and machine learning. Comput. Electron. Agric. 2017, 142, 50–58. [Google Scholar] [CrossRef]
- Wade, B.S.C.; Joshi, S.H.; Gutman, B.A.; Thompson, P.M. Machine learning on high dimensional shape data from subcortical brain surfaces: A comparison of feature selection and classification methods. Pattern Recognit. 2017, 63, 731–739. [Google Scholar] [CrossRef]
- Georganos, S.; Grippa, T.; Vanhuysse, S.; Lennert, M.; Shimoni, M.; Kalogirou, S.; Wolff, E. Less is more: Optimizing classification performance through feature selection in a very-high-resolution remote sensing object-based urban application. Gisci. Remote Sens. 2018, 55, 221–242. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Y.; Bao, Y.; Luo, J.; Jin, X.; Xu, X.; Song, X.; Yang, G. Exploring the Best Hyperspectral Features for LAI Estimation Using Partial Least Squares Regression. Remote Sens. 2014, 6. [Google Scholar] [CrossRef]
- Kabir Yunus, P.; Onisimo, M.; Riyad, I. Does simultaneous variable selection and dimension reduction improve the classification of Pinus forest species? J. Appl. Remote Sens. 2014, 8, 1–16. [Google Scholar] [CrossRef]
- Farrés, M.; Platikanov, S.; Tsakovski, S.; Tauler, R. Comparison of the variable importance in projection (VIP) and of the selectivity ratio (SR) methods for variable selection and interpretation. J. Chemom. 2015, 29, 528–536. [Google Scholar] [CrossRef]
- Maimaitiyiming, M.; Ghulam, A.; Bozzolo, A.; Wilkins, J.L.; Kwasniewski, M.T. Early Detection of Plant Physiological Responses to Different Levels of Water Stress Using Reflectance Spectroscopy. Remote Sens. 2017, 9, 745. [Google Scholar] [CrossRef]
- Eriksson, L.; Byrne, T.; Johansson, E.; Trygg, J.; Wikström, C. Multi- and Megavariate Data Analysis Basic Principles and Applications, 3rd ed.; Umetrics Academy: Umeå, Sweden, 2001; p. 500. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Hariharan, S.; Mandal, D.; Tirodkar, S.; Kumar, V.; Bhattacharya, A.; Lopez-Sanchez, J.M. A Novel Phenology Based Feature Subset Selection Technique Using Random Forest for Multitemporal PolSAR Crop Classification. IEEE J.-Stars 2018, 11, 4244–4258. [Google Scholar] [CrossRef]
- Rami, A.-R.; Abdallah, S.; Mohamed, B.G.; Bahareh, K. Multi-scale correlation-based feature selection and random forest classification for LULC mapping from the integration of SAR and optical Sentinel images. In Proceedings of the SPIE Remote Sensing Technologies and Applications in Urban Environments IV, Strasbourg, France, 9–12 September 2019. [Google Scholar]
- Singh, A.; Ganapathysubramanian, B.; Singh, A.K.; Sarkar, S. Machine Learning for High-Throughput Stress Phenotyping in Plants. Trends Plant Sci. 2016, 21, 110–124. [Google Scholar] [CrossRef]
- Hasegawa, T. Principal Component Regression and Partial Least Squares Modeling. In Handbook of Vibrational Spectroscopy; Chalmers, J.M., Griffiths, P.R., Eds.; Wiley: Hoboken, NJ, USA, 2006. [Google Scholar] [CrossRef]
- Helland, I. Partial Least Squares Regression. Wiley StatsRef: Stat. Ref. Online 2014. [Google Scholar] [CrossRef]
- Rodriguez-Galiano, V.; Mendes, M.P.; Garcia-Soldado, M.J.; Chica-Olmo, M.; Ribeiro, L. Predictive modeling of groundwater nitrate pollution using Random Forest and multisource variables related to intrinsic and specific vulnerability: A case study in an agricultural setting (Southern Spain). Sci. Total Environ. 2014, 476–477, 189–206. [Google Scholar] [CrossRef] [PubMed]
- Gleason, C.J.; Im, J. Forest biomass estimation from airborne LiDAR data using machine learning approaches. Remote Sens. Environ. 2012, 125, 80–91. [Google Scholar] [CrossRef]
- Cristianini, N.; Shawe-Taylor, J. An Introduction to Support Vector Machines and Other Kernel-Based Learning Methods; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Kooistra, L.; Clevers, J.G.P.W. Estimating potato leaf chlorophyll content using ratio vegetation indices. Remote Sens. Lett. 2016, 7, 611–620. [Google Scholar] [CrossRef]
- Cui, S.C.; Zhou, K.F. A comparison of the predictive potential of various vegetation indices for leaf chlorophyll content. Earth Sci. Inform. 2017, 10, 169–181. [Google Scholar] [CrossRef]
- Lu, S.; Lu, F.; You, W.Q.; Wang, Z.Y.; Liu, Y.; Omasa, K. A robust vegetation index for remotely assessing chlorophyll content of dorsiventral leaves across several species in different seasons. Plant Methods 2018, 14. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Duan, A.W.; Ata-Ul-Karim, S.T.; Liu, Z.D.; Chen, Z.F.; Gong, Z.H.; Zhang, J.Y.; Xiao, J.F.; Liu, Z.G.; Qin, A.Z.; et al. Exploring new spectral bands and vegetation indices for estimating nitrogen nutrition index of summer maize. Eur. J. Agron. 2018, 93, 113–125. [Google Scholar] [CrossRef]
- Hunt, E.R.; Horneck, D.A.; Spinelli, C.B.; Turner, R.W.; Bruce, A.E.; Gadler, D.J.; Brungardt, J.J.; Hamm, P.B. Monitoring nitrogen status of potatoes using small unmanned aerial vehicles. Precis. Agric. 2018, 19, 314–333. [Google Scholar] [CrossRef]
- Xu, M.Z.; Liu, R.G.; Chen, J.M.; Liu, Y.; Shang, R.; Ju, W.M.; Wu, C.Y.; Huang, W.J. Retrieving leaf chlorophyll content using a matrix-based vegetation index combination approach. Remote Sens. Environ. 2019, 224, 60–73. [Google Scholar] [CrossRef]
- Caturegli, L.; Gaetani, M.; Volterrani, M.; Magni, S.; Minelli, A.; Baldi, A.; Brandani, G.; Mancini, M.; Lenzi, A.; Orlandini, S.; et al. Normalized Difference Vegetation Index versus Dark Green Colour Index to estimate nitrogen status on bermudagrass hybrid and tall fescue. Int. J. Remote Sens. 2020, 41, 455–470. [Google Scholar] [CrossRef]
- Croft, H.; Arabian, J.; Chen, J.M.; Shang, J.; Liu, J. Mapping within-field leaf chlorophyll content in agricultural crops for nitrogen management using Landsat-8 imagery. Precis. Agric. 2019. [Google Scholar] [CrossRef]
- Kwiatkowska, E.J.; Fargion, G.S. Application of machine-learning techniques toward the creation of a consistent and calibrated global chlorophyll concentration baseline dataset using remotely sensed ocean color data. IEEE Trans. Geosci. Remote Sens. 2003, 41, 2844–2860. [Google Scholar] [CrossRef]
- Haigang, Z.; Ping, S.; Chuqun, C. Retrieval of oceanic chlorophyll concentration using support vector machines. IEEE Trans. Geosci. Remote Sens. 2003, 41, 2947–2951. [Google Scholar] [CrossRef]
- Ji, L.; Peters, A.J. Forecasting vegetation greenness with satellite and climate data. IEEE Geosci. Remote Sens. Lett. 2004, 1, 3–6. [Google Scholar] [CrossRef]
- Vapnik, V.N. Controlling the Generalization Ability of Learning Processes. In The Nature of Statistical Learning Theory; Vapnik, V.N., Ed.; Springer: New York, NY, USA, 2000; pp. 93–122. [Google Scholar] [CrossRef]
| VI | Equation | Reference |
|---|---|---|
| ARI1 | [77] | |
| ARI2 | [77] | |
| Cart1 | [78] | |
| Cart2 | [78] | |
| Cart3 | [78] | |
| Cart4 | [78] | |
| Cart5 | [78] | |
| CCI | [79] | |
| Datt1 | [80] | |
| Datt2 | [80] | |
| Datt3 | [80] | |
| EVI | [81,82] | |
| GNDVI1 | [83] | |
| GNDVI2 | [83] | |
| MCARI1 | [84] | |
| MCARI2 | [85] | |
| mNDVI | [80,86] | |
| mSR | [80,86] | |
| MTCI | [87] | |
| MTVI1 | [85] | |
| NDCI | [88] | |
| NDVI | [89] | |
| PRI | [90] | |
| PSRI | [91] | |
| REP | [92] | |
| RIdb | [93] | |
| SIPI | [94] | |
| SPVI1 | [95,96] | |
| SPVI2 | [95] | |
| SR440/690 | [97] | |
| SR700/670 | [98] | |
| SR750/550 | [98] | |
| SR750/700 | [99] | |
| SR750/710 | [100] | |
| SR752/690 | [100] | |
| SR800/680 | [86] | |
| SRPI | [101] | |
| TCARI | [18] | |
| TCARI2 | [20] | |
| TVI | [102] | |
| VOG1 | [103] | |
| VOG2 | [103] | |
| VOG3 | [103] |
| Sample Size | Maximum | Minimum | Mean | SD | CV (%) | |
|---|---|---|---|---|---|---|
| LCC (µg/cm2) | 349 | 70.30 | 30.80 | 50.26 | 7.54 | 15.00 |
| Ord. | Metrics | Reflectance-based | VI-based | ||||||
|---|---|---|---|---|---|---|---|---|---|
| PLSR | RFR | SVR | ELR | PLSR | RFR | SVR | ELR | ||
| 0.0 | R2 | 0.671 | 0.443 | 0.676 | 0.558 | 0.673 | 0.618 | 0.717 | 0.744 |
| RMSE | 4.493 | 5.842 | 4.459 | 5.207 | 4.477 | 4.841 | 4.169 | 3.964 | |
| RMSE% | 9.035 | 11.747 | 8.966 | 10.471 | 9.002 | 9.734 | 8.382 | 7.971 | |
| Features | VIP-75 | MDI-50 | VIP-75 | VIP-50 | VIP-30 | MDI-10 | PCC-25 | PCC-15 | |
| 0.2 | R2 | 0.701 | 0.509 | 0.706 | 0.548 | 0.714 | 0.625 | 0.708 | 0.698 |
| RMSE | 4.279 | 5.486 | 4.249 | 5.265 | 4.187 | 4.794 | 4.231 | 4.306 | |
| RMSE% | 8.603 | 11.032 | 8.543 | 10.588 | 8.418 | 9.639 | 8.509 | 8.658 | |
| Features | VIP-75 | MDI-75 | VIP-175 | VIP-50 | PCC-10 | VIP-30 | PCC-15 | PCC-10 | |
| 0.4 | R2 | 0.653 | 0.654 | 0.720 | 0.704 | 0.674 | 0.696 | 0.651 | 0.579 |
| RMSE | 4.616 | 4.605 | 4.142 | 4.261 | 4.468 | 4.320 | 4.623 | 5.081 | |
| RMSE% | 9.281 | 9.259 | 8.330 | 8.567 | 8.984 | 8.686 | 9.295 | 10.217 | |
| Features | VIP-25 | MDI-125 | MDI-100 | VIP-25 | PCC-20 | MDI-10 | PCC-15 | PCC-15 | |
| 0.6 | R2 | 0.653 | 0.661 | 0.680 | 0.608 | 0.672 | 0.675 | 0.678 | 0.650 |
| RMSE | 4.614 | 4.560 | 4.427 | 4.901 | 4.482 | 4.464 | 4.445 | 4.624 | |
| RMSE% | 9.278 | 9.169 | 8.902 | 9.855 | 9.012 | 8.975 | 8.938 | 9.296 | |
| Features | VIP-50 | MDI-50 | MDI-175 | MDI-25 | PCC-15 | MDI-20 | VIP-15 | VIP-15 | |
| 0.8 | R2 | 0.621 | 0.648 | 0.729 | 0.589 | 0.670 | 0.672 | 0.660 | 0.640 |
| RMSE | 4.820 | 4.649 | 4.078 | 5.018 | 4.499 | 4.483 | 4.566 | 4.697 | |
| RMSE% | 9.692 | 9.347 | 8.201 | 10.090 | 9.047 | 9.014 | 9.182 | 9.445 | |
| Features | VIP-200 | MDI-50 | MDI-25 | MDI-25 | PCC-15 | MDI-5 | PCC-10 | VIP-10 | |
| 1.0 | R2 | 0.632 | 0.683 | 0.734 | 0.578 | 0.655 | 0.616 | 0.555 | 0.644 |
| RMSE | 4.747 | 4.409 | 4.041 | 5.086 | 4.596 | 4.850 | 5.226 | 4.673 | |
| RMSE% | 9.546 | 8.865 | 8.125 | 10.227 | 9.241 | 9.753 | 10.508 | 9.397 | |
| Features | VIP-200 | MDI-75 | MDI-75 | PCC-25 | PCC-10 | MDI-20 | PCC-20 | VIP-10 | |
| 1.2 | R2 | 0.528 | 0.673 | 0.708 | 0.573 | 0.526 | 0.514 | 0.543 | 0.494 |
| RMSE | 5.380 | 4.480 | 4.235 | 5.119 | 5.393 | 5.461 | 5.296 | 5.572 | |
| RMSE% | 10.818 | 9.009 | 8.515 | 10.294 | 10.844 | 10.981 | 10.649 | 11.203 | |
| Features | VIP-175 | VIP-75 | VIP-150 | VIP-50 | MDI-5 | MDI-15 | MDI-5 | MDI-5 | |
| 1.4 | R2 | 0.536 | 0.602 | 0.662 | 0.492 | 0.056 | 0.286 | 0.282 | 0.249 |
| RMSE | 5.332 | 4.937 | 4.550 | 5.579 | 7.607 | 6.614 | 6.633 | 6.786 | |
| RMSE% | 10.721 | 9.927 | 9.149 | 11.219 | 15.295 | 13.299 | 13.337 | 13.645 | |
| Features | VIP-200 | MDI-25 | PCC-150 | MDI-25 | VIP-15 | MDI-15 | MDI-5 | PCC-5 | |
| 1.6 | R2 | 0.446 | 0.588 | 0.573 | 0.420 | −0.020 | 0.066 | −0.023 | 0.075 |
| RMSE | 5.830 | 5.028 | 5.119 | 5.962 | 7.906 | 7.567 | 7.919 | 7.530 | |
| RMSE% | 11.724 | 10.110 | 10.294 | 11.988 | 15.898 | 15.215 | 15.924 | 15.141 | |
| Features | VIP-175 | PCC-25 | VIP-150 | PCC-50 | MDI-10 | MDI-10 | MDI-5 | MDI-5 | |
| 1.8 | R2 | 0.281 | 0.339 | 0.457 | 0.109 | −0.065 | −0.028 | −0.087 | −0.296 |
| RMSE | 6.637 | 6.368 | 5.771 | 7.393 | 8.082 | 7.940 | 8.164 | 8.915 | |
| RMSE% | 13.347 | 12.805 | 11.605 | 14.865 | 16.251 | 15.966 | 16.417 | 17.926 | |
| Features | PCC-200 | MDI-25 | VIP-150 | MDI-25 | PCC-5 | MDI-25 | PCC-10 | VIP-10 | |
| 2.0 | R2 | 0.128 | 0.035 | 0.116 | 0.166 | −0.280 | −0.239 | −0.089 | −0.040 |
| RMSE | 7.311 | 7.691 | 7.361 | 7.151 | 8.860 | 8.715 | 8.173 | 7.986 | |
| RMSE% | 14.701 | 15.465 | 14.802 | 14.380 | 17.816 | 17.525 | 16.434 | 16.058 | |
| Features | VIP-150 | MDI-75 | VIP-100 | VIP-50 | MDI-5 | MDI-10 | MDI-5 | VIP-30 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhadra, S.; Sagan, V.; Maimaitijiang, M.; Maimaitiyiming, M.; Newcomb, M.; Shakoor, N.; Mockler, T.C. Quantifying Leaf Chlorophyll Concentration of Sorghum from Hyperspectral Data Using Derivative Calculus and Machine Learning. Remote Sens. 2020, 12, 2082. https://doi.org/10.3390/rs12132082
Bhadra S, Sagan V, Maimaitijiang M, Maimaitiyiming M, Newcomb M, Shakoor N, Mockler TC. Quantifying Leaf Chlorophyll Concentration of Sorghum from Hyperspectral Data Using Derivative Calculus and Machine Learning. Remote Sensing. 2020; 12(13):2082. https://doi.org/10.3390/rs12132082
Chicago/Turabian StyleBhadra, Sourav, Vasit Sagan, Maitiniyazi Maimaitijiang, Matthew Maimaitiyiming, Maria Newcomb, Nadia Shakoor, and Todd C. Mockler. 2020. "Quantifying Leaf Chlorophyll Concentration of Sorghum from Hyperspectral Data Using Derivative Calculus and Machine Learning" Remote Sensing 12, no. 13: 2082. https://doi.org/10.3390/rs12132082
APA StyleBhadra, S., Sagan, V., Maimaitijiang, M., Maimaitiyiming, M., Newcomb, M., Shakoor, N., & Mockler, T. C. (2020). Quantifying Leaf Chlorophyll Concentration of Sorghum from Hyperspectral Data Using Derivative Calculus and Machine Learning. Remote Sensing, 12(13), 2082. https://doi.org/10.3390/rs12132082









