Extraction of Information about Individual Trees from High-Spatial-Resolution UAV-Acquired Images of an Orchard
Abstract
1. Introduction
2. Materials and Methods
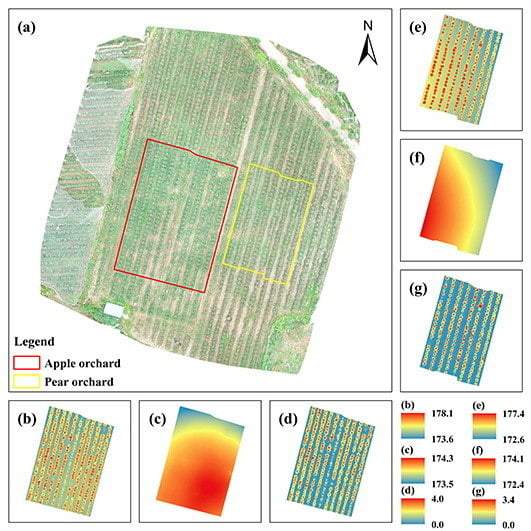
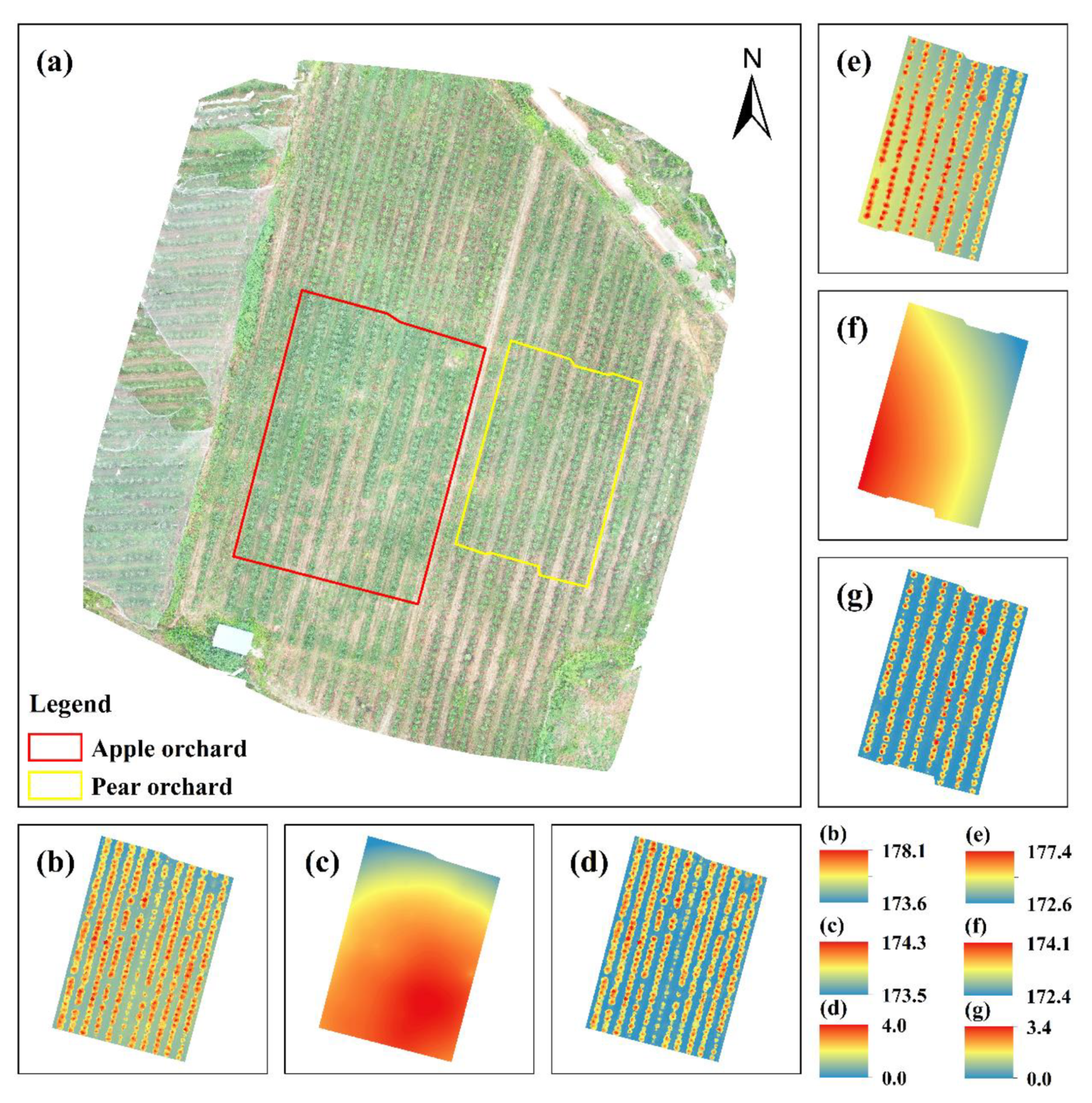
2.1. Study Areas
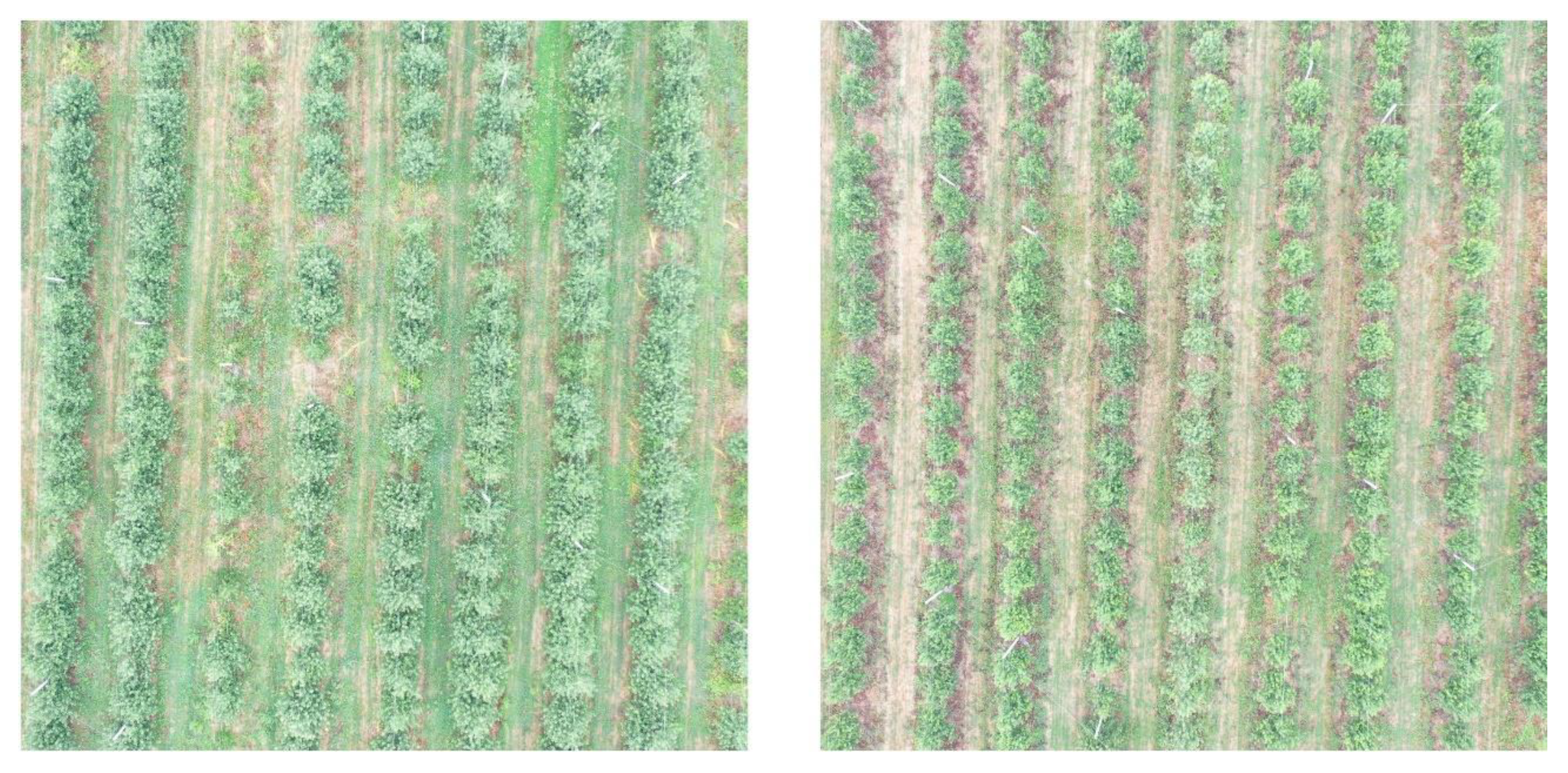
2.2. Acquisition of Images by UAV
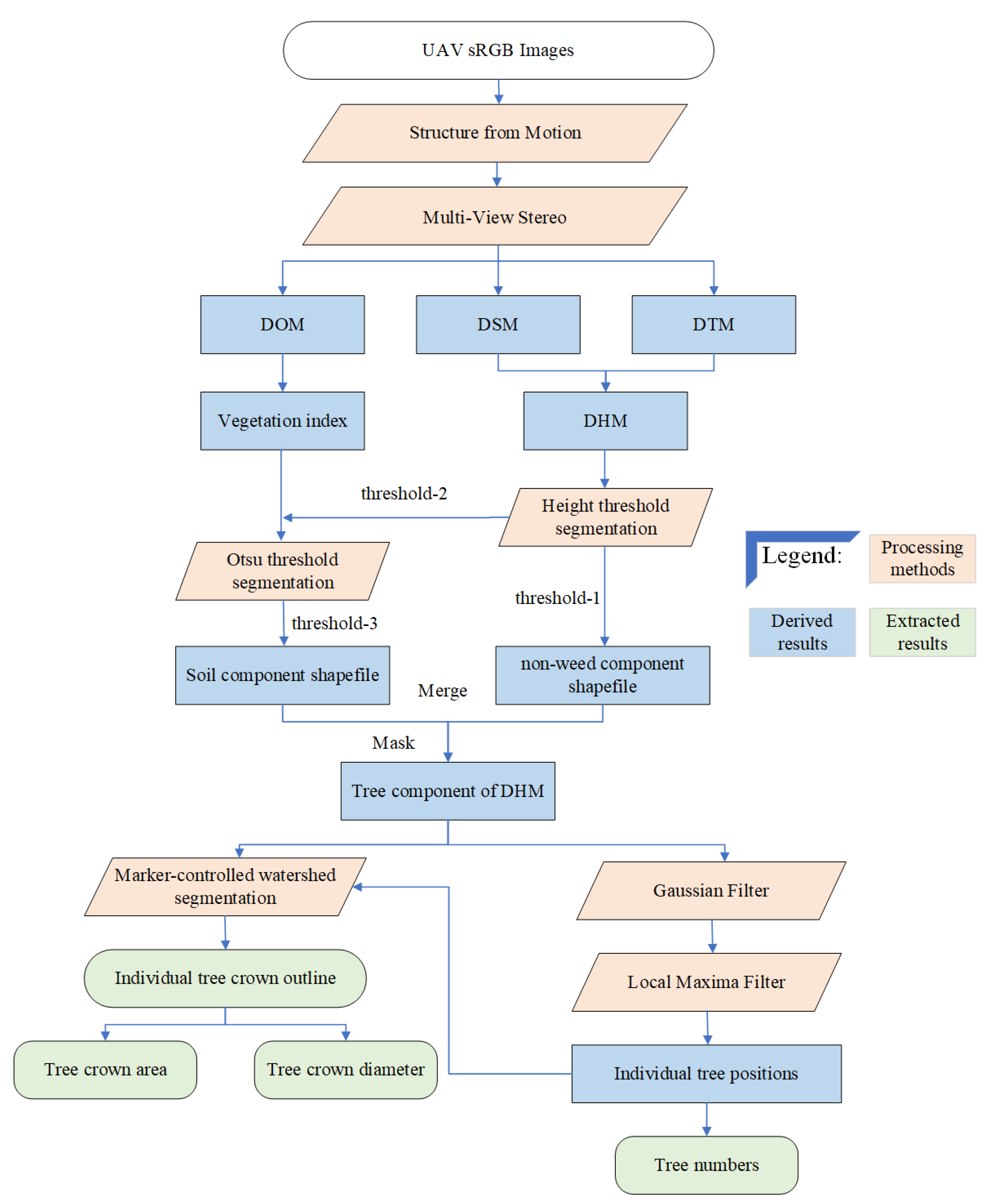
2.3. Proposed Method
2.3.1. Image Processing
2.3.2. Separation of Vegetation and Soil
2.3.3. Detection of Individual Trees
2.3.4. Extraction of Tree Components from the DHM
2.3.5. Delineation of Individual Tree-Crowns
2.4. Validation
2.4.1. Number of Detected Trees
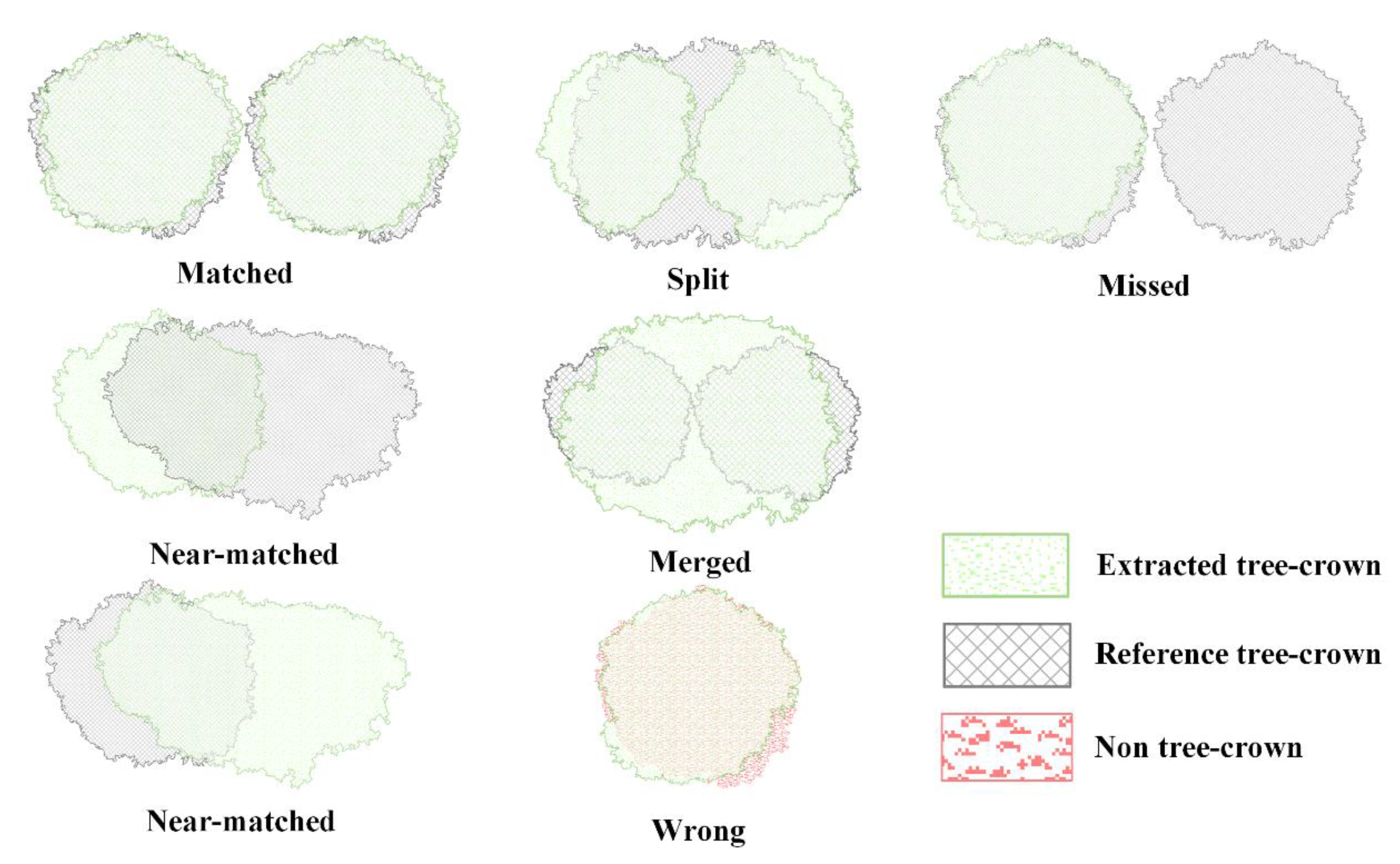
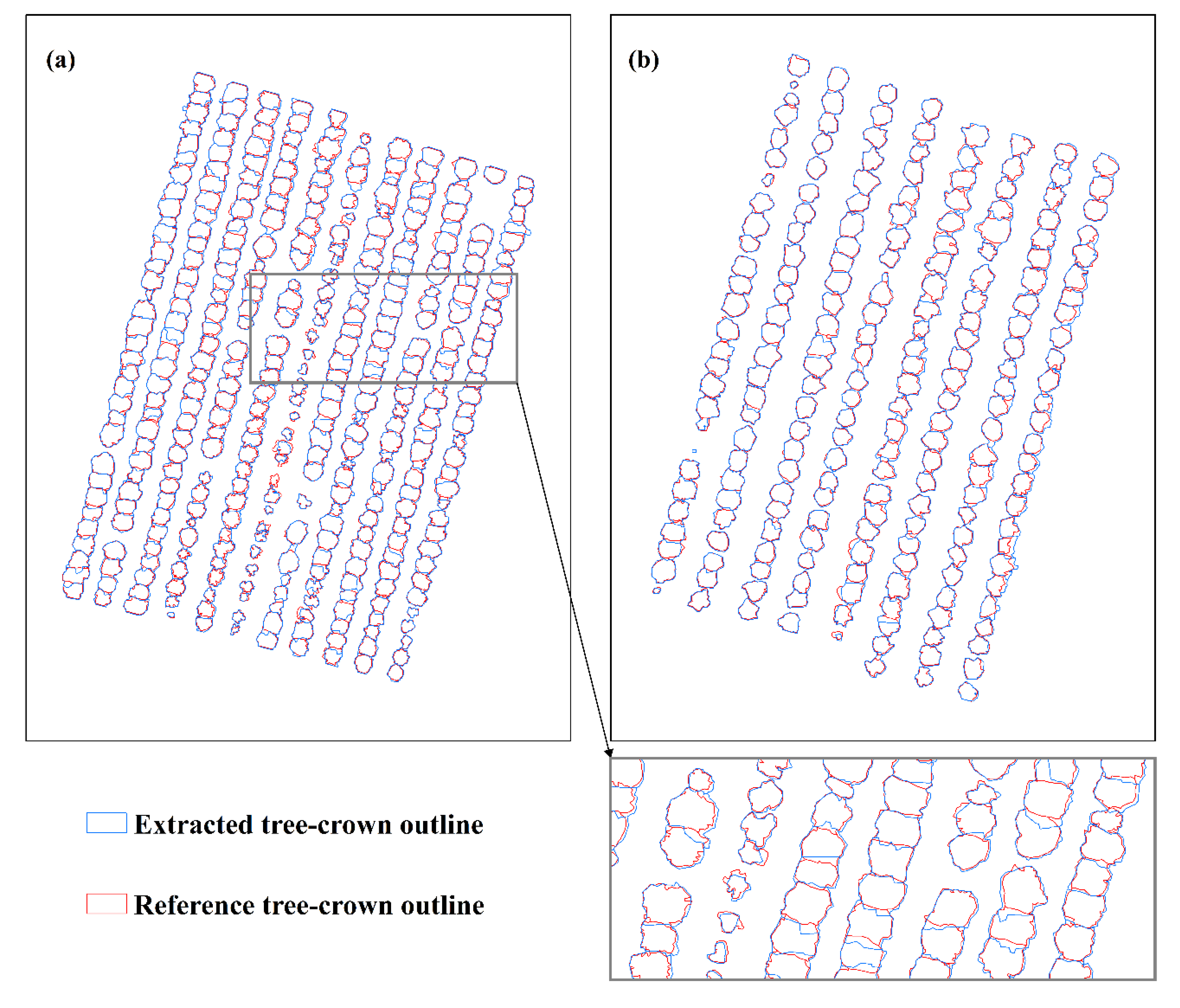
2.4.2. Delineation of Tree-Crowns
- Matched. If more than 50% of a reference tree-crown overlaps with only one extracted tree-crown element in the extracted tree-crown shapefile, and the center the reference tree-crown also overlaps with the extracted tree-crown, we considered the extracted tree-crown to have been matched correctly;
- Near-matched. If a reference tree-crown and an extracted tree-crown overlap by more than 50% of the tree-crown area on only one side of the extracted tree-crown, and the center of the reference tree-crown also overlaps with the extracted tree-crown, the reference tree-crown was classified as near-matched;
- Merged. If more than one reference tree-crown overlapped with one extracted tree-crown, and the centers of the reference tree-crowns were also covered by the extracted tree-crown, these reference tree-crowns were all considered to be merged tree-crowns;
- Split. If more than 50% of a reference tree-crown is occupied by more than one extracted tree-crown, this reference crown was considered to be a split tree-crown;
- Missed. If less than 50% of a reference tree-crown overlaps with any extracted tree-crowns, this reference tree-crown was considered to be missed;
- Wrong. If an extracted tree-crown was misidentified (i.e., an FP), this extracted tree-crown was classified as wrong.
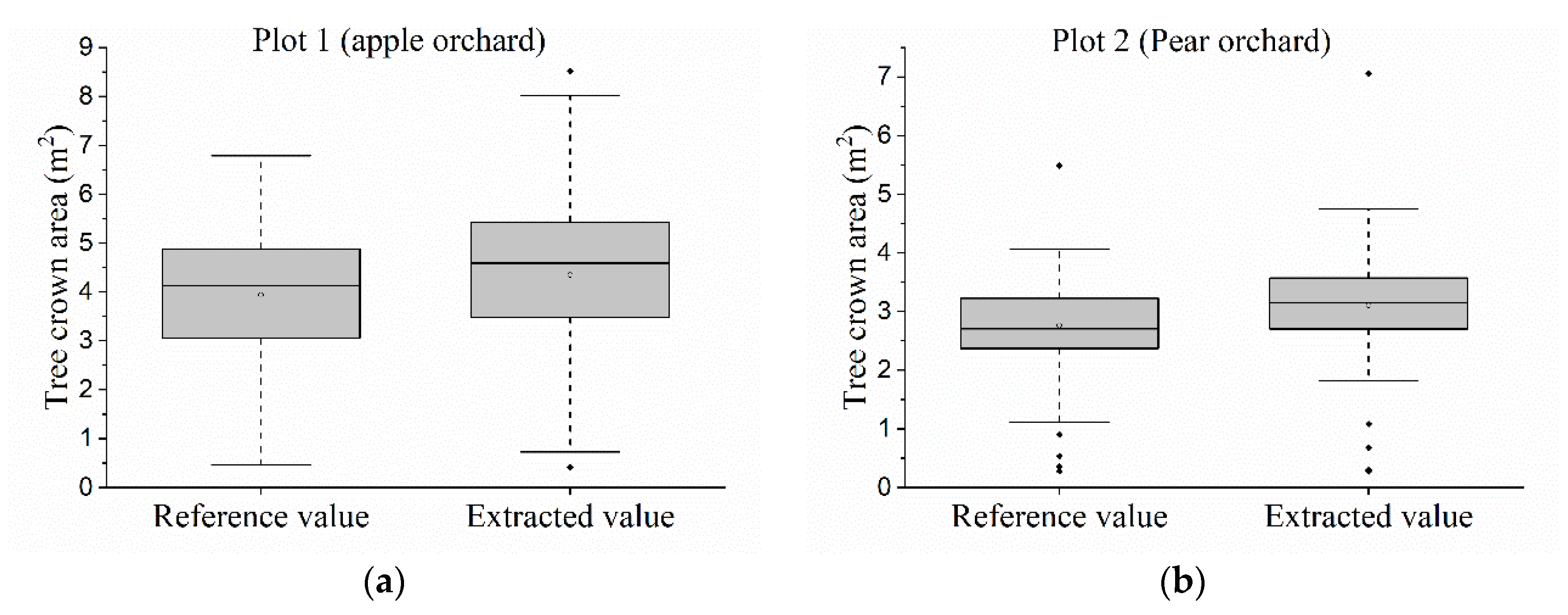
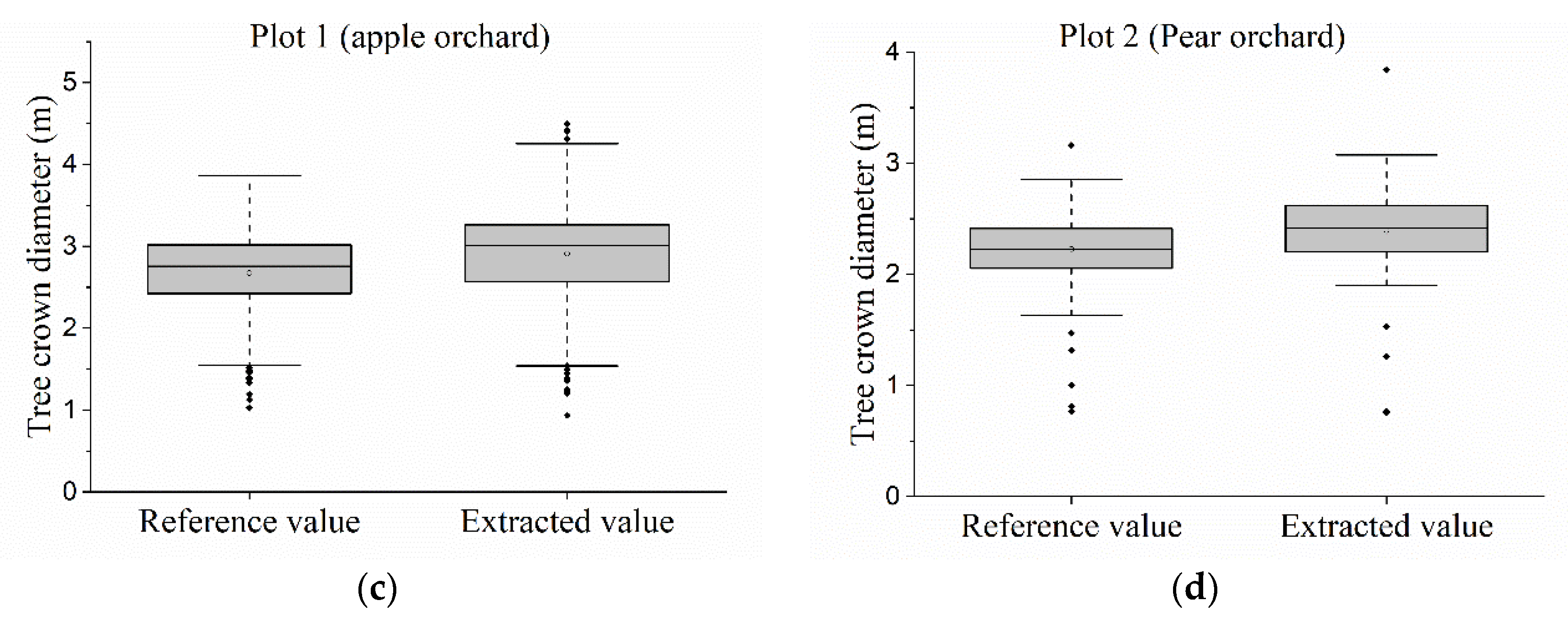
2.4.3. Estimation of Tree-Crown Area and Tree-Crown Diameter
3. Results
3.1. DOM, DSMs, DTMs, and DHMs
3.2. Estimation of the Numbers of Individual Trees
3.3. Delineation of Tree-Crowns
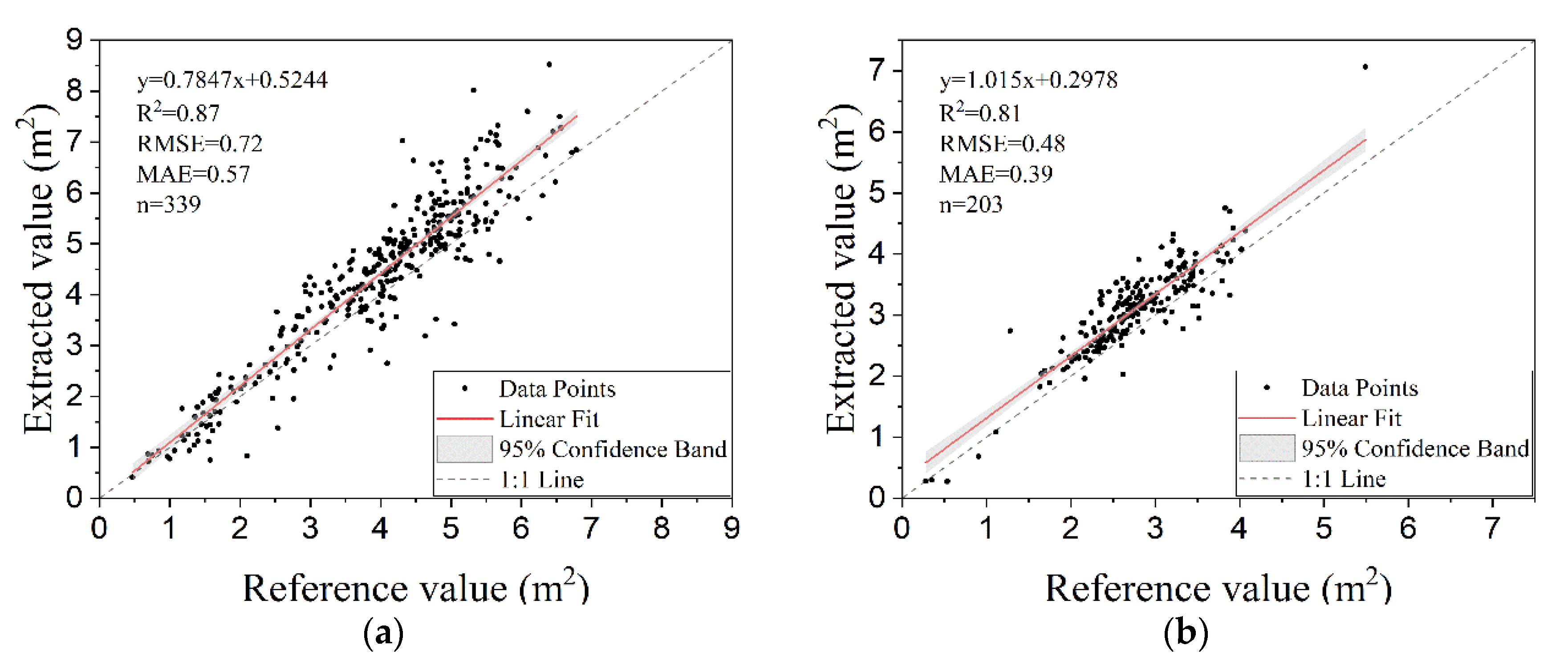
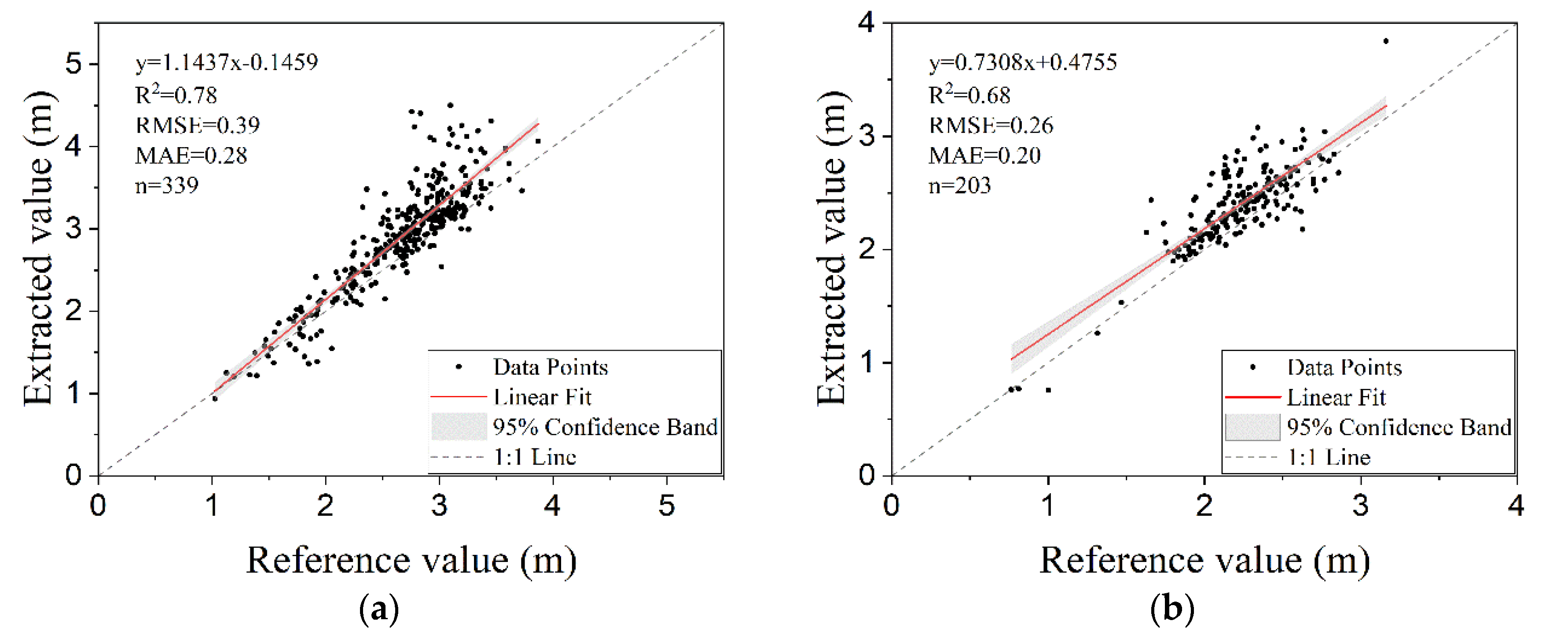
3.4. Estimation of Areas and Diameters of Tree-Crowns
4. Discussion
4.1. Determination of the Thresholds in the Proposed Method
4.2. Detection of Individual Trees
4.3. Delineation of Individual Tree-Crowns and Estimation of Tree-Crown Parameters
4.4. Limitations of the Proposed Method
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Panda, S.S.; Hoogenboom, G.; Paz, J.O. Remote sensing and geospatial technological applications for site-specific management of fruit and nut crops: A review. Remote Sens. 2010, 2, 1973–1997. [Google Scholar] [CrossRef]
- Das, U. Precision Farming a Promising Technology in Horticulture: A Review. Int. J. Pure Appl. Biosci. 2018, 6, 1596–1606. [Google Scholar] [CrossRef]
- Gebbers, R.; Adamchuk, V.I. Precision agriculture and food security. Science 2010, 327, 828–831. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, M.A.; Vallario, A.; Aguilar, F.J.; Lorca, A.G.; Parente, C. Object-based greenhouse horticultural crop identification from multi-temporal satellite imagery: A case study in Almeria, Spain. Remote Sens. 2015, 7, 7378–7401. [Google Scholar] [CrossRef]
- Singh, V.; Patel, A.N.; Dalwadi, A.; Kathota, J.; Suthar, J.; Kalubarme, M.H. Horticultural Fruit Crop Plantations Mapping using Geo-informatics Technology in Gujarat State India. Int. J. Adv. Remote Sens. GIS 2017, 6, 2033–2049. [Google Scholar] [CrossRef][Green Version]
- Paul, N.C.; Sahoo, P.M.; Ahmad, T.; Sahoo, R.N.; Krishna, G.; Lal, S.B. Acreage estimation of mango orchards using hyperspectral satellite data. Hortic. Soc. India 2018, 75, 27–33. [Google Scholar] [CrossRef]
- Roth, S.I.B.; Leiterer, R.; Volpi, M.; Celio, E.; Schaepman, M.E.; Joerg, P.C. Automated detection of individual clove trees for yield quantification in northeastern Madagascar based on multi-spectral satellite data. Remote Sens. Environ. 2019, 221, 144–156. [Google Scholar] [CrossRef]
- Csillik, O.; Cherbini, J.; Johnson, R.; Lyons, A.; Kelly, M. Identification of Citrus Trees from Unmanned Aerial Vehicle Imagery Using Convolutional Neural Networks. Drones 2018, 2, 39. [Google Scholar] [CrossRef]
- Morales, G.; Kemper, G.; Sevillano, G.; Arteaga, D.; Ortega, I.; Telles, J. Automatic segmentation of Mauritia flexuosa in unmanned aerial vehicle (UAV) imagery using deep learning. Forests 2018, 9, 736. [Google Scholar] [CrossRef]
- Mubin, N.A.; Nadarajoo, E.; Shafri, H.Z.M.; Hamedianfar, A. Young and mature oil palm tree detection and counting using convolutional neural network deep learning method. Int. J. Remote Sens. 2019, 40, 7500–7515. [Google Scholar] [CrossRef]
- Colaço, A.F.; Molin, J.P.; Rosell-Polo, J.R.; Escolà, A. Application of light detection and ranging and ultrasonic sensors to high-throughput phenotyping and precision horticulture: Current status and challenges. Hortic. Res. 2018, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kwak, D.A.; Lee, W.K.; Lee, J.H.; Biging, G.S.; Gong, P. Detection of individual trees and estimation of tree height using LiDAR data. J. For. Res. 2007, 12, 425–434. [Google Scholar] [CrossRef]
- Maes, W.H.; Steppe, K. Perspectives for Remote Sensing with Unmanned Aerial Vehicles in Precision Agriculture. Trends Plant Sci. 2019, 24, 152–164. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, M.P.; Laursen, M.S.; Jørgensen, R.N.; Skovsen, S.; Gislum, R. Designing and testing a UAV mapping system for agricultural field surveying. Sensors 2017, 17, 2703. [Google Scholar] [CrossRef] [PubMed]
- Ballesteros, R.; Ortega, J.F.; Hernandez, D.; Moreno, M.A. Onion biomass monitoring using UAV-based RGB imaging. Precis. Agric. 2018, 19, 840–857. [Google Scholar] [CrossRef]
- Tu, Y.H.; Johansen, K.; Phinn, S.; Robson, A. Measuring canopy structure and condition using multi-spectral UAS imagery in a horticultural environment. Remote Sens. 2019, 11, 269. [Google Scholar] [CrossRef]
- Jorge, J.; Vallbé, M.; Soler, J.A. Detection of irrigation inhomogeneities in an olive grove using the NDRE vegetation index obtained from UAV images. Eur. J. Remote Sens. 2019, 52, 169–177. [Google Scholar] [CrossRef]
- Jiménez-Brenes, F.M.; López-Granados, F.; Castro, A.I.; Torres-Sánchez, J.; Serrano, N.; Peña, J.M. Quantifying pruning impacts on olive tree architecture and annual canopy growth by using UAV-based 3D modelling. Plant Methods 2017, 13, 55. [Google Scholar] [CrossRef]
- Ampatzidis, Y.; Partel, V.; Meyering, B.; Albrecht, U. Citrus rootstock evaluation utilizing UAV-based remote sensing and artificial intelligence. Comput. Electron. Agric. 2019, 164, 104900. [Google Scholar] [CrossRef]
- Maillard, P.; Gomes, M.F. Detection and counting of orchard trees from vhr images using a geometrical-optical model and marked template matching. In Proceedings of the ISPRS Annals of the Photogrammetry, Remote Sensing and Spatial Information Sciences, Prague, Czech Republic, 12–19 July 2016; Volumn III-7, pp. 75–82. [Google Scholar]
- Mu, Y.; Fujii, Y.; Takata, D.; Zheng, B.; Noshita, K.; Honda, K.; Ninomiya, S.; Guo, W. Characterization of peach tree crown by using high-resolution images from an unmanned aerial vehicle. Hortic. Res. 2018, 5, 74. [Google Scholar] [CrossRef]
- Gongal, A.; Silwal, A.; Amatya, S.; Karkee, M.; Zhang, Q.; Lewis, K. Apple crop-load estimation with over-the-row machine vision system. Comput. Electron. Agric. 2016, 120, 26–35. [Google Scholar] [CrossRef]
- Caruso, G.; Tozzini, L.; Rallo, G.; Primicerio, J.; Moriondo, M.; Palai, G.; Gucci, R. Estimating biophysical and geometrical parameters of grapevine canopies (‘Sangiovese’) by an unmanned aerial vehicle (UAV) and VIS-NIR cameras. Vitis 2017, 56, 63–70. [Google Scholar]
- Johansen, K.; Raharjo, T.; McCabe, M.F. Using multi-spectral UAV imagery to extract tree crop structural properties and assess pruning effects. Remote Sens. 2018, 10, 854. [Google Scholar] [CrossRef]
- Caruso, G.; Zarco-Tejada, P.J.; González-Dugo, V.; Moriondo, M.; Tozzini, L.; Palai, G.; Rallo, G.; Hornero, A.; Primicerio, J.; Gucci, R. High-resolution imagery acquired from an unmanned platform to estimate biophysical and geometrical parameters of olive trees under different irrigation regimes. PLoS ONE 2019, 14, e0210804. [Google Scholar] [CrossRef]
- Salamí, E.; Gallardo, A.; Skorobogatov, G.; Barrado, C. On-the-fly olive tree counting using a UAS and cloud services. Remote Sens. 2019, 11, 316. [Google Scholar] [CrossRef]
- Ok, A.O.; Ozdarici-Ok, A. Detection of citrus trees from UAV DSMs. In Proceedings of the ISPRS Annals of the Photogrammetry, Remote Sensing and Spatial Information Sciences, Hannover, Germany, 6–9 June 2017; Volumn 4, pp. 27–34. [Google Scholar]
- Torres-Sánchez, J.; López-Granados, F.; Serrano, N.; Arquero, O.; Peña, J.M. High-throughput 3-D monitoring of agricultural-tree plantations with Unmanned Aerial Vehicle (UAV) technology. PLoS ONE 2015, 10, e0130479. [Google Scholar] [CrossRef]
- Fawcett, D.; Azlan, B.; Hill, T.C.; Kho, L.K.; Bennie, J.; Anderson, K. Unmanned aerial vehicle (UAV) derived structure-from-motion photogrammetry point clouds for oil palm (Elaeis guineensis) canopy segmentation and height estimation. Int. J. Remote Sens. 2019, 40, 1–23. [Google Scholar] [CrossRef]
- Sarron, J.; Malézieux, É.; Sané, C.A.B.; Faye, É. Mango yield mapping at the orchard scale based on tree structure and land cover assessed by UAV. Remote Sens. 2018, 10, 1900. [Google Scholar] [CrossRef]
- Marques, P.; Pádua, L.; Adão, T.; Hruška, J.; Peres, E.; Sousa, A.; Sousa, J.J. UAV-Based Automatic Detection and Monitoring of Chestnut Trees. Remote Sens. 2019, 11, 855. [Google Scholar] [CrossRef]
- Gevaert, C.M.; Persello, C.; Sliuzas, R.; Vosselman, G. Informal settlement classification using point-cloud and image-based features from UAV data. ISPRS J. Photogramm. Remote Sens. 2017, 125, 225–236. [Google Scholar] [CrossRef]
- Neto, J.C. A Combined Statistical-Soft Computing Approach for Classification and Mapping Weed Species in Minimum—Tillage Systems. Ph.D. Thesis, University of Nebraska—Lincoln, Lincoln, NE, USA, August 2004. [Google Scholar]
- Bendig, J.; Yu, K.; Aasen, H.; Bolten, A.; Bennertz, S.; Broscheit, J.; Gnyp, M.L.; Bareth, G. Combining UAV-based plant height from crop surface models, visible, and near infrared vegetation indices for biomass monitoring in barley. Int. J. Appl. Earth Obs. Geoinf. 2015, 39, 79–87. [Google Scholar] [CrossRef]
- Pérez, A.J.; López, F.; Benlloch, J.V.; Christensen, S. Colour and shape analysis techniques for weed detection in cereal fields. Comput. Electron. Agric. 2000, 25, 197–212. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Yang, W.; Wang, S.; Zhao, X.; Zhang, J.; Feng, J. Greenness identification based on HSV decision tree. Inf. Process. Agric. 2015, 2, 149–160. [Google Scholar] [CrossRef]
- Meyer, G.E.; Neto, J.C. Verification of color vegetation indices for automated crop imaging applications. Comput. Electron. Agric. 2008, 63, 282–293. [Google Scholar] [CrossRef]
- Lv, J.; Wang, F.; Xu, L.; Ma, Z.; Yang, B. A segmentation method of bagged green apple image. Sci. Hortic. 2019, 246, 411–417. [Google Scholar] [CrossRef]
- Huang, H.; Li, X.; Chen, C. Individual Tree Crown Detection and Delineation from Very-High-Resolution UAV Images Based on Bias Field and Marker-Controlled Watershed Segmentation Algorithms. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2018, 11, 2253–2262. [Google Scholar] [CrossRef]
- Marker-Controlled_Watershed. Available online: https://imagej.net/Marker-controlled_Watershed (accessed on 23 October 2019).
- Bala, A. An Improved Watershed Image Segmentation Technique using MATLAB. Int. J. Sci. Eng. Res. 2012, 3, 1–4. [Google Scholar]
- Mohan, M.; Silva, C.A.; Klauberg, C.; Jat, P.; Catts, G.; Cardil, A.; Hudak, A.T.; Dia, M. Individual tree detection from unmanned aerial vehicle (UAV) derived canopy height model in an open canopy mixed conifer forest. Forests 2017, 8, 340. [Google Scholar] [CrossRef]
- Ozdarici-Ok, A. Automatic detection and delineation of citrus trees from VHR satellite imagery. Int. J. Remote Sens. 2015, 36, 4275–4296. [Google Scholar] [CrossRef]
- Yin, D.; Wang, L. How to assess the accuracy of the individual tree-based forest inventory derived from remotely sensed data: A review. Int. J. Remote Sens. 2016, 37, 4521–4553. [Google Scholar] [CrossRef]
- Jing, L.; Hu, B.; Li, J.; Noland, T. Automated delineation of individual tree crowns from lidar data by multi-scale analysis and segmentation. PERS Photogramm. Eng. Remote Sens. 2012, 78, 1275–1284. [Google Scholar] [CrossRef]
- Fang, F.; Im, J.; Lee, J.; Kim, K. An improved tree crown delineation method based on live crown ratios from airborne LiDAR data. GISci. Remote Sens. 2016, 53, 402–419. [Google Scholar] [CrossRef]
- Pohl, C.; Van Genderen, J. Remote Sensing Image Fusion: A Practical Guide; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Ok, A.O.; Ozdarici-Ok, A. 2-D delineation of individual citrus trees from UAV-based dense photogrammetric surface models. Int. J. Digit. Earth 2018, 11, 583–608. [Google Scholar] [CrossRef]
- Díaz-Varela, R.A.; de la Rosa, R.; León, L.; Zarco-Tejada, P.J. High-resolution airborne UAV imagery to assess olive tree crown parameters using 3D photo reconstruction: Application in breeding trials. Remote Sens. 2015, 7, 4213–4232. [Google Scholar] [CrossRef]



| Parameters | Specifications | |
|---|---|---|
| Aircraft | Takeoff weight | 0.907 kg |
| Battery | Lithium-Polymer 4S battery: 3850 mAh | |
| Maximum flight time | 31 minutes (at a consistent 25 kph) | |
| Camera | Sensor | 1″ (CMOS) |
| Camera lens | FOV 77° 28 mm | |
| Effective pixels | 20 Megapixels | |
| Pixel size | 2.4 μm ∗ 2.4 μm | |
| Still image size | 5472 × 3648 (pixels) | |
| Image format | JPEG |
| Processing Steps | Parameters | Setting |
|---|---|---|
| Initial processing | Keypoints Image Scale | Full |
| Matching Image Pairs | Aerial Grid or Corridor | |
| Targeted Number of Keypoints | Automatic | |
| Calibration Method | Standard | |
| Point Cloud and Mesh | Image Scale | Multiscale, 1/2 (Half image size, Default) |
| Point Density | Optimal | |
| Minimum Number of Matches | 3 | |
| Generate 3D Textured Mesh | Yes | |
| 3D Textured Mesh Settings | Medium Resolution (default) Color Balancing: no | |
| DSM, Orthomosaic, and index | Resolution | 1*GSD (1.14 (cm/pixel)) |
| DSM Filters | Noise Filtering: yes Surface Smoothing: yes, Type: Sharp | |
| Raster DSM | Generated: yes Method: Inverse Distance Weighting Merge Tiles: yes | |
| Orthomosaic | Generated: yes Merge Tiles: yes | |
| Raster DTM | Generated: yes Resolution: 5*GSD (1.14 (cm/pixel)) |
| Evaluate Indicators | Plot 1 | Plot 2 |
|---|---|---|
| TP | 345 | 206 |
| FP | 1 | 1 |
| FN | 6 | 2 |
| PA (%) | 98.3 | 99.0 |
| UA (%) | 99.7 | 99.5 |
| F (%) | 99.0 | 99.3 |
| Categories | Plot 1 | Plot 2 |
|---|---|---|
| Ema | 336 | 201 |
| Enm | 3 | 2 |
| Eme | 0 | 0 |
| Esp | 6 | 2 |
| Emi | 0 | 0 |
| Ewr | 0 | 1 |
| PA (%) | 97.4 | 98.0 |
| Categories | Plot 1 | Plot 2 |
|---|---|---|
| Rma | 336 | 201 |
| Rnm | 3 | 2 |
| Rme | 12 | 4 |
| Rsp | 0 | 0 |
| Rmi | 1 | 0 |
| UA (%) | 95.5 | 97.1 |
| Statistical Parameters | Plot 1 | Plot 2 | ||
|---|---|---|---|---|
| Ae(m2) | Ar (m2) | Ae(m2) | Ar(m2) | |
| Min | 0.412 | 0.470 | 0.274 | 0.280 |
| Q1 | 3.479 | 3.058 | 2.701 | 2.368 |
| Median | 4.592 | 4.130 | 3.149 | 2.707 |
| Q3 | 5.434 | 4.877 | 3.572 | 3.227 |
| Max | 8.521 | 6.789 | 7.061 | 5.492 |
| Mean | 4.351 | 3.939 | 3.105 | 2.766 |
| Statistical Parameters | Plot 1 | Plot 2 | ||
|---|---|---|---|---|
| De(m) | Dr(m) | De(m) | Dr(m) | |
| Min | 0.938 | 1.026 | 0.756 | 0.766 |
| Q1 | 2.574 | 2.424 | 2.201 | 2.055 |
| Median | 3.008 | 2.759 | 2.416 | 2.228 |
| Q3 | 3.264 | 3.018 | 2.621 | 2.414 |
| Max | 4.498 | 3.866 | 3.839 | 3.162 |
| Mean | 2.914 | 2.675 | 2.395 | 2.226 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, X.; Zhang, Z.; Yu, R.; Tian, Q.; Zhu, X. Extraction of Information about Individual Trees from High-Spatial-Resolution UAV-Acquired Images of an Orchard. Remote Sens. 2020, 12, 133. https://doi.org/10.3390/rs12010133
Dong X, Zhang Z, Yu R, Tian Q, Zhu X. Extraction of Information about Individual Trees from High-Spatial-Resolution UAV-Acquired Images of an Orchard. Remote Sensing. 2020; 12(1):133. https://doi.org/10.3390/rs12010133
Chicago/Turabian StyleDong, Xinyu, Zhichao Zhang, Ruiyang Yu, Qingjiu Tian, and Xicun Zhu. 2020. "Extraction of Information about Individual Trees from High-Spatial-Resolution UAV-Acquired Images of an Orchard" Remote Sensing 12, no. 1: 133. https://doi.org/10.3390/rs12010133
APA StyleDong, X., Zhang, Z., Yu, R., Tian, Q., & Zhu, X. (2020). Extraction of Information about Individual Trees from High-Spatial-Resolution UAV-Acquired Images of an Orchard. Remote Sensing, 12(1), 133. https://doi.org/10.3390/rs12010133